Abstract
The study’s purpose is to investigate the clinical characteristics and research progress of PURA syndrome. It will also provide new ideas and methods for the diagnosis of neonatal hypotonia etiology. A case of PURA syndrome admitted to Shenzhen Hospital of Peking University was analyzed retrospectively. The keywords “PURA”, “PURα”, “PURA syndrome”, and “5q31” were used to search the Chinese periodical full-text database and Wanfang database. The keywords “PURA”, “PURα”, “Pur-alpha”, “PURA syndrome”, and “5q31” were used to search the biomedical literature database (PubMed). The Web of Science database and Proquest database were used to find works of literature from the establishment of the database to November 10, 2019. By analyzing the 72 cases of PURA syndrome reported in ten Chinese and international studies, it was found that 57% (21/37) of the patients had a gestational age greater than 41 weeks. Neonatal patients exhibited hypotonia (82%, 59/72), feeding difficulties (97%, 64/66), apnea or primary hypoventilation (57%, 41/72), intrauterine excessive hiccupping (55%, 6/11), and drowsiness (51%, 24/47). After the neonatal period, the pediatric patients demonstrated moderate to severe mental retardation (100%), epilepsy (54%, 29/54), progressive hip dysplasia (17%, 7/42), scoliosis (48%, 11/23), dysphagia and salivation (69%, 25/36), and constipation (60%, 21/35). The clinical manifestations of the present case were consistent with those in the literature reports. It was the first confirmed case at Shenzhen Hospital in the neonatal period and had a de novo mutation. It was difficult to diagnose PURA syndrome in the neonatal period, which might affect multiple systems. In newborns with obvious hypotonia, the evaluation should be expanded to consider other symptoms. Additionally, targeted gene detection should be completed to achieve early diagnosis and intervention, improve the prognosis, and perform genetic counseling.
Keywords: Hypotonia, purine-rich binding element protein alpha (PURα), PURA syndrome, newborn
Introduction
PURA syndrome is caused by the mutation of the purine-rich binding element protein alpha (PURα) gene in chromosome 5q31.2–q31.3. It is a congenital developmental abnormality recognized only in recent years. The clinical manifestations include obvious hypotonia, feeding difficulties, abnormal breathing patterns, seizures, and severe mental retardation (1-3). With the improvement and popularization of genetic testing technology, PURA syndrome diagnosis has increased. However, there are still only a few reported cases, especially in newborns. In the present study, the clinical data of a pediatric patient diagnosed with PURA syndrome in the neonatal period are reported, together with a review of the relevant literature. The present study aimed to discuss the clinical characteristics of PURA syndrome and strengthen the clinicians’ understanding of the disease.
We present the following article/case in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/tp-20-248).
Case presentation
Data of the present case
Clinical materials
A male neonate, 4 days after birth, was hospitalized for “crying, eating, and moving less”. The patient was a first pregnancy and birth to the mother. The gestational age was 42 weeks. The birth weight was 3,900 g, and there was no asphyxia. After birth, the patient was breastfed, with minimal milk intake, weak sucking, and little crying and moving. There was no irritability, screaming, or fever. Jaundice began to appear on the third day after birth. The fetal feces were excreted within 24 hours, and the quantity of feces was small. There were no unique features noted in the regular maternal prenatal examinations or family history.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Peking University Shenzhen Hospital committee (20200610) and with the Helsinki Declaration (as revised in 2013). The patient’s guardian provided written informed consent.
Physical examination at admission
The patient’s vitals were: T: 36.7 °C, P: 140 beats/min, R: 43 beats/min, body weight 3,610 g, body height 49 cm.
There was no deformity in the appearance and little reaction (i.e., the patient frowned after stimulation but exhibited little other reaction). The following were observed: drowsiness, moderate to severe yellowing of skin with the yellow sclera, the anterior fontanelle was flat and soft, and there were no cardiac or pulmonary abnormalities in auscultation. The abdomen was flat and soft, with no enlargement of the liver or the spleen on palpation. No abnormalities in the external genitalia or warm acromegaly were noted. The muscle tension of limbs was low and soft. The feeding reflex was weak. Sucking, hugging, and holding reflexes were not evoked.
Laboratory examinations
The results of the myocardial enzymes, hepatic and renal functions, and electrolytes were: total bilirubin 279.9 µmol/L, non-binding bilirubin 235.5 µmol/L. There were no abnormalities in the remaining results. The blood routine test, C-reactive protein, and procalcitonin were all normal. The TORCH-IgM was negative. The monitoring of blood glucose and blood gas was normal. The examination found no abnormality in the cardiac color ultrasonography or chest X-ray. The five items of thyroid function were standard. Serum ammonia of 38.6 µmol/L and lactic acid of 1.86 mmol/L were both within the normal range. There was no abnormality in the analysis of blood, amino acid, or urine organic acid.
Diagnosis and treatment
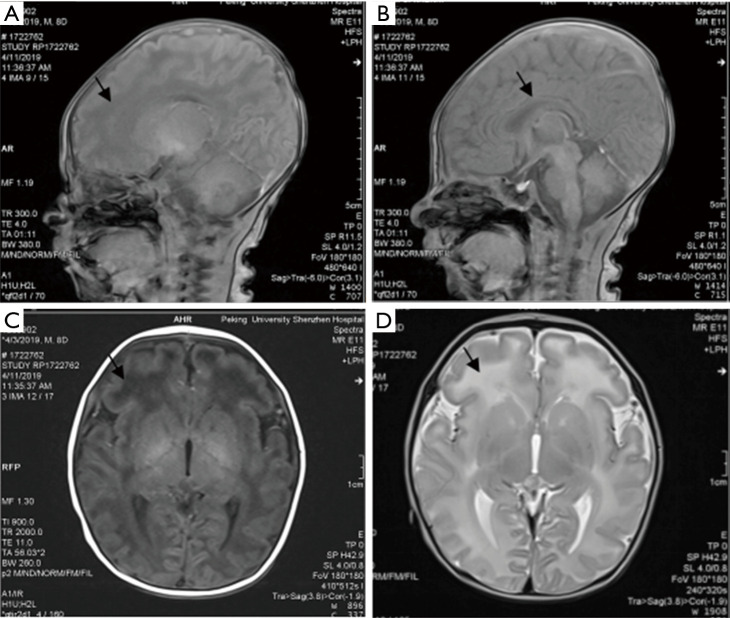
After admission, jaundice subsided quickly without rebound after treatment with blue light. The patient demonstrated minimal crying, movement, and weak sucking; thus, tube feeding and intravenous nutrition support were applied. The patient had apnea, although no shortness of breath or dyspnea was noted. Nasal oxygen inhalation was administered. There was no opisthotonus, fever, or binocular gaze throughout the disease. On the seventh day after admission, the plain scanning of cerebral magnetic resonance imaging (MRI) reported that the frontal lobe showed slightly low signal on T1-weighted imaging (T1WI) and slightly hyperintense on T2-weighted imaging (T2WI), with thin corpus callosum. No abnormal signal opacity was found in additional brain parenchyma. No enlargement or compression of the ventricular system was noted, with the midline structure appearing in the middle. There was no abnormality in the cerebellum, brainstem, or pituitary. As shown in Figure 1, the hearing screening was routine. The whole-exome sequencing (WES) of hereditary disease was performed on the patient and his parents. The results are illustrated in Table 1. The parents asked to leave the hospital. At the telephone follow-up 3 months later, it was reported that the patient had died suddenly at home 1 month after discharge. The cause of death was not disclosed.
Figure 1.
Images of the head MRI in the patient (arrow indicates the lesion site): (A,C) low signal on T1; (B) high signal on T2; (D) the thinned corpus callosum.
Table 1. The manifestations of PURA syndrome in the neonatal period.
| Clinical presentation | In this paper | Literature report | |
|---|---|---|---|
| Cases | Proportion (%) | ||
| Gestational age >41 weeks | Yes | 21/37 | 57 |
| Hypotonia | Yes | 64/66 | 97 |
| Feeding difficulties | Yes | 59/72 | 82 |
| Gastroesophageal reflux | Suspicious | 14/53 | 26 |
| Breathing problem (apnea/hypoventilation) | Yes | 41/72 | 57 |
| Hypersomnia | Yes | 24/47 | 51 |
| Startle hyperresponsiveness | No | 20/52 | 38 |
| Hypothermia | No | 11/31 | 35 |
| Excessive hiccup in utero | Yes | 6/11 | 55 |
Results of gene sequencing
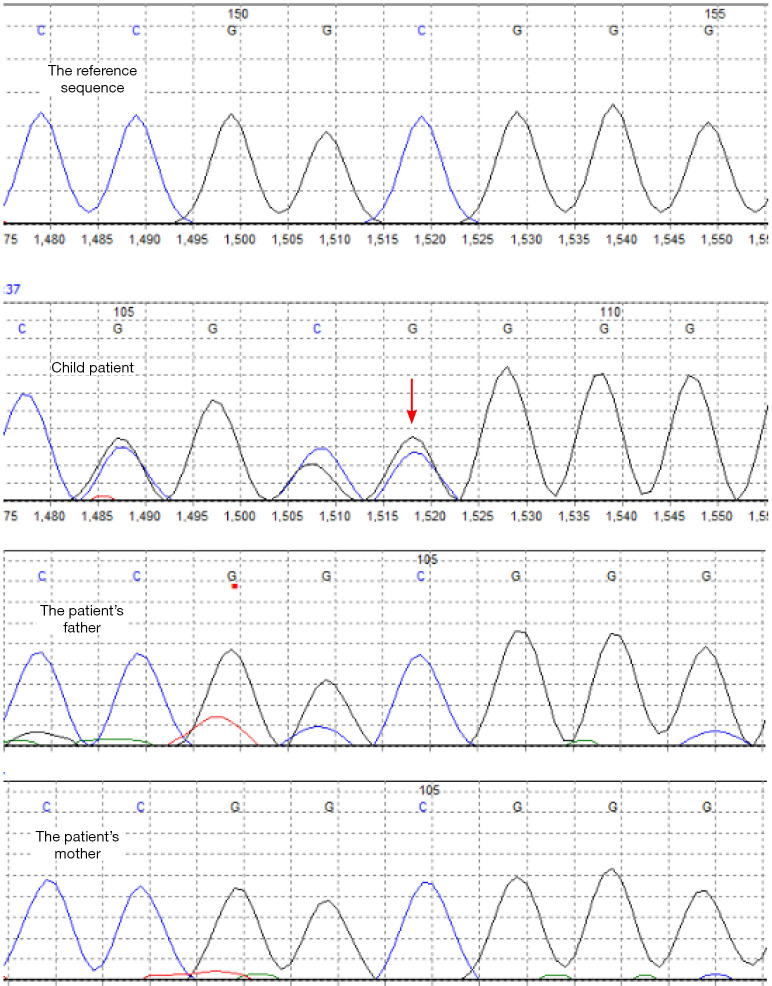
The WES (Guangzhou Jinyu Medical Laboratory Center) results were that heterozygous pathogenic variation of the PURA gene existed in the patient. Based on the patient’s and his parents’ test results, the heterozygous pathogenic variation of the PURA gene might be a de novo mutation, and the possibility that the parents were chimeric carriers of germ cells could not be ruled out. The mutation is located in 5q31 exon1c.98dupG (p.Gly34fs) (NM_005859.4), as shown in Figure 2. The present mutation was a frameshift mutation [i.e., the coding protein began to shift from glycine (Gly) at position 34, which resulted in the early termination of protein translation]. It was speculated that the encoded protein would be truncated and lose normal function. The present test covered 20,000 genes, and there were no reports in the HGMD database or recorded in the ESP6500siv2_ALL, 1000 Genomes (1000g2015aug_ALL), and the dbSNP147 databases. It was widely considered that the mutation was pathogenic and inherited in an autosomal dominant manner. Patients with heterozygous variants might have a 50% chance of transmitting the pathogenic variant to their offspring.
Figure 2.
The whole exome sequencing of hereditary disease of the pedigree (the arrow indicates the mutation location, PURα gene, 5q31region, sequence NM_005859.4, Exon1, c.98dupG (p.Gly34fs), heterozygous pathogenic variation, the parents were normal).
Literature review
The keywords “PURA”, “PURα”, “PURA syndrome”, and “5q31” were used to search the Chinese periodical full-text database and Wanfang database in literature collected from the establishment of the database to November 10, 2019. This search found no relevant literature. The keywords “PURA”, “PURα”, “Pur-alpha”, “PURA syndrome”, and “5q31” were used to search the biomedical literature database (PubMed), Web of Science database, and Proquest database to search for literature from the establishment of the database to November 10, 2019. Ten articles were retrieved, and 72 cases of PURA syndrome were reported (3-12), with the patients’ ages ranging from 6 months to 27 years. The present case was the first case reported with a definite diagnosis in the neonatal period. All the manifestations of PURA syndrome in the neonatal period were summarized. Of the 64 cases, 97% (64/66) had hypotonia, and 82% had resultant feeding difficulties. More than half of the patients (57%) were born at a gestational age greater than 41 weeks. Of the patients, 57% had apnea or primary hypoventilation, 55% had intrauterine excessive hiccupping, and 51% had drowsiness. See Table 1 for details. The most frequently mentioned MRI imaging change was delayed myelination. Other imaging changes included white matter abnormalities (thinning, dysplasia), significant periventricular space, mild parenchymal dysplasia, lateral ventricle widening, and corpus callosum dysplasia. Cortical development was normal. Most patients showed a decreased volume of white matter, a slight enlargement of lateral ventricles, and subarachnoid cysts. The case examined in the present study was consistent with the common symptoms of PURA syndrome reported in the literature. The MRI findings of the present case were mainly white matter dysplasia (especially in the frontal lobe) and corpus callosum thinning.
After the neonatal period, patients with PURA syndrome would have moderate to severe mental retardation, accompanied by severe language disorders and motor retardation (100%, 72/72). Dystonia, which persists from birth, was more pronounced in the trunk. A few patients had limb spasms after maturing, such as the positive Babinski sign, usually with gait instability and rigid hand movements (some cases described Rett syndrome). Dyskinesia, including dystonia, chorea-like movement, epileptiform movement [with normal electroencephalogram (EEG)], and ataxia-like movement, were observed in 20% (6/30) of the patients. Among the pediatric patients, 54% (29/54) were diagnosed with epilepsy. The age of convulsion ranged from 6 months to 15 years (mostly between 2 to 4 years). Lennox-Gastaut syndrome could develop. Most of the epilepsy episodes in these patients were refractory epilepsy. In the pediatric patients, 51% (26/51) had eye movement and vision problems. Strabismus (mostly esotropia) was the most frequently reported problem, usually accompanied by strabismus-related ametropia (mainly hyperopia). There was progressive dysplasia of the hip (17%, 7/42) and scoliosis (48%, 11/23) in the bone system. The causes might be chronic trunk muscle hypotonia (with or without limb muscle dystonia), joint laxity, and slow or incomplete motor development. Short stature [five cases with a height of ≤ 2.5 standard deviation (SD), and six cases with a height between −2 to −2.5 SD] was reported in some studies, and flat-footedness was observed in 11 cases. Hypotonia caused dysphagia and salivation to occur in 69% (25/36) of the cases. Constipation occurred in 60% (21/35) of the patients who were dependent on laxatives since childhood. Other multiple endocrine abnormalities were reported, with vitamin D deficiency (42%, 8/19) in most cases. Some studies also reported abnormal cortical response and abnormal thyroxine secretion. However, an endocrine examination was not conducted in all cases. Thus, the incidence rate might be underestimated. Generally, there are no characteristic facial features in PURA syndrome, but it may manifest as myopathy, high anterior hairline, amygdaloid eye cleft, and buccal fullness.
The most frequently reported mutation site was c.697_699del (p.Phe233del), which was reported in seven cases. There were three cases with the c.812_814del (p.Phe271del) mutation, two cases with the c.289A > G (p.Lys97Glu) mutation, and two cases with the c.734G > C (p.Arg245Pro) mutation. The other mutation sites were only reported in a single incident, as shown in Table 2. The present case’s mutation site was located in c.98dupg (p.gly34fs), which was not found in the previous reports. The mutations in all the patients were de novo mutations; their parents were healthy, and the patients were heterozygous. WES of the family trios was performed in the same family pedigree in 25 cases, and the autosomal dominant inheritance was verified.
Table 2. Genetic examination results of PURA syndrome.
| No. | cDNA | Amino acid | Monitoring method | Genetic characteristics | Cases |
|---|---|---|---|---|---|
| 1 | c.697_699del | p.Phe233del | WES | De novo | 4 |
| 2 | c.697_699del | p.Phe233del | WES (trio) | AD, de novo | 3 |
| 3 | c.812_814del | p.Phe271del | WES | AD, de novo | 3 |
| 4 | c.289A>G | p.Lys97Glu | WES | AD, de novo | 2 |
| 5 | c.734G>C | p.Arg245Pro | WES (patient + mother) | De novo | 1 |
| 6 | c.734G>C | p.Arg245Pro | WES (trio) | AD, de novo | 1 |
| 7 | c.235C>T | p.Gln79* | WES | De novo | 1 |
| 8 | c.220T>C | p.Tyr74His | WES | De novo | 1 |
| 9 | c.675_676insA | p.Val226Serfs*68 | WES | De novo | 1 |
| 10 | c.25G>T | p.Glu9* | WES | De novo | 1 |
| 11 | c.802G>T | p.Gly268* | WES | De novo | 1 |
| 12 | c.572C>T | p.Pro191Leu | WES | De novo | 1 |
| 13 | c.677_678del | p.Val226Glyfs*67 | WES (patient + mother) | De novo | 1 |
| 14 | c.338_341dupACCT | p.Gly115Profs*87 | Single gene analysis | AD, de novo | 1 |
| 15 | c.746_749dupTGAA | p.Lys250Asnfs*45 | WES (trio) | AD, de novo | 1 |
| 16 | c.158_159delGG | p.Gly53Alafs*147 | WES (trio) | AD, de novo | 1 |
| 17 | c.351dupC | p.Ile118Hisfs*83 | WES (trio) | AD, de novo | 1 |
| 18 | c.771_776del | p.Ile257_Val259delinsMet | WES (trio) | AD, de novo | 1 |
| 19 | c.340delC | p.Leu114TrpfsTer111 | WES (trio) | AD, de novo | 1 |
| 20 | c.488_489 insGCGCGGCCGCTTCCT | p.Gly165_Arg169dup | WES (trio) | AD, de novo | 1 |
| 21 | c.127-130delAGTG | p.Ser43Alafs*34 | WES | De novo | 1 |
| 22 | c.382C>T | p.Gln128* | WES | De novo | 1 |
| 23 | c.153delA | p.Leu54Cysfs*24 | WES | Unknown | 1 |
| 24 | c.616_618delATC | p.Ile206del | WES | AD, de novo | 1 |
| 25 | c.478A>T | p.Leu160* | WES | AD, de novo | 1 |
| 26 | c.711dupC | p.Asn238Glnfs*56 | WES | AD, de novo | 1 |
| 27 | c.135_138dup | p.Gly47Argfs*155 | WES | AD, de novo | 1 |
| 28 | c.808_809delAC | p.T270LfsX23 | WES | Not in mother | 1 |
| 29 | c.155delG | p.Leu54CysfsTer24 | WES (trio) | AD, de novo | 1 |
| 30 | c.685A>T | p.Lys229* | WES (trio) | AD, de novo | 1 |
| 31 | c.1A>T | p.Met1 | WES | De novo | 1 |
| 32 | c.4_8delGCGGA | p.Ala2Profs*197 | WES | De novo | 1 |
| 33 | c.307_308delTC | p.Ser103Hisfs*97 | WES | De novo | 1 |
| 34 | c.556C>T | p.Gln186* | WES | De novo | 1 |
| 35 | c.299T>C | (p.Leu100Pro) | WES | De novo | 1 |
| 36 | c.363C>G | (p.Tyr121*) | WES | De novo | 1 |
| 37 | c.783C>G | (p.Tyr261*) | WES | De novo | 1 |
| 38 | c.470T>A | (p.Met157Lys) | WES | De novo | 1 |
| 39 | c.265G>C | (p.Ala89Pro) | WES | De novo | 1 |
| 40 | c.263_265delTCG | (p.Ile88_Ala89delinsThr) | WES | De novo | 1 |
| 41 | c.596G>C | (p.Arg199Pro) | WES | De novo | 1 |
| 42 | c.302_310delCTCTCTCCA | p.Thr101_Ser103del | WES | De novo | 1 |
| 43 | c.331_342del | p.Arg111_Leu114del | WES | De novo | 1 |
| 44 | c.768dupC | p.Ile257Hisfs*37 | WES | De novo | 1 |
| 45 | c.563 T>C | p.Ile188Thr | WES | De novo | 1 |
| 46 | c.726_727delGT | p.Phe243Tyrfs*50 | WES (trio) | AD, de novo | 1 |
| 47 | C616A>T | p.Ile206Phe | WES (trio) | AD, de novo | 1 |
| 48 | c.847delG | p.Glu283Arg fs*45 | WES (trio) | AD, de novo | 1 |
| 49 | c.596G>C | p.Arg199Pro | WES | De novo | 1 |
| 50# | c.683A>G | p.Asp228Ser | WES | De novo | 1 |
| c.796A>T | p.Lys266* | WES | De novo | 1 | |
| 51 | c.593dupT | – | WES | De novo | 1 |
| 52 | c.419G>C | p.Arg140Pro | WES | De novo | 1 |
| 53 | c.502del | p.Leu168Cysfs* | WES | De novo | 1 |
| 54 | c.458G>C | p.Arg153Pro | WES | De novo | 1 |
| 55 | c.264delC | p.Ile188Metfs*137 | WES | De novo | 1 |
| 56 | c.218T>C | p.Phe73Ser | WES | De novo | 1 |
| 57 | c.382C>T | p.Gln128* | WES | De novo | 1 |
| 58 | c.745delG | p.Val249* | WES | De novo | 1 |
| 59 | c.759T | ,p.Tyr253Lys | WES | De novo | 1 |
| 60 | c.159_182dup | p.Gln55Alafs*147 | WES | De novo | 1 |
| 61 | c.7_11delGACCG | p.Asp3Argfs*196 | WES | De novo | 1 |
| 62 | chr5:139494213-139494221 GCGCGAGAA > G | p.Arg150Profs*48 | WES | De novo | 1 |
| 63 | c.367C>T | p.Gln123* | WES | De novo | 1 |
| 64 | c.159delG | p.Leu54Cysfs*24 | WES | De novo | 1 |
| 65 | c.98dupG | p.(Gly34fs) | WES | De novo | 1 |
#, this patient has two mutation sites. WES, whole exome sequencing, exon check; WES trio, three generations of the same family WES; de novo, new mutation; AD, autosomal dominant.
Discussion
PURA syndrome is a congenital developmental abnormality caused by the truncation of the PURA protein encoded by the mutation of the PURα gene in the region of chromosome 5q31.2–q31.3, resulting in the loss of its normal function. It is a clinical syndrome characterized by severe growth retardation, clearly diminished muscle tone, difficulty in feeding, abnormal breathing patterns, and convulsions. The development of genetic testing methods has led to the gradual recognition of PURA syndrome. The human PURA is a gene located on chromosome 5q31 with a length of 11,640 bp. It is highly conserved during evolution. It encodes the purine-rich DNA and RNA binding protein (PURA protein), containing 322 amino acids. The protein’s relative molecular weight is 34.911×103. It is located primarily in the nucleus and partly in the cytoplasm and mitochondria (13,14). The PURA protein is a member of the PUR protein family and a multi-effect transcription factor. It can directly or indirectly combine with DNA to play the role of a transcription factor, promoting or inhibiting the transcription of downstream genes. It also plays an essential role in the process of DNA replication and RNA translation by binding with mRNA and influencing its transport and translation (4,15). The PUR family proteins play an essential role in bone marrow-derived cell development, muscle development, brain development, and mRNA transportation to neuron dendrites. Deletion, mutation, and distortion of the PUR family proteins are closely related to human diseases (16).
The present case’s clinical manifestations were obvious hypotonia, feeding difficulty, less crying, less moving, drowsiness, and respiratory insufficiency. The MRI of the patient’s head showed abnormal white matter and dysplasia of the corpus callosum (i.e., slightly hypointense on T1WI and T2WI of the frontal lobe and thinned corpus callosum). Those symptoms are consistent with the clinical manifestations and imaging changes of PURA syndrome. Along with the WES results, which indicated a de novo PURα mutation (heterozygous pathogenic variant), the symptoms were consistent with the genetic characteristics of PURA syndrome of the autosomal dominant inheritance. According to the genetic variation classification standard set in the Chinese version of the expert consensus guide of the American College of Medical Genetics and Genomics (ACMG) (17), the mutation was consistent with the evidence of pvsPSV1 (highly pathogenic variant) and PS2 (de novo mutation). With that, as well as the consistent clinical symptoms, the pathogenicity could be classified as “pathogenic”. In conclusion, the diagnosis of the present case was PURA syndrome.
Therefore, the common problems of PURA syndrome in the neonatal period are hypotonia, the resulting feeding difficulties, and apnea or primary hypoventilation. Feeding and respiratory problems are often the leading causes of hospitalization. The main MRI findings include delayed myelination, white matter dysplasia, lateral ventricle enlargement, and corpus callosum dysplasia. Generally, there were no characteristic facial features in patients with PURA syndrome. The prognosis generally results in moderate to severe mental retardation, severe language impairment, motor retardation, and peculiar gait and movement abnormalities. Sometimes the patients are diagnosed with cerebral palsy. More than half of the patients may have epilepsy; most of them develop into refractory epilepsy.
Meanwhile, the patients may demonstrate multi-system abnormalities. There is currently no specific treatment for PURA syndrome, with symptomatic treatment and rehabilitation therapy as the primary treatment. Because of the diversity in sites and forms of PURα gene mutation and the form of autosomal dominant inheritance, early diagnosis and early intervention may improve the prognosis and benefit genetic counseling. The phenotype of the de novo heterozygous mutation of the PURA gene in humans has not been completely determined. Whether there are differences in the clinical manifestations caused by different gene variants needs to be further explored.
Differentia diagnosis should be performed in neonatal dystonia with other hereditary diseases. Zadeh et al. (18) summarized the related genetic diseases and corresponding detection methods. The details were as follows: Prader-Willi syndrome: methylation analysis or SNRPN expression, and then high-resolution chromosome analysis. Myotonic dystrophy: detection of the DMPK gene trinucleotide repeats. Spinal muscular atrophy: target mutation analysis of SMN1 and SMN2. Congenital muscular dystrophy: gene sequencing of LAMA2, POMT1, POMT2, POMGNT1, or Fukutin. Nemaline myopathy: ACTA1 sequencing and NEB deletion or duplication, muscle biopsy. Congenital myelodysplastic neuropathy: gene sequencing of MPZ, PMP22, or EGR2. Congenital disorders of glycosylation: detection of carbohydrate-deficient transferrin by mass spectrometry followed by PMM2 gene mutation analysis. Pompeii disease: gene sequencing of GAA or enzyme activity assay in cultured skin fibroblast. After analyzing the present study, gene analysis of PURA syndrome and the corresponding PURα gene should be added.
In summary, PURA syndrome is challenging to diagnose clinically. In cases of decreased muscle tension in neonatal, together with breathing and feeding problems, drowsiness, hypothermia, and other manifestations, further testing is warranted to rule out PURA syndrome. In combination with the imaging changes, early targeted gene testing for PURA syndrome should be performed to achieve early diagnosis. A PURA gene examination should be performed for pediatric patients with epilepsy, mental retardation, neuromuscular dysfunction, and motor retardation. With the diagnosis of PURA syndrome, it is necessary to take active measures to deal with the symptoms, pay attention to all the patient’s systems, evaluate the possible long-term complications, and perform genetic counseling.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to acknowledge the hard and dedicated work of all the staff that implemented the intervention and evaluation components of the study.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Peking University Shenzhen Hospital committee (20200610) and with the Helsinki Declaration (as revised in 2013). The patient’s guardian provided written informed consent.
Footnotes
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-248
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-248). The authors have no conflicts of interest to declare.
References
- 1.Brown N, Burgess T, Forbes R, et al. 5q31.3 Microdeletion syndrome: clinical and molecular characterization of two further cases. Am J Med Genet A 2013;161A:2604-8. 10.1002/ajmg.a.36108 [DOI] [PubMed] [Google Scholar]
- 2.Bonaglia MC, Zanotta N, Giorda R, et al. Long-term follow-up of a patient with 5q31.3 microdeletion syndrome and the smallest de novo 5q31.2q31.3 deletion involving PURA. Mol Cytogenet 2015;8:89. 10.1186/s13039-015-0193-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reijnders MRF, Janowski R, Alvi M, et al. PURA syndrome: clinical delineation and genotype-phenotype study in 32 individuals with review of published literature. J Med Genet 2018;55:104-13. 10.1136/jmedgenet-2017-104946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee BH, Reijnders MRF, Abubakare O, et al. Expanding the neurodevelopmental phenotype of PURA syndrome. Am J Med Genet A 2018;176:56-67. 10.1002/ajmg.a.38521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimojima K, Isidor B, Le Caignec C, et al. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A 2011;155A:732-6. Erratum in: Am J Med Genet A. 2011 Nov;155A(11):2903. 10.1002/ajmg.a.33891 [DOI] [PubMed] [Google Scholar]
- 6.Hosoki K, Ohta T, Natsume J, et al. Clinical phenotype and candidate genes for the 5q31.3 microdeletion syndrome. Am J Med Genet A 2012;158A:1891-6. 10.1002/ajmg.a.35439 [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld JA, Drautz JM, Clericuzio CL, et al. Deletions and duplications of developmental pathway genes in 5q31 contribute to abnormal phenotypes. Am J Med Genet A 2011;155A:1906-16. 10.1002/ajmg.a.34100 [DOI] [PubMed] [Google Scholar]
- 8.Rezkalla J, Von Wald T, Hansen KA. Premature Thelarche and the PURA Syndrome. Obstet Gynecol 2017;129:1037-9. 10.1097/AOG.0000000000002047 [DOI] [PubMed] [Google Scholar]
- 9.Hunt D, Leventer RJ, Simons C, et al. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet 2014;51:806-13. 10.1136/jmedgenet-2014-102798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lalani SR, Zhang J, Schaaf CP, et al. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet 2014;95:579-83. 10.1016/j.ajhg.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka AJ, Bai R, Cho MT, et al. De novo mutations in PURA are associated with hypotonia and developmental delay. Cold Spring Harb Mol Case Stud 2015;1:a000356. 10.1101/mcs.a000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto N, Nakao H, Niihori T, et al. Patient with a novel purine-rich element binding protein A mutation. Congenit Anom (Kyoto) 2017;57:201-4. 10.1111/cga.12214 [DOI] [PubMed] [Google Scholar]
- 13.Li W, Guo ZM, Zhao XH. Advances in the structure and function of PURA gene. Chinese Bulletin of Life Sciences 2015;27:1160-7. [Google Scholar]
- 14.Lezon-Geyda K, Najfeld V, Johnson EM. Deletions of PURA, at 5q31, and PURB, at 7p13, in myelodysplastic syndrome and progression to acute myelogenous leukemia. Leukemia 2001;15:954-62. 10.1038/sj.leu.2402108 [DOI] [PubMed] [Google Scholar]
- 15.Bae CH, Kim DS, Jun YL, et al. Proteomic analysis of the effect of acupuncture on the suppression of kainic Acid-induced neuronal destruction in mouse hippocampus. Evid Based Complement Alternat Med 2013;2013:436315. 10.1155/2013/436315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel DC, Johnson EM. PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene 2018;643:133-43. 10.1016/j.gene.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang QJ, Shen YP, Wu LQ, et al. Standards and guidelines for the interpretation of sequence variants. Sci Sin Vitae 2017;47:668-88. [Google Scholar]
- 18.Zadeh N, Hudgins L. The genetic approach to hypotonia in the neonate. NeoReviews 2009;10:e600-7. 10.1542/neo.10-12-e600 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as