Abstract
Background
The usefulness of the Oncomine Dx Target test (Oncomine Dx), a next‐generation sequencing (NGS) test, has already been proven in clinical trials. However, NGS requires high‐quality tumor samples and takes a long time to generate results. The feasibility of NGS for use in advanced non‐small cell lung cancer (NSCLC) patients in clinical practice has not yet been determined.
Methods
Patients serially diagnosed with advanced NSCLC were evaluated in our hospital. The Oncomine Dx, Cobas EGFR mutation test (Cobas EGFR), and ALK‐IHC were performed. The patients were divided into four sets: the full analysis set (FAS) that referred to patients diagnosed with NSCLC, the intent to perform companion diagnostics (CDx) set (IPS) that referred to patients in which CDx had been ordered regardless of sample quality, the per‐performed CDx set (PPS) that referred to patients who could undergo CDx regardless of the results, and the per‐completed CDx set (CCS) that referred to patients in which informative results were received from the CDx.
Results
The total number of patients analyzed in the study was 167. The IPS/FAS of Oncomine Dx (80.2%) was lower than that of the ALK‐IHC (85.0%) and Cobas EGFR (92.8%). The CCS/FAS of Oncomine Dx (65.9%) was lower than that of the ALK‐IHC (82.0%) and Cobas EGFR (92.2%). PPS/IPS and CCS/PPS of the Oncomine Dx with nonsurgical biopsy ranged between 78.6% and 90.9%, which was lower than those patients who underwent surgical resection (95.0% and 100%).
Conclusions
The feasibility of Oncomine Dx in clinical practice was lower than the other CDx. The feasibility of Oncomine Dx will increase by improving the biopsy procedure.
Key points
Significant study findings
The usefulness of a next‐generation sequencing (NGS) test has been proven in clinical trials.
The feasibility of NGS is lower than other diagnostics in clinical practice especially with regard to nonsurgical biopsy.
What this study adds
It is necessary to improve the feasibility of NGS in clinical practice.
To improve NGS feasibility, turnaround time must be shortened, and larger samples must be obtained during surgical procedures.
Keywords: ALK‐immunohistochemistry, Cobas EGFR mutation test, Oncomine Dx Target test, real‐world analysis, turnaround time
The feasibility of Oncomine Dx in clinical practice is relatively low compared with that of the ALK‐IHC and Cobas EGFR.

Introduction
For several decades, progression has been made in the treatment of advanced non‐small cell lung cancer (NSCLC). The discovery of actionable gene mutations and development of molecular‐targeted drugs have improved long‐term survival in NSCLC patients. For example, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) and anaplastic lymphoma kinase (ALK) TKIs have been shown to prolong progression‐free survival compared with cytotoxic chemotherapy which has led to longer overall survival in NSCLC patients with EGFR‐activating mutations and ALK rearrangement. 1 , 2 , 3 Recently, several actionable genes (ROS1, BRAF, etc) have been identified, and new molecular‐targeted drugs for each gene have been developed. 4 , 5 Therefore, the diagnosis of actionable gene mutations is becoming an important issue.
To use molecular targeted drugs, detection of the actionable gene mutation with companion diagnostics (CDx) approved by the Japanese administration is necessary in Japan. However, various types of CDx are used even if the same actionable gene mutation is targeted. For example, the Therascreen and Cobas EGFR mutation tests have been approved as CDx for first‐ or second‐generation EGFR‐TKIs, while the Cobas EGFR mutation test v2 has been approved for osimertinib for third‐generation EGFR‐TKI. As for ALK rearrangement, the Vysis ALK Break Apart FISH probe kit has been approved as a CDx for crizotinib, while the Histofine ALK iAEP kit has been approved for alectinib. Moreover, several molecular‐targeted drugs and different CDx for ROS1, BRAF, NTRK, and MET have also been approved. Therefore, performing several singleplex CDx became necessary to select the precise treatment for each patient. This led to increasing tissue consumption and longer turnaround time (TAT) before starting first‐line chemotherapy. 6 , 7
Multiplex diagnostics, which are next‐generation sequencing (NGS)‐based tests (Oncomine Dx Target test [Oncomine Dx], Foundation one, etc), have been developed and are used in clinical practice to address this problem. Oncomine Dx was approved in June 2019 as a CDx for four drugs targeting four driver genes (EGFR, ALK, ROS1, and BRAF) in Japanese clinical practice. Oncomine Dx can detect not only four but also 42 gene mutations for research use. The usefulness and accuracy of Oncomine Dx has been proven in clinical trials.
However, some problems have been encountered with the use of NGS‐based tests, including Oncomine Dx, in clinical practice. First, the necessary tissue samples for NGS testing may be different from those for PCR‐based testing. NGS needs more tumor samples with a higher proportion of tumor cells than PCR‐based CDx for testing. 8 , 9 However, it is difficult to obtain large biopsy samples with commonly used biopsy procedures such as transbronchial biopsy (TBB). Therefore, the number of patients that can be tested with Oncomine Dx remains unknown. Second, it takes longer to obtain results using NGS than PCR‐based tests. 10 Longer TAT may also be harmful to some patients with advanced NSCLC. Moreover, early detection of EGFR mutations is important in Japanese clinical practice because these mutations are common in Japan. 11
The Oncomine Dx as CDx was used in the hospital of this study in advanced NSCLC patients from September 2019. In addition, the Cobas EGFR mutation test (Cobas EGFR) and ALK immunohistochemistry (IHC) were concurrently used with Oncomine Dx. This study compared the feasibility of each CDx (Oncomine Dx, Cobas EGFR, and ALK‐IHC) in a real‐world setting. Moreover, the feasibility of Oncomine Dx with each biopsy procedure was analyzed.
Methods
Patients serially diagnosed with advanced NSCLC in the Cancer Institute Hospital of the Japanese Foundation for Cancer Research from September 2019 to July 2020 were retrospectively evaluated. The Oncomine Dx as CDx was used instead of singleplex CDx during the study period. The performance of Oncomine Dx was covered by health insurance. Moreover, Cobas EGFR and ALK‐IHC were used for research to avoid the oversight of gene mutations with Oncomine Dx. This study was approved by the Institutional Review Board of the Cancer Institute Hospital of the Japanese Foundation for Cancer Research. The patients signed their informed consent for a comprehensive agreement, and those who requested to opt‐out of this study were able to do so through the hospital website.
Sample preparations
The samples were obtained by TBB, endobronchial ultrasound‐guided transbronchial needle aspiration (EBUS‐TBNA), echo‐ or computed tomography (CT)‐guided needle biopsy (CTNB), surgical resection, or others (transesophageal biopsy, thoracentesis, or plasma liquid biopsy). An experienced pathologist checked the tissue samples before performing the Oncomine Dx, and samples with ≥200 tumor cells and ≥20% tumor cell content were submitted to the laboratory. The samples were not submitted if they did not meet the aforementioned criteria. A total of 10 slides, each 5 μm thick, and FFPE sections were used to extract the nucleic acid in one patient until December 2019, while 20 slides were used from January 2020. Oncomine Dx was performed at LSI Medience Corporation, a Japanese commercial laboratory. ALK‐IHC was ordered for all patients if the biopsy sample contained cancer cells. Moreover, ALK‐IHC was performed in house using the ALK antibody clone 5A4 (not an approved CDx IHC test kit). Cobas EGFR was ordered for all patients if cytology or tissue samples contained cancer cells. Cobas EGFR was performed at the LSI Medience Corporation (other PCR‐based methods were performed instead of the Cobas EGFR test in two patients).
Definition of patient sets
Patient sets were established to evaluate the feasibility of each CDx (Fig 1). The full analysis set (FAS) referred to patients diagnosed with NSCLC. The intent to perform CDx set (IPS) referred to patients in which CDx was ordered regardless of tissue sample quality. The per‐performed CDx set (PPS) referred to patients who could undergo CDx regardless of the results. The per‐completed CDx set (CCS) referred to patients in which informative results could be provided from CDx regardless of whether the results were positive or negative. The mutation‐positive set (MPS) referred to patients in which there were some gene mutations. The IPS/FAS, CCS/FAS, MPS/FAS, and CCS/PPS were calculated to evaluate in clinical practice the actual ordering rate of CDx, the actual feasibility rate of CDx, the actual mutation detection rate of CDx, and the actual success rate of CDx, respectively. TAT was defined as the day from the date of CDx ordering until the date of CDx results recorded in the medical chart. Moreover, IPS/FAS, CCS/FAS, MPS/FAS, CCS/PPS, and TAT were compared between each CDx. PPS/IPS and CCS/PPS in each biopsy procedure were also evaluated to evaluate the characteristics of each biopsy procedure. In addition, PPS/IPS was calculated to evaluate the actual sufficient sample preparation rate of CDx. Furthermore, the results of the CDx were directly compared for each patient to evaluate the concordance of each CDx.
Figure 1.
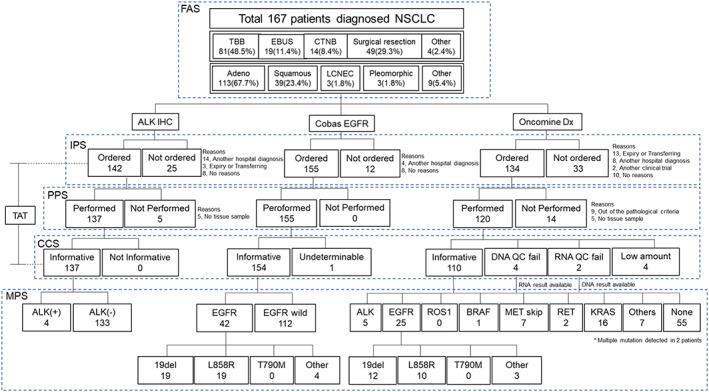
The flowchart of each diagnostic method.
ALK rearrangement analysis
The ALK expression was tested by RT‐PCR and gel electrophoresis. The primers used on PCR are F: 5′‐GACCTCCTCCATCAGTGACCT‐3′ and R: 5′‐CAGCGTCTTCACAGCCACTTG‐3′ ((ALK ex20‐22); F: 5′‐GCAACATCAGCCTGAAGACA‐3′ and R: 5′‐GCCTGTTGAGAGACCAGGAG‐3′ (ALK ex28‐29). Split fluorescence in situ hybridization (FISH) assays were carried out on unstained slides (4 μm thick) using bacterial artificial chromosome (BAC) clone‐derived DNA probes for ALK. The names of the BAC clone used are available upon request.
Results
Diagnostic method flowchart
A total of 167 patients were analyzed (FAS). The flowchart of each diagnostic method is shown in Fig 1. The characteristics of the FAS and IPS patients in each CDx are shown in Table 1. ALK‐IHC was ordered in 142 patients (IPS of ALK‐IHC). The ALK‐IHC was not ordered in 25 patients because 14 were diagnosed at another hospital, three had died or had been transferred to another hospital, and eight for no known reason. ALK‐IHC was performed in 137 patients (PPS of ALK‐IHC). In five patients, ALK‐IHC was not carried out because no tissue samples were obtained. Informative results (CCS of ALK‐IHC) were received for 137 patients. Cobas EGFR was ordered in 155 patients (IPS of Cobas EGFR). Cobas EGFR was not performed in 12 patients due to the following reasons: four were diagnosed at another hospital and eight for no known reason. Cobas EGFR was conducted in 155 patients (PPS of Cobas EGFR). Informative results (CCS of Cobas EGFR) were received for a total of 154 patients. Oncomine Dx was ordered in 134 patients (IPS of Oncomine Dx). Meanwhile, Oncomine Dx was not ordered in some patients because 13 had died or had been transferred to another hospital, eight were diagnosed at another hospital, two participated in another clinical trial, and there was no known reason in 10 patients. Oncomine Dx was examined in 120 patients (PPS of Oncomine Dx). Oncomine Dx was not examined in some patients because nine samples did not meet the pathological criteria, and no tissue samples were obtained in five. Informative results from Oncomine Dx (CCS of Oncomine Dx) were received for 110 patients. The reasons for failing to receive informative results from Oncomine Dx were because four patients had low nucleic acid, four had poor DNA quality, and two had poor RNA quality.
Table 1.
Patient characteristics of FAS and IPS of each companion diagnostic (CDx)
| Total (FAS) | ALK IHC (IPS) | Cobas EGFR (IPS) | Oncomine Dx (IPS) | |
|---|---|---|---|---|
| N = 167 | N = 142 | N = 155 | N = 134 | |
| Age (median, range) | 68 (40–84) | 67 (40–84) | 68 (40–84) | 67 (40–84) |
| Sex (male/female) | 108/59 | 92/50 | 106/49 | 90/44 |
| Smoking status (never/ex‐current) | 49/118 | 40/102 | 46/109 | 39/95 |
| Stage (I/II/III/IV/Rec) | 1/6/34/88/38 | 1/5/25/76/35 | 1/6/28/83/37 | 1/4/27/73/29 |
| Pathology (Ad/Sq/other) | 113/39/15 | 102/29/11 | 108/32/15 | 91/29/14 |
| Diagnostic procedure | ||||
| TBB | 81 | 71 | 75 | 64 |
| EBUS‐TBNA | 19 | 16 | 18 | 14 |
| CTNB | 14 | 9 | 12 | 13 |
| Surgical resection | 49 | 43 | 47 | 40 |
| Others | 4 | 3 | 3 | 3 |
Ad, adenoma; CTNB, computed tomography (CT)‐guided needle biopsy; EBUS‐TBNA, endobronchial ultrasound‐guided transbronchial needle aspiration; Sq, squamous; TBB, transbronchial biopsy.
Feasibility of each CDx
IPS/FAS, CCS/FAS, MPS/FAS, CCS/PPS, and TAT were compared between each CDx to evaluate the practical feasibility of each CDx (Table 2). The IPS/FAS of Oncomine Dx (80.2%) was lower than that of ALK‐IHC (85.0%) and Cobas EGFR (92.8%). This finding indicates that the ordering rate of Oncomine Dx was lower than that of ALK‐IHC and Cobas EGFR; the CCS/FAS of Oncomine Dx (65.9%) was lower than that of ALK‐IHC (80.2%) and Cobas EGFR (92.2%); the CCS/PPS of Oncomine Dx (91.7%) was lower than that of ALK‐IHC (100%) and Cobas EGFR (99.4%). These findings indicate that the feasibility of Oncomine Dx is lower than that of ALK‐IHC and Cobas EGFR. The median TAT of ALK‐IHC, Cobas EGFR, and Oncomine Dx was 1 (range: 1–3), six (range: 5–18), and 13 (range: 9–29) days, respectively. The TAT of Oncomine Dx was longer than that of ALK‐IHC and Cobas EGFR.
Table 2.
IPS/FAS, CCS/FAS, MPS/FAS, CCS/PPS and TAT of each companion diagnostic (CDx)
| ALK IHC | Cobas EGFR | Oncomine Dx | |
|---|---|---|---|
| IPS/FAS | 142/167 (85.0%) | 155/167 (92.8%) | 134/167 (80.2%) |
| CCS/FAS | 137/167 (80.2%) | 154/167 (92.2%) | 110/167 (65.9%) |
| MPS/FAS | 4/167 (2.4%) | 42/167 (25.1%) | 63/167 (37.7%) |
| TAT (days) median range | 1, 1–3 | 6, 5–18 | 13, 9–29 |
| CCS/PPS | 137/137 (100%) | 154/155 (99.4%) | 110/120 (91.7%) |
| MPS for ALK/FAS | 4/167 (2.4%) | −/− | 5/167 (3.0%) |
| MPS for EGFR/FAS | −/− | 42/167 (25.1%) | 25/167 (15.0%) |
| Other gene MPS/FAS | −/− | −/− | 33/167 (19.8%) |
CCS, per‐completed CDx set; FAS, full analysis set; IPS, intent to perform companion diagnostics (CDx) set; MPS, mutation positive set; PPS, per‐performed CDx set; TAT, turnaround time.
CDx mutation detection
There were 4 patients who had positive results for ALK‐IHC. ALK fusion genes were detected in five patients who underwent Oncomine Dx. EGFR mutations were detected in 42 patients with Cobas EGFR (19, EGFR exon 19 deletions; 19, L858R; and four, other mutations). EGFR mutations were detected in 25 patients with Oncomine Dx (12, EGFR exon 19 deletions; 10, L858R; and three, other mutations). BRAF V600E, MET exon 14 skippings, KRAS mutation, and RET fusion were detected with Oncomine Dx in one, seven, 16, and two patients, respectively. The MPS for ALK/FAS of Oncomine Dx (3.0%) was higher than that of ALK‐IHC (2.4%). The MPS for EGFR/FAS of Oncomine Dx (15.0%) was lower than that of Cobas EGFR (25.1%). The MPS/FAS for other genes of Oncomine Dx was 19.8%. The concordance between each CDx is shown in Table 3. The positive, negative, and overall percentage agreement of ALK rearrangement were 100%, 98.9%, and 99.0%, respectively, excluding patients who did not receive informative results. The positive, negative, and overall percent agreement of EGFR mutations were all 100%, excluding patients in whom informative results had not been received. The diagnostic accuracy of Oncomine Dx would be sufficient if each CDx was precisely performed. One case had a discordant result on ALK rearrangement (IHC negative and Oncomine Dx positive). The case was reanalyzed with FISH, ALK‐IHC CDx test kit, and kinase domain sequence with RT‐PCR. All examinations showed a negative result on ALK rearrangement. Moreover, treatment response with alectinib and ALK‐TKI were not confirmed (Fig 2).
Table 3.
Concordance between each companion diagnostic (CDx)
| ALK‐IHC | |||
|---|---|---|---|
| Positive (N = 4) | Negative (N = 133) | Unknown (N = 30) | |
| Oncomine Dx | |||
| Positive (N = 5) | 4 | 1 | 0 |
| Negative (N = 109) | 0 | 94 | 15 |
| Not informative (N = 6) | 0 | 6 | 0 |
| Not performed (N = 47) | 0 | 32 | 15 |
| Cobas EGFR | |||
|---|---|---|---|
| Positive (N = 42) | Negative (N = 112) | Unknown (N = 13) | |
| Oncomine Dx | |||
| Positive (N = 25) | 25 | 0 | 0 |
| Negative (N = 87) | 0 | 78 | 9 |
| Not informative (N = 8) | 1 | 6 | 1 |
| Not performed (N = 47) | 16 | 28 | 3 |
Figure 2.

The case with a discordant result on ALK rearrangement. (a) The FFPE sample analyzed with Oncomine Dx (p40‐IHC positive and ALK‐IHC negative). (b) FISH using green (3′) and red (5′) ALK probes showed a negative pattern on ALK rearrangement. (c) The CT images before and after alectinib treatment. The mediastinal lymph node metastasis (#7) did not shrink after two months of alectinib treatment.
Influence of biopsy procedures
The PPS/IPS and CCS/PPS of each CDx in every biopsy procedure were analyzed to evaluate the influence of biopsy procedures on CDx (Table 4). The PPS/IPS of Oncomine Dx, ALK‐IHC, and Cobas EGFR in TBB were 90.6%, 97.2%, and 100%, respectively. The PPS/IPS of Oncomine Dx, ALK‐IHC, and Cobas EGFR in EBUS‐TBNA were 78.6%, 100%, and 100%, respectively. The PPS/IPS of Oncomine Dx, ALK‐IHC, and Cobas EGFR in CTNB were 84.6%, 100%, and 100%, respectively. The PPS/IPS of Oncomine Dx, ALK‐IHC, and Cobas EGFR in surgical resection were all 100%. The PPS/IPS of Oncomine Dx in every biopsy procedure except for surgical resection was lower than that of ALK‐IHC and Cobas EGFR. This finding indicates that biopsy procedures except for surgical resection are not usually appropriate methods for obtaining sufficient samples for Oncomine Dx. The CCS/PPS of Oncomine Dx of Oncomine Dx, ALK‐IHC, and Cobas EGFR in TBB were 89.7%, 100%, and 98.7%, respectively. The CCS/PPS of Oncomine Dx of Oncomine Dx, ALK‐IHC, and Cobas EGFR in EBUS‐TBNA were 90.9%, 100%, and 100%, respectively. The CCS/PPS of Oncomine Dx of Oncomine Dx, ALK‐IHC, and Cobas EGFR in CTNB were 90.9%, 100%, and 100%, respectively. The CCS/PPS of Oncomine Dx, ALK‐IHC, and Cobas EGFR in surgical resection were 95.0%, 100%, and 100%, respectively. These findings indicate that the biopsy procedures except for surgical resection are sometimes not the appropriate methods for obtaining high‐quality tumor samples for Oncomine Dx.
Table 4.
PPS/IPS and CCS/PPS of each companion diagnostic (CDx) in each biopsy procedure
| FAS | TBB (n = 81) | EBUS‐TBNA (N = 19) | CTNB (N = 14) | Surgical resection (N = 49) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALK IHC | Cobas EGFR | Oncomine Dx | ALK IHC | Cobas EGFR | Oncomine Dx | ALK IHC | Cobas EGFR | Oncomine Dx | ALK IHC | Cobas EGFR | Oncomine Dx | |
| IPS | 71 | 75 | 64 | 16 | 18 | 14 | 9 | 12 | 13 | 43 | 47 | 40 |
| PPS | 69 | 75 | 58 | 16 | 18 | 11 | 9 | 12 | 11 | 43 | 47 | 40 |
| CCS | 69 | 74 | 52 | 16 | 18 | 10 | 9 | 12 | 10 | 43 | 47 | 38 |
| MPS | 0 | 17 | 27 | 1 | 5 | 5 | 1 | 3 | 5 | 2 | 14 | 24 |
| PPS/IPS | 69/71 (97.2%) | 75/75 (100%) | 58/64 (90.6%) | 16/16 (100%) | 18/18 (100%) | 11/14 (78.6%) | 9/9 (100%) | 12/12 (100%) | 11/13 (84.6%) | 43/43 (100%) | 47/47 (100%) | 40/40 (100%) |
| CCS/PPS | 69/69 (100%) | 74/75 (98.7%) | 52/58 (89.7%) | 16/16 (100%) | 18/18 (100%) | 10/11 (90.9%) | 9/9 (100%) | 12/12 (100%) | 10/11 (90.9%) | 43/43 (100%) | 47/47 (100%) | 38/40 (95.0%) |
| ALK MPS/FAS | 0/81 (0%) | 1/81 (1.2%) | 1/19 (5.3%) | 1/19 (5.3%) | 1/14 (7.1%) | 1/14 (7.1%) | 2/49 (4.1%) | 2/49 (4.1%) | ||||
| EGFR MPS/FAS | 17/81 (21.0%) | 12/81 (14.8%) | 5/19 (26.3%) | 2/19 (10.5%) | 3/14 (21.4%) | 2/14 (14.3%) | 14/49 (28.6%) | 7/49 (14.3%) | ||||
| Other gene MPS/FAS | 14/81 (17.3%) | 2/19 (10.5%) | 2/14 (14.3%) | 13/49 (26.5%) | ||||||||
CCS, per‐completed CDx set; FAS, full analysis set; IPS, intent to perform companion diagnostics (CDx) set; MPS, mutation positive set; PPS, per‐performed CDx set.
Discussion
This study found that Oncomine Dx had low feasibility in clinical practice. The IPS/FAS (actual ordering rate) and CCS/FAS (actual feasibility rate) of Oncomine Dx were lower than those of ALK‐IHC and Cobas EGFR. This is one of the challenges encountered when using Oncomine Dx, especially when diagnosing NSCLC patients with EGFR mutations. EGFR mutations are common in Japan. This is an essential piece of information in deciding on first‐line chemotherapy. EGFR mutations may be overlooked in many patients if Oncomine Dx is performed alone, and Cobas EGFR is not conducted. The MPS for EGFR/FAS of Oncomine Dx was lower than that of Cobas EGFR (15.0% vs. 25.1%).
We know that most surgical samples were successfully analyzed with Oncomine Dx, which would ensure the feasibility of Oncomine Dx. However, not all patients were able to undergo surgical resection, and biopsies were performed in most advanced NSCLC patients with TBLB or other procedures in clinical practice. Therefore, the influence of biopsy procedures on the feasibility of Oncomine Dx was evaluated. The PPS/IPS (actual sample preparation rate) and CCS/PPS of each CDx were compared in each biopsy procedure to evaluate the difference between biopsy procedures. The PPS/IPS and CCS/PPS of Oncomine Dx in TBB, EBUS‐TBNA, and CTNB in this study ranged between 78.6% and 90.9%. The PPS/IPS of Oncomine Dx in EBUS‐TBNA was particularly low (78.6%). Therefore, a nonsurgical biopsy procedure, especially EBUS‐TBNA, may not be appropriate for Oncomine Dx. Surgical resection should be considered as a biopsy procedure if sufficient samples cannot be obtained using other procedures to improve the feasibility of Oncomine Dx.
In addition, individual reasons for not ordering Oncomine Dx included patients “diagnosed at another hospital.” This may have been avoided if the patients had been diagnosed at our hospital. Until recently, PCR‐based single plex CDx has been used in Japan, and practitioners may have little motivation to obtain large tissue samples for NGS. As mentioned above, it will be possible to improve the feasibility of Oncomine Dx if the biopsy procedure to obtain large samples is improved.
The long TAT is also a disadvantage of Oncomine Dx in the diagnosis of EGFR mutations. The median TAT from ordering Oncomine Dx was longer than that of Cobas EGFR (13 vs. 6 days). Moreover, it takes more time before ordering Oncomine Dx because a pathological check is still required to assess the quality of tissue samples, unlike Cobas EGFR. A previous study reported the median time from the confirmation of NSCLC diagnosis to the initiation of first‐line treatment was 19 days. 7 Therefore, 13 days as the median TAT of Oncomine Dx is relatively long. As a result, several problems need to be solved if Oncomine Dx is going to be routinely used as a CDx only for EGFR and ALK, which are the most important molecular targets in Japan at present. However, other actionable genes such as ROS‐1 BRAF MET as well as EGFR and ALK have been identified and effective molecular target agents approved which are now available in Japanese clinical practice. If all actionable gene mutations are examined with each single‐plex CDx, the TAT for all target genes would be significantly longer. Therefore, the usefulness of Oncomine Dx will increase in the near future. Automated systems such as Genexus are currently being developed, 12 and these systems will resolve the problems of the long TAT of Oncomine Dx.
This study has some limitations. First, some bias could not be eliminated because of the retrospective nature of this study. Second, this study was conducted in a single institution. Hence, some biases may exist, especially the rate of CDx ordering. Third, the number of patients with positive gene mutations was not sufficient to analyze ALK gene fusion. Therefore, the analysis performed, especially on the concordance between each CDx, would not be enough.
In conclusion, the feasibility of Oncomine Dx in clinical practice was relatively lower than that of ALK‐IHC and Cobas EGFR. However, it will hopefully be possible to improve the feasibility of Oncomine Dx if the biopsy procedure to obtain larger samples is refined.
Disclosure
K.U. reports employment/leadership position/advisory role of Daiichi Sankyo. N.Y. reports employment/leadership position/ advisory role of Chugai Pharmaceutical. K.T. reports grants and personal fees from Kyowa Kirin, personal fees from Chugai, MSD, Takeda, Janssen, Eizai, Cellgene, Yakult and Taiho, outside the submitted work. M.N. reports grants and nonfinancial support from F. Hoffmann‐La Roche, during the conduct of the study; grants and personal fees from Ono Pharmaceutical, Bristol Myers Squibb, Pfizer, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, AstraZeneca, MSD and Novartis, personal fees from Boehringer‐Ingelheim, Sankyo Healthcare, Taiho Pharmaceutical and from Merck Serono, grants from Astellas, outside the submitted work. All the other authors have stated that they have no conflicts of interest.
References
- 1. Mitsudomi T, Morita S, Yatabe Y et al Gefitinib versus cisplatin plus docetaxel in patients with non‐small‐cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol 2010; 11 (2): 121–8. [DOI] [PubMed] [Google Scholar]
- 2. Solomon BJ, Mok T, Kim D‐W et al First‐line crizotinib versus chemotherapy in ALK‐positive lung cancer. N Engl J Med 2014; 371 (23): 2167–77. [DOI] [PubMed] [Google Scholar]
- 3. Kris MG, Johnson BE, Berry LD et al Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014; 311 (19): 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shaw AT, Ou S‐HI, Bang Y‐J et al Crizotinib in ROS1‐rearranged non‐small‐cell lung cancer. N Engl J Med 2014; 371 (21): 1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Planchard D, Besse B, Groen HJM et al Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)‐mutant metastatic non‐small cell lung cancer: An open‐label, multicentre phase 2 trial. Lancet Oncol 2016; 17 (7): 984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu TM, Morrison C, Gold EJ, Tradonsky A, Layton AJ. Multiple biomarker testing tissue consumption and completion rates with single‐gene tests and investigational use of Oncomine dx target test for advanced non–small‐cell lung cancer: A single‐center analysis. Clin Lung Cancer 2019; 20 (1): 20–29.e8. [DOI] [PubMed] [Google Scholar]
- 7. Shimizu J, Masago K, Saito H et al Biomarker testing for personalized, first‐line therapy in advanced nonsquamous non‐small cell lung cancer patients in the real world setting in Japan: A retrospective, multicenter, observational study (the BRAVE study). Ther Adv Med Oncol 2020; 12: 175883592090452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Einaga N, Yoshida A, Noda H et al Assessment of the quality of DNA from various formalin‐fixed paraffin‐embedded (FFPE) tissues and the use of this DNA for next‐generation sequencing (NGS) with no artifactual mutation. PLOS One 2017; 12 (5): e0176280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arreaza G, Qiu P, Pang L et al Pre‐analytical considerations for successful next‐generation sequencing (NGS): Challenges and opportunities for formalin‐fixed and paraffin‐embedded tumor tissue (FFPE) samples. Int J Mol Sci 2016; 17 (9): 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding PN, Becker T, Bray V et al Plasma next generation sequencing and droplet digital PCR‐based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent‐line osimertinib. Thorac Cancer 2019; 10 (10): 1879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kohno T, Nakaoku T, Tsuta K et al Beyond ALK‐RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015; 4 (2): 156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Low S‐KK, Uchibori K, Hayashi R et al Evaluation of Genexus system that automates specimen‐to‐report for cancer genomic profiling within a day using liquid biopsy. J Clin Oncol 2020; 38 (15_suppl): 3538.32795225 [Google Scholar]


