Abstract
Type 1 diabetes (T1D) is the third most common autoimmune disease which develops due to genetic and environmental risk factors. Based on the World Health Organization (WHO) report from 2014 the number of people suffering from all types of diabetes ascended to 422 million, compared to 108 million in 1980. It was calculated that this number will double by the end of 2030. In 2015 American Diabetes Association (ADA) announced that 30.3 million Americans (that is 9.4% of the overall population) had diabetes of which only approximately 1.25 million had T1D. Nowadays, T1D represents roughly 10% of adult diabetes cases total. Multiple genetic abnormalities at different loci have been found to contribute to type 1 diabetes development. The analysis of genome-wide association studies (GWAS) of T1D has identified over 50 susceptible regions (and genes within these regions). Many of these regions are defined by single nucleotide polymorphisms (SNPs) but molecular mechanisms through which they increase or lower the risk of diabetes remain unknown. Genetic factors (in existence since birth) can be detected long before the emergence of immunological or clinical markers. Therefore, a comprehensive understanding of the multiple genetic factors underlying T1D is extremely important for further clinical trials and development of personalized medicine for diabetic patients. We present an overview of current studies and information about regions in the human genome associated with T1D. Moreover, we also put forward information about epigenetic modifications, non-coding RNAs and environmental factors involved in T1D development and onset.
Keywords: non-coding RNA, genetic factors, type 1 diabetes, genome-wide association studies, linkage, epigenetic factors
Introduction
Type 1 diabetes (T1D) is one of the most common auto-immune diseases which develops due to genetic and environmental risk factors [1]. This disease is also known as insulin-dependent, juvenile or childhood-onset diabetes [2]. T1D is based on the immune-mediated loss of pancreatic β cells. This deprivation leads to insulin deficiency, which means that people suffering from this disease do not produce enough insulin and are characterized by the clinical need for it. Insulin-secreting cells can be destroyed by genetic and non-genetic factors [1]. Clinical symptoms occur when β cell deficiency exceeds 40% of their overall amount. This is combined with detectable decrease in concentration a marker of pancreatic insulin production, plasma C-peptide [3]. What is more, most of T1D patients exhibit a presence of disease-associated serum autoantibodies [1]. Nowadays, T1D diagnosis is partially based on the detection of islet cells autoantibodies. The most often detected are the following antibodies: 1) insulin autoantibodies (IAA), 2) anti-glutamic acid decarboxylase (GAD) autoantibodies, 3) protein tyrosine phosphatase-related islet antigen 2 (IA-2) autoantibodies (also known as ICA512 autoantibodies) [4] and 4) zinc transporter 8 (ZnT8) auto-antibodies [3]. It has been already proven that specific human leukocyte antigen (HLA) haplotypes are associated with islet primary autoantibodies, which are observed at seroconversion. For example, HLA-DR4-DQ8 is associated with IAA and HLA-DR3-DQ2 is associated with GAD [5]. These circulating autoantibodies are characteristic of T1D type 1A. A much less common type (roughly 10% of cases) of T1D is type 1B (alias idiopathic T1D). Patients suffering from this type of diabetes are autoantibody-negative, which might suggest that they suffer from 1) a rare (about 6% of T1D cases) monogenic form of diabetes or 2) a lack of measurable autoantibodies responses to common antigens [6, 7].
T1D occurs in genetically susceptible patients that were subjected to one or more triggering environmental factors which induced an autoimmune process leading to destruction of β cells in islets of Langerhans [8-11]. The preclinical period of antibody production and islet destruction can last several months or even years and it is crucial in terms of medical intervention focused on slowing down or stopping this process [12-14]. Early identification of people with high genetic predisposition to T1D extends the time for prevention or treatment and helps to decelerate the disease [15-17]. Maybe in the future we will be able to stop the changes in the immune system and the development of autoantibodies that take part in the destruction process of insulin-producing cells [1].
Polymorphisms of multiple genes are reported to modify the risk and course of T1D [17-20]. Among them, numerous variants of genes from the HLA region are factors playing a very important role in T1D. They can affect the presence of autoreactive antigen T lymphocytes during a diabetic inflammatory process [21-24]. Coexistence of various haplotypes affecting autoimmune response and balance between them might determine the onset time, the process and dynamics of the disease or its clinical form [20, 21, 25].
Identification of factors implicated in diabetes and mechanisms of interaction between those factors should lead to a better understanding of the disease pathogenesis and help in the development of T1D prevention strategies.
Genetic studies in diabetes
T1D is a member of the multifactorial disease group [22]. One of its most important factors are variants of HLA region. First data suggesting the significance of associations in the HLA region were published in the 1970s [23, 24]. Many studies have been performed since then identifying associations not only in genes constituting the HLA region but also outside this region [25]. The single nucleotide polymorphism (SNPs), which are associated with islet autoimmunity and high risk of T1D, are placed in such non-HLA genes as PTPN22 (protein tyrosine phosphatase non-receptor type 22), UBASH3A (ubiquitin associated and SH3 domain containing A), ERBB3 (Erb-B2 receptor tyrosine kinase 3), CLEC16A (C-type lectin domain containing 16A), IL27 (interleukin 27), CTRB (chymotrypsinogen B1), C14orf64 [also known as LINC01550 (long intergenic non-protein coding RNA 1550)], GSDM (gasdermin A) and HORMAD2 [HORMA (Hop1, Rev7 and MAD2) domain containing 2] [4]. Three technical approaches are used for detection of risk loci in T1D: linkage studies, association studies and genome-wide association studies (GWAS).
High risk loci identified by these three approaches explain almost 80% of the heritability of T1D. To get a better understanding of T1D, further genetic studies are required to explain how genetic variants change β-cell response to environmental factors or inflammatory mediators. This can become possible with the use of new technologies like next generation sequencing (NGS).
Linkage studies
Linkage studies are capable of identifying genomic regions which are shared more frequently between affected relatives, and they generally examine sibling pairs [26]. This approach is mostly used when effect sizes of the risk factors are relatively large (compared to GWAS) while analyzed markers are present in a genome in small density. And for this reason it is worth analyzing those markers further because they may play a much more important role in T1D. The HLA region (6p21) is a good example and an evidence of linkage with T1D and therefore it proves the importance of additional research [27, 28].
Association studies
Association studies allow us to detect alleles that are rather coon but they have a modest effect on a disease risk [26]. In the simplified form, association studies compare frequency of an allele/a genotype of a specific variant between disease cases and a control group [29]. They are generally focused on candidate genes [30]. T1D association studies identified 4 non-HLA genes with established risk loci [HLA, INS (insulin), CTLA4 (cytotoxic T-lymphocyte antigen 4), PTPN22] [4].
Genome-wide association studies
Emergence of GWAS revolutionized genetic studies and genome examination in search of risk factors. GWAS lets us survey a genome in search of causal variants i.e. variants that contribute to a disease but may not be a sufficient reason for this disease in isolation [31]. Analyses performed by The Welcome Trust Case Control Consortium (WTCCC) [32] and Type 1 Diabetes Genetics Consortium [33, 34] have revealed more than 50 genetic variants associated with T1D development [19]. These studies, next to well-established non-HLA loci, have reported many new associations with T1D, among them: GLIS3 (GLIS family zinc finger 3), LMO7 (LIM Domain Only 7), FUT2 (fuco-syltransferase 2) [35-37]. Additionally, genome-wide genotyping approach of nonsynonymous SNPs (nsSNP), a direct precursor of GWAS, uncovered the sixth gene ever associated with T1D – IFIH1 (interferon induced with helicase C domain 1) [38, 39].
Candidate genes for type 1 diabetes
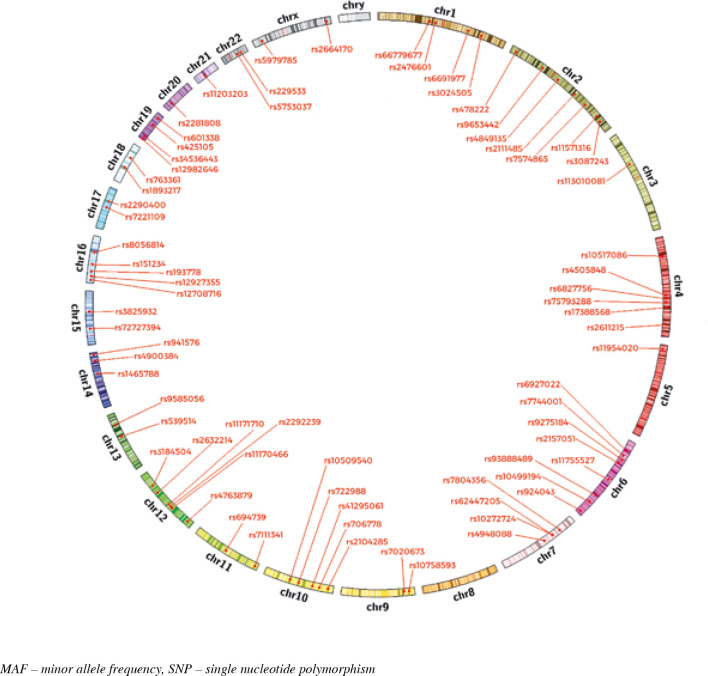
Susceptibility to T1D is strongly associated with a genetic background. GWAS contributed to the discovery of a number of T1D susceptibility genes. Before GWAS only five loci were determined as associated with T1D. They were (in that order): HLA region, INS, CTLA4, PTPN22 and IL2RA. GWAS meta-analyses have identified more than 50 T1D susceptibility loci (Fig. 1). Some of these genes can act as modulators of an immune-related process through both antigen presentation and other modifications of the immune function. A number of candidate genes involved in T1D are known as modulators of β-cell apoptosis, viral infection or islet inflammation. Some of the main factors causing changes in susceptibility to T1D are SNPs and variations in gene expression (genetic variants in regulatory elements of genes can result in alteration of transcription and then gene expression). In addition, most of the identified genes are expressed both in immune competent cells and in pancreatic β-cells, which suggests a genetically modulated dialogue between these two components of T1D. Below we present recently examined genes associated with T1D development according to GWAS study with the statistical threshold for genome-wide significance, P-values < 5 × 10–8 (P-values calculated using Fisher’s combined p-value method implemented in Haploview) [25]. In Table 1 we characterised genes which have been indicated as ones associated with T1D in studies performed so far. Their SNPs positions on chromosomes are illustrated in Figure 1.
Fig. 1.
SNPs positions on chromosomes
Table 1.
Type 1 diabetes associated loci according to ImmunoBas [91] and the latest studies
| Region | Gene | SNP | HGVS nomenclature [92] | MAF | Coding protein | Role | Ref. |
|---|---|---|---|---|---|---|---|
| 1p13.2 |
PTPN22 PHTF1 |
rs2476601 rs6679677 |
NC_000001.11:g.113834946A>G NC_000001.10:g.114303808C>A |
A = 2.74% A = 2.58% |
Protein tyrosine phosphatase, non-receptor type 22 Putative homeodomain transcription factor 1 |
Lymphoid protein tyrosine phosphate is a key molecule regulating TCR signaling and the SNP in these gene could leads to higher T-cell activation PHTF1 plays role in transcription regulation |
[91] |
| 1q32.1 | CAMSAP2 | rs6691977 | NC_000001.10:g.200814959T>C | C = 42.45% | Calmodulin regulated spectrin associated protein family member 2 | Not applicable | [93] |
| 1q32.1 | IL10 | rs3024505 | NC_000001.10:g.206939904G>A | A = 8.63% | Interleukin 10 | IL-10 is an anti-inflammatory cytokine | [91] |
| 2q24.2 | IFIH1 | rs2111485 | NC_000002.11:g.163110536A>G | G = 33.93% | Interferon induced with helicase C domain 1 | The protein acts as cytoplasmic sensor of viral nucleic acids and plays role in sensing viral infection and in the activation of cascade of antiviral responses by induces of type I interferons and proinflammatory cytokines. SNP in these gene may lead to increase viral infection and innate immune response | [93] |
| 2q32.3 | STAT4 | rs7574865 | NC_000002.11:g.191964633T>G | T = 25.54% | Signal transducer and activator of transcription 4 | STAT4 plays role as a signal transduction and activation of transcription. SNP in these gene is associated with several autoimmune disorders but there is no know exactly role of these mutation in developing of disease | [65] |
| 2q33.2 | CTLA4 | rs11571316 rs3087243 |
NC_000003.11:g.102220660A>G NC_000002.11:g.204738919G>A |
A = 47.1% A = 36.9% |
Cytotoxic T-lymphocyte associated protein 4 | CTLA4 is an protein receptor and downregulates immune reaction | [25, 91] |
| 2q13 | ACOXL | rs4849135 | NC_000002.11:g.111615079T>G | T = 30.7% | acyl-CoA oxidase like | Not applicable | [93] |
| 2p23.3 | EFR3B | rs478222 | NC_000002.11:g.25301755A>T | T = 36.74% | EFR3 homolog B | The protein is involved in signaling processes | [25] |
| 2q11.2 | AFF3 | rs9653442 | NC_000002.11:g.100825367C>T | T = 43.11% | AF4/FMR2 family member 3 | Transcriptional activator, involved in lymphoid development and oncogenesis | [94] |
| 3p21.31 | CCR5 | rs113010081 | NC_000003.11:g.46457412T>C | C = 2.84% | C-C chemokine receptor 5 | The CCR5 affects the immune cell function, mutation in this gene influences the function of immune cells response | [93] |
| 4q32.3 |
AC080079.1 or NOL8P1 |
rs2611215 | NC_000004.11:g.166574267A>G | A = 18.17% | Tripartite motif-containing 38 (TRIM38) pseudogene or nucleolar protein 8 pseudogene 1 |
Not applicable | [93] |
| 4q27 |
ADAD1 IL21 IL2 |
rs6827756 rs4505848 rs17388568 |
NC_000004.12:g.122263256T>A NC_000004.12:g.122211337A>G NC_000004.12:g.122408207G>A |
T = 40.83% G = 29.63% A = 14.06% |
Adenosine deaminase domain containing 1 Interleukin 21 Interleukin 2 |
ADA1 plays role in spermatogenesis and binds to RNA IL-21 binds to receptors on B and T lymphocyte and NK cells IL-2 is the most important growth factor for developing T lymphocytes |
[32, 94] |
| 4p15.2 | LINC02357 | rs10517086 | NC_000004.11:g.26085511G>A | A = 18.59% | Long intergenic non-protein coding RNA 2357 | Not applicable | [91] |
| 5p13.2 | IL7R | rs11954020 | NC_000005.9:g.35883251C>G | G = 39.92% | Interleukin 7 receptor | This receptor plays a key role in developing lymphocytes | [93] |
| 6q22.32 | CENPW | rs9388489 | NC_000006.11:g.126698719A>T | A = 44.81% | Centromere protein W | One of the components of the CENPA-NAC complex which plays key role in assembly of kinetochore protein | [95] |
| 6q15 | BACH2 | rs11755527 | NC_000006.11:g.90958231C>G | G = 36.66% | BTB domain and CNC homolog 2 | Plays role in coordinating transcription activation and repression. Induces apoptosis in response to oxidative stress | [96] |
| 6q27 |
AL596442.1 or AL049612.1 |
rs924043 | NC_000006.11:g.170379025T>C | T = 41.47% | Novel transcript | Not applicable | [25] |
| 6q23.3 | TNFAIP3 | rs6920220 rs10499194 |
NC_000006.11:g.138006504G>A NC_000006.11:g.138002637C>T |
A = 9.44% T = 19.15% |
TNF-α induced protein 3 | Involved in immune and inflammatory responses signaled by cytokine such as TNF-α or by pathogens | [73] |
| 6p21.3 | HLA class II | rs6927022 rs2157051 rs9275184 rs7744001 |
NC_000006.11:g.32612397A>G NC_000006.11:g.32658624G>A NC_000006.11:g.32654714T>C NC_000006.11:g.32626086G>C |
G = 35.76% G = 39.08% C = 8.71% A = 38.92% |
Major histocompatibility complex | Binds peptide derived from antigens and presents them on the cell surface for recognition by the CD4 T-cells | [97] |
| 7p15.2 | SKAP2 | rs7804356 | NC_000007.13:g.26891665T>C | C = 17.83% | Src kinase associated phosphoprotein 2 | Not applicable | [91] |
| 7p12.1 | COBL | rs4948088 | NC_000007.13:g.51027194A>C | A = 3.99% | Cordon-Bleu WH2 repeat protein | Involved in the reorganization of the actin cytoskeleton and plays key role in morphogenetic processes of the central nervous system | [98] |
| 7P12.2 | IKZF1 | rs10272724 rs62447205 |
NC_000007.13:g.50477213T>C NC_000007.13:g.50465830A>G |
C = 21.33% G = 22.62% |
IKAROS family zinc finger 1 | This protein binds to DNA and plays role as a transcriptional regulator of hematopoietic cell differentiation | [93, 99] |
| 9p24.2 | GLIS3 | rs7020673 rs10758593 |
NC_000009.11:g.4291747C>G NC_000009.11:g.4292083G>A |
C = 35.82% A = 47.92% |
GLIS family zinc finger 3 | Plays a role in generation of pancreatic beta cells and in insulin gene expression | [100] |
| 10p11.22 | NRP1 | rs722988 | NC_000010.10:g.33426147T>C | T = 44.95% | Neuropilin 1 | Involved in axonal growth and guidance and in physiological angiogenesis | [101] |
| 10p15.1 | IL2RA | rs41295061 rs2104285 rs706778 |
NC_000010.11:g.6072697C>A NC_000010.11:g.5868542G>A NC_000010.11:g.6056986C>T |
A = 2.8% T = 32.61% C = 49.92% |
Interleukin 2 receptor subunit α | The receptor plays role in the immune tolerance by controlling regulatory T cells activity | [60, 102] |
| 10q23.31 | RNLS | rs10509540 | NC_000010.11:g.88263276T>C | C = 24.74% | Renalase FAD dependent anime oxidase | This enzyme degrades catecholamines such as adrenaline in the blood circulation and may regulates blood pressure | [103] |
| 11p15.5 | INS | rs7111341 | NC_000011.10:g.2191936C>T | T = 23.86% | Insulin | Decreases blood glucose concentration | [104] |
| 11q13.1 | BAD | rs694739 | NC_000011.10:g.64329761A>G | G = 21.17% | BCL2 associated agonist of cell death | Promotes cell death by initiating apoptosis | [101] |
| 12q13.13 | ITGB7 | rs11170466 | NC_000012.11:g.53585859C>T | T = 6.43% | Integrin subunit β7 | Adhesion molecule, mediates lymphocyte migration and homing to gut-associated lymphoid tissue (GALT). Interacts with fibronectin, an extracellular matrix component | [101] |
| 12q13.2 |
ERBB3 DGKA |
rs2292239 rs11171710 |
NC_000012.11:g.56482180T>G NC_000012.11:g.56368078G>A |
T = 6.43% A = 35.18% |
Erb-B2 receptor tyrosine kinase 3 Diacylglycerol kinase α |
ERBB3 plays role as cell surface receptor for molecules involved in developing of nervous system and embryogenesis DGKA converts a second messenger diacylglycerol into phosphatidate |
[101, 105] |
| 12q21.2 | ZDHHC17 | rs2632214 | NC_000012.11:g.77032629T>C | C = 25.66$ | Zinc finger DHHC-type containing 17 | Probably involved in the sorting of critical proteins involved in the initiating endocytosis at the plasma membrane. | [36] |
| 12q24.13 | SH2B3 | rs3184504 | NC_000012.11:g.111884608T>C | T = 14.74% | SH2B adaptor protein 3 | This protein functions as a regulator in signaling pathways relating to inflammation, hematopoiesis and cell migration | [104] |
| 12p13.31 | CD69 | rs4763879 | NC_000012.11:g.9910164G>A | A = 31.29% | CD69 molecule | Involved in lymphocyte proliferation and functions as a signal-transmitting receptor in lymphocytes | [91] |
| 13q22.2 | LMO7 | rs539514 | NC_000013.10:g.76326282A>C | A = 26.78% | LIM domain 7 | Probably LMO7 is involved in protein-protein interaction | [25] |
| 13q32.3 | GPR183 | rs9585056 | NC_000013.10:g.100081766C>T | C = 28.23% | G protein-coupled receptor 183 | The protein is expressed in lymphocytes and acts as chemotactic receptor for B-cells, T-cells, splenic dendritic cells | [93] |
| 14q32.2 |
AL163932.1 or LINC01550 |
rs4900384 | NC_000014.9:g.98032614A>G | G = 49.76% | Novel transcript or long intergenic non-protein coding RNA 1550 |
Not applicable | [91] |
| 14q24.1 |
ZFP36L1 or MAGOH3P |
rs1465788 | NC_000014.9:g.68796882T>C | T = 28.81% | ZFP36 ring finger protein like 1 or mago homolog 3, pseudogene |
Not applicable | [91] |
| 14q32.2 | DLK1 | rs941576 | NC_000014.9:g.100839708A>G | G = 37.84% | Delta like non-canonical notch ligand 1 | May have a role in an neuroendocrine differentiation. | [106] |
| 15q25.1 | CTSH | rs3825932 | NC_000015.9:g.79235446T>C | C = 36.8% | Cathepsin H | It is a lysosomal cysteine proteinase important for the overall degradation of proteins in lysosomes | [91] |
| 15q14 | RASGPR1 | rs72727394 | NC_000015.9:g.38847022C>T | T = 13.78% | RAS guanyl releasing protein 1 | It regulates T-cells and B-cells development, homeostasis and differentiation | [93] |
| 16p11.2 | IL27 | rs151234 | NC_000016.9:g.28505660G>C | C = 13.20% | Interleukin 27 | It has pro- and anti-inflam- matory properties that, regulate T-helper cell development, suppress T-cell proliferation and has diverse influence on innate immune cells | [93] |
| 16p13.13 |
CLEC16A DEXI |
rs12708716 rs12927355 rs193778 |
NC_000016.9:g.11179873A>G NC_000016.9:g.11194771C>T NC_000016.9:g.11351211A>G |
G = 35.08% T = 27.16% G = 14.88% |
C-type lectin domain containing 16A Dexamethasone-induced protein |
Regulator of mitophagy, a selective form of autophagy necessary for mitochondrial quality control DEXI functions as an anti-inflammatory and immunosuppressant |
[91, 93] |
| 16q23.1 | BCAR1 | rs8056814 | NC_000016.9:g.75252327G>A | A = 18.09% | Breast cancer anti-estrogen resistance 1 | Plays a role in coordinating for tyrosine kinase-based signaling related to cell adhesion. Involved in induction of cell migration | [93] |
| 17q21.2 | SMARCE1 | rs7221109 | NC_000017.11:g.40614034T>C | T = 29.59% | SWI/SNF related, matrix associated, actin dependent regulator of chro- matin, subfamily e, member 1 or TEC |
Not applicable | [91, 107] |
| 17q12 | ORMDL3 | rs2290400 | NC_000017.10:g.38066240T>C | C = 42.05% | ORMDL sphingolipid biosynthesis regulator 3 | Negative regulator for sphingolipid synthesis and could plays role in regulate endoplasmic reticulum-mediated Ca2+ signaling | [91] |
| 18q22.2 | CD226 | rs763361 | NC_000018.9:g.67531642T>C | C = 46.94% | CD226 molecule | Involved in intracellular adhesion, lymphocyte signaling, cytotoxicity, stimulates T-cell proliferation and cytokine production | [91] |
| 18p11.21 | PTPN2 | rs1893217 | NC_000018.9:g.12809340A>G | G = 11.96% | Protein tyrosine phosphatase, non-receptor type 2 | Negatively regulates signaling pathways and biological processes such as hematopoiesis, inflammatory reactions, cell proliferation and glucose homeostasis | [91] |
| 19q13.33 | FUT2 | rs601338 | NC_000019.9:g.49206674G>A | A = 32.17% | Fucosyltransferase 2 | The protein is Golgi stack membrane protein that plays role in the creation of a precursor of antigen H. It mediates the transfer of fucose to the terminal galactose on glycan chains of cell surface glycoproteins and glycolipids | [37] |
| 19q13.32 | PRKD2 | rs425105 | NC_000019.9:g.47208481T>C | C = 14.26% | Protein kinase D2 | Not applicable | [91] |
| 19p13.2 | TYK2 | rs34536443 | NC_000019.9:g.10463118G>C | C = 1.02% | Tyrosine kinase 2 | Involved in intracellular signal transduction, may plays role in anti-viral immunity | [108] |
| 19p13.3 |
CDC34 MADCAM1 |
rs12982646 | NC_000019.9:g.499978G>A | A = 15.73% | Ubiquitin conjugating enzyme E2 R1 Mucosal vascular addressin cell adhesion molecule 1 |
CDC34 catalyzes the covalent attachment of ubiquitin to other proteins, is involved in degradation of cell cycle G1 regulators MADCAM1 is a cell adhesion leukocyte receptor expressed by mucosal venules and helps lymphocytes traffic into mucosal tissues |
[101] |
| 20p13 | SIRPG | rs2281808 | NC_000020.10:g.1610551T>A | T = 21.29% | Signal regulatory protein gamma | Not applicable | [91] |
| 21q22.3 | UBASH3A | rs11203203 | NC_000021.9:g.42416077G>A | A = 19.79% | Ubiquitin associated and SH3 domain containing A | Promotes accumulation of activated receptors, like T-cell receptors on the cell surface | [91] |
| 22q12.2 | AC002378.1 | rs5753037 | NC_000022.10:g.30581722C>T | T = 27.88% | novel transcript | Not applicable | [91] |
| 22q12.3 |
C1QTNF6 RAC2 |
rs229533 | NC_000022.10:g.37587111A>C | A = 44.59% | C1q and TNF related 6 Ras-related C3 botulinum toxin substrate 2 |
Rac2 is a signaling G protein, regulates diverse cellular processes, controls cell growth and activation of protein kinases | [93] |
| Xp22.2 | TLR7/8 | rs5979785 | NC_000023.10:g.12971524C>T | C = 42.62% | Toll like receptor 7/8 | TLR family plays a role in pathogen recognition and activation of immune responses | [71] |
| Xq28 | GAB3 | rs2664170 | NC_000023.10:g.153945602G>A | G = 35.26% | GRB2 associated binding protein 3 | Not applicable | [91] |
The HLA region involved in progress of type 1 diabetes
The first connection between genetics and T1D that came into view was the HLA region located on chromosome 6p21. It explains around 50% of the overall heritability of the disease, therefore it is the strongest determinant for T1D. Genes from that region play multiple roles during immune response and they are the first checkpoints in its activation. Genetic polymorphisms encoding different amino acid residues in the peptide-binding pockets of HLA molecules are the main connection between HLA molecules and T1D. Moreover, the binding repertoire and affinity of peptides can be presented on T-cells [6, 40]. There are 2 classes of HLA genes: 1) class I – HLA-A, HLA-B and HLA-C, 2) class II – HLA-DR, HLA-DQ and HLA DP (HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, HLA-DPB1) [5].
The strongest association with T1D is located within the HLA class II region genes that encode highly polymorphic β-chains (HLA-DRB1, -DQA1 and -DQB1) [41]. There are two main high-risk haplotypes “DR4-DQ8” (DR4-DQA1*03:01-DQB1*03:02) and “DR3-DQ2” (DRB1*03:01-DQA1*05:01-DQB1*02:01). Around 90% of T1D patients carry DR4-DQ8 or DR3-DQ2 and roughly 30% patients carry the combination of both of those haplotypes (DR4-DQ8/DR3-DQ2). This group confers the highest risk of T1D development (OR = 16) [6, 42].
HLA class I alleles are also responsible for T1D but less strongly. The most associated genes from that group are HLA-A and HLA-B [43]. The presence of the HLA-B*39 allele has been shown as a significant risk factor for the disease. Moreover, it is connected with T1D diagnosis at young age [44]. In addition, HLA-A*02 also increases the likelihood of T1D development and is one of the most frequent class I alleles, with frequency of > 60% in T1D patients [45].
The non-HLA genes associated with type 1 diabetes
The insulin gene (INS) encoded on chromosome 11p15 was the second locus linked to T1D. One of the main reasons for insulin gene association with the disease is the existence of the VNTR (variable number tandem repeat) polymorphisms. Polymorphisms of this region have been divided into three classes according to the amount of nucleotide repeats: class I (26-63 repeats), class II (around 85 repeats) and class III (120-170 repeats) [46]. All these polymorphisms are responsible for the amount of insulin mRNA in the thymus. Class I VNTRs (the shortest) present the highest risk of T1D while class III VNTRs are believed to protect against T1D [47-49].
The next recognized locus was the cytotoxic T-lymphocyte antigen 4 (CTLA4 gene) encoded on chromosome 2q33.2, which belongs to the immunoglobulin superfamily and binds to the B7 molecule on antigen-presenting cells. A study showed that the A49G polymorphism [A to G transition at position 49 of the first exon NC_000002.12:g.203867991A>G (rs231775)] is the most associated with T1D (from all CTLA4 SNPs) [50, 51]. There are two more CTLA4 gene variants discovered: 1) C to T transition at position – 319 of the promoter region and 2) (AT)n dinucleotide repeat polymorphism at position 642 of the 3’ UTR. Both of those polymorphic markers did not show as strong association with T1D as NC_000002.12:g.203867991A>G in the meta-analysis [52].
PTPN22 is another gene involved in T1D, strongly correlated with its development. It is located on chromosome 1p13. This gene encodes tyrosine-protein phosphatase non-receptor type 22 (also known as lymphoid tyrosine phosphatase – LYP), a negative regulator of T-cells receptor signaling. A SNP at position 1858 [NC_000001.11:g.113834946A>G (rs2476601)] encoding Arg620Trp is associated with the induction of islet autoantibodies and diabetes development [53, 54]. Its association with T1D was confirmed after observing its role in disruption of T-cell deactivation mechanism that leads to expansion of autoreactive T-cells [35].
The interleukin 2 receptor α (IL2RA; also known as CD25) gene on chromosome 10p15 is the fifth discovered T1D locus [55]. Protein encoded by this gene forms one subunit of the receptor for interleukin 2. This receptor takes part in controlling TREGs (regulatory T-cells) activity, whereas TREGs are responsible for the activation and expansion of autoreactive T-cells [56]. First study investigating IL2RA locus association with T1D was performed using a tag single nucleotide polymorphism approach [55]. Together with GWAS five SNPs were predominantly determined and presented a significant T1D association: NC_000010.11:g.6056986C>T (rs706778), NC_000010.11:g.6059750T>C (rs3118470) [57], NC_000010.11:g.6072697C>A (rs41295061), NC_000010.11:g.6080046T>A (rs11594656) [58] and NC_000010.11:g.6057082T>C (rs2104286) [59]. All those SNPs are considered T1D risk factors, though the strongest association was distinguishable for NC_000010.11:g.6080046T>A, NC_000010.11 :g.6057082T>C and NC_000010.11:g.6072697C>A, all of which were proved to be separately linked to the circulating concentration of IL2RA soluble form [58, 60].
The sixth identified gene and the first in the GWAS era was IFIHI (early type I interferon β-responsive gene) on chromosome 2q24.3. It was one of four genes in 2q.24 region with revealed association but it turned out to be the most noteworthy because of the protein it encodes. IFIH1 encodes interferon-induced helicase C domain-containing protein 1 (also called MDA5 – mela-noma differentiation-associated protein 5), which is an intracellular pathogen receptor. This discovery posed an interesting link between viral infections and T1D development and made IFIH1a particularly functional candidate for T1D disease [9, 38, 61]. The first association of IFIH1 was discovered through an interim analysis of a genome-wide nsSNP (direct precursor of GWAS) – NC_000002.12:g.162267541C>T (rs1990760) that leads to the A946T substitution [38]. Further research into this genetic variation led to the discovery postulating that A®T substitution in IFIH1 protein takes part in limiting viral infection (higher expression of type 1 interferons) but at the same time it promotes the risk of autoimmunity [62]. In the later years four rare variants of IFIH1 were discovered – NC_000002.12:g.162268127T>C (rs35667974), NC_000002.12:g.162279995C>G (rs35337543), NC_000002.12:g.162268086C>T (rs35732034) and NC_000002.12:g.162277580C>G (rs35744605) – which were proven to lower the risk of T1D independently of each other [63]. Moreover, another group of researchers showed three more SNPs with a distinct T1D association: one in the coding region [NC_000002.12:g.162272314T>C (rs3747517)] and two in the 3’ intergenic region of IFIH1 [NC_000002.12:g.162243749G>A (rs13422767) and NC_000002.12:g.162254026A>G (rs2111485)] [64].
The signal transducer and activator of transcription (STAT4) is a central mediator involved in generating inflammation during protective immune responses and immune-mediated disease. This gene is located on chromosome 2q32.2-q32.3 and studies showed that NC_000002.12:g.191099907T>G (rs7574865) polymorphism is associated with diabetes risk, especially T1D [65]. This polymorphism might affect STAT4 production or phosphorylation processes but this question has yet to be investigated.
The ubiquitin associated and SH3 domain containing A (UBASH3A) is a gene located on chromosome 21q22.3 and it encodes one of two family members belonging to the T-cell ubiquitin ligand (TULA) family that can negatively regulate T-cell signaling. Genetic variants of this gene have been proven to be involved in T1D. Studies demonstrated that SNPs, including NC_000021.9:g.42416077G>A (rs11203203) and NC_000021.9:g.42415901T>C (rs80054410), display significant associations with T1D [66]. These two risk variants in UBASH3A regulate expression of UBASH3A and IL2 in human primary CD4+ T cells.
The SH2B adapter protein 3 (SH2B3) gene located on locus 12q24 encodes a protein involved in signaling activities through growth factors and cytokine receptors. The nonsynonymous SNP NC_000012.12:g.111446804T>G (rs3184504) is a gene variant the most associated with T1D development (among all SH2B gene variants) [67]. This polymorphism can lead to novel SH2B3 isoforms that affect β-cell inflammation and death in vivo or modulate the immune system, however it is still unclear. Nonetheless, new insights into SH2B adapter protein 3’s function were shown recently. It was proved that it is responsible for controlling the homeostasis in adipose tissue. What is more it reduces the risk of diabetes through regulation of IL-15-dependent adipose G1-ILC [68].
The TLR7 and TLR8 genes (toll-like receptor 7 and 8) are located on one region of X chromosome (Xp22.2). They encode type I transmembrane proteins belonging to Toll-like receptor family and play a fundamental role in pathogen recognition and activation of innate immunity. NC_000023.11:g.12953405C>T (rs5979785) SNP was located 30 kb centromeric of those two genes and it presents some level of association with T1D [69-71]. It is highly probable that this SNP may modify the expression of both TLR7 and TLR8, which are functional T1D candidate genes based on their role as pathogen recognition receptors. Moreover, TLR7 overexpression has been associated with murine autoimmune disease directly [72].
The TNFAIP3 (Tumor necrosis factor α-induced protein 3) gene located on chromosome 6q23 with its two SNPs [NC_000006.12:g.137685367G>A (rs6920220) and NC_000006.12:g.137681500C>T (rs10499194)] is also associated with T1D disease. Both of those SNPs were shown to be linked to T1D independently of each other [73]. TNFAIP3 encodes a ubiquitin-editing enzyme (A20) which inhibits NF-κB activation and TNF-mediated apoptosis and acts as an anti-apoptotic protein in specific cell types [74].
The Huntingtin-interacting protein gene (HIP14/ZDHHC17) expressed in β-cells is required for β-cells survival and is a target of proinflammatory cytokines which contribute to β-cells destruction in T1D. HIP14 is a palmitoyltransferase specific for a subset of neuronal proteins and is important for intracellular trafficking and exocytosis in neurons [75]. A study showed that decreased level of HIP14 in β-cells led to increased cell apoptosis and disease development. Knockdown of HIP14 causes a decrease in release of insulin and therefore it suggests that HIP14 is important for β-cell insulin release [36].
The FUT2 gene located on chromosome 19q13.33 encodes a protein α-1,2-fucosyltransferase which is responsible for the creation of H antigen, that is required for the final step in the soluble A and B antigen synthesis. Studies demonstrated that a single nucleotide polymorphism in this gene [NC_000019.10:g.48703417G>A (rs60133)] encodes the “non-secretor” variant which is associated with T1D [37].
The LMO7 (LIM Domain Only 7) gene located on locus 13q22 encodes a protein containing a calponin homo-logy (CH) domain, a PDZ domain [PSD-95 (post-synaptic density protein 95); Dlg1 (Drosophila disc large tumor suppressor)]; ZO-1 (zona occludens 1)] and a LIM domain [Lin11 (Protein lin-11); Isl-1 (Insulin gene enhancer binding protein); Mec-3 (Mechanosensory protein 3)], and it may be involved in protein-protein interactions. This gene is expressed in pancreatic islets and thus it is a candidate for a gene associated with T1D. Studies showed that the SNP NC_000013.11:g.75752146A>T (rs539514) is involved in the development of disease [25].
The EFR3B (protein EFR3 homology B) gene is located on chromosome 2p23. Its protein is a component of a complex required to localize phosphatidylinositol 4-kinase (PIL4) on the plasma membrane and thus EFR3B presumably plays a role in membrane-anchoring [76]. EFR3B, like LMO7, is a newly discovered gene modulating susceptibility to T1D. The factor involved in the progress of T1D is SNP NC_000002.12:g.25078886A>T (rs478222) [25].
The Kruppel-like zinc finger protein Gli-similar (GLIS3) is a candidate gene involved in T1D. GLIS3 is located on 9p24.2 locus and encodes a nuclear protein with five C2H2-type zinc finger domains. This protein functions as a transcription activator and repressor and is also entangled in development of pancreatic β-cells. Mutations in this transcription factor cause neonatal diabetes and hypothyroidism [77]. GWAS studies showed that SNPs in this gene are associated both with T1D and T2D. What is more, knockdown of GLIS3 leads to increased apoptosis of islet cells both in basal condition and following an exposure to proinflammatory cytokines.
Non-coding RNAs in type 1 diabetes
A considerable part of the genome is transcribed but only a little portion of DNA encodes proteins. The non-protein-coding part of the genome is transcribed to create a wide spectrum of non-coding RNAs including tRNAs (transfer RNA), rRNAs (ribosomal RNA), micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs) [78].
Long non-coding RNA
Long non-coding RNAs are 200 nucleotides or longer. They are capped, polyadenylated and spliced, in the same way as protein coding transcripts. The studies have shown that lncRNAs play a significant role in a variety of biological processes such as transcription, splicing, translation and apoptosis [79]. A number of mammalian lncRNAs are expressed in a cell-type specific way. The role of these parts of the genome as transcription factors suggests that lncRNAs could be mediators of lineage-specific differentiation or specialized cellular functions. Mutations in lncRNAs could be a potential factor in disease development and cell-specific regulatory lncRNAs might provide therapeutic targets. The latest studies have demonstrated that human islet lncRNAs are highly cell-type specific and they mediate biological processes in islet cells such as β-cell differentiation, insulin biosynthesis and insulin secretion [80].
So far, GWAS studies have identified a number of disease-associated single nucleotide polymorphisms not limited to protein-coding genes. The study from 2015 has shown that lncRNAs play a role in the autoimmune process and autoimmune diseases [81]. The analysis of non-coding portion of the genome is important to identify novel biomarkers of disease development. According to transcriptome profiling studies of pancreatic islets, there are 1000 islet-specific lncRNAs, which are common for human and mouse islets. SNPs within lncRNA regions might modify their secondary structure and change regulatory functions, which could lead to an increased risk of developing diabetes. It has been presented that HI-LNC25 lncRNA [also known as LINC01370 (long intergenic non-protein coding RNA 1370)] is a regulator of GLIS3 mRNA [80]. Knockdown of HI-LNC25 caused reduced mRNA levels of GLIS3, a gene encoding an islet transcription factor. Genetic variations of this gene can lead to T2D development. Another study indicated a link between lncRNAs and phenotype-associated loci within type 1 diabetes candidate loci. The SNP of NONHSAG044354 lncRNA located within BACH2 gene [NC_000006.12:g.90247744C>T (rs3757247)] was associated with T1D [82]. A further analysis showed that lncRNAs often regulate genes associated with clusters of islet enhancers and together with transcription factors regulate common genes. One example is lncRNA named PLUTO (PDX1 locus upstream transcript), which regulates a key pancreatic β-cell transcriptional regulator, PDX1 (pancreatic and duodenal homeobox 1). PLUTO controls PDX1 by regulating the 3D architecture of the enhancer cluster in PDX1 locus in human islets. Knockdown of PLUTO is associated with downregulation of PDX1 in primary islet cells and it could be associated with developing both type 1 and type 2 dia-betes [83]. According to a study of immune-mediated disease genes, more than 90% of disease-associated SNPs are placed within non-coding regions of genomes such as ncRNA genes and around 10% of the autoimmune disease associated SNPs are present within lncRNas. The role of lncRNAs in T1D has not yet been profoundly investigated but this is an attractive field to explore. It could help people to better understand the genetic basis for the development of diabetes and to improve diagnostic tests that can detect the disease at its beginning stadium.
MicroRNA
MicroRNAs (miRNAs) are molecules of the size between 18 and 22 nucleotides. They bind to 3’UTR (untranslated regions) of mRNA and cause their degradation or inhibition of translation [84, 85]. Nowadays, more and more studies present miRNA changes in the pathogenesis of diabetes. It has been showed that miRNAs (such as miR-150, miR-146a and miR-424) take part in the formation of autoimmunity and β-cells dysfunctions. Another example are miR-146a NC_000005.10:g.160485411C>G (rs2910164) and miR-155 NC_000021.9:g.25572410T>A (rs767649) variants, which are less common in T1D patients compared to healthy individuals. Moreover, the most common miRNA in pancreatic islets is miR-375 which is a β-cells death biomarker85studies are focusing on other factors that may contribute to the pathogenesis of diabetes, such as epigenetics, a term \”traditionally\” encompassing changes to the DNA that do not alter sequence and are heritable (primary methylation and histone modification. Increase in its amount cause the decrease of β-cells mass. β-cells can release miR-375 into the extracellular environment. This type of miRNA regulates insulin secretion and its expression might be altered in β-cells concomitantly to its circulating level. Furthermore, miR-375 level was downregulated in human pancreatic islets treated with high glucose [86].
Epigenetic modifications in type 1 diabetes
Next to the genetic factors of diseases there are also non-genetic factors which are less clear-cut. These factors are called epigenetic factors and they execute changes in gene expression without altering DNA sequence directly [87]. Epigenetic mechanisms above all include DNA methy-lation and histone modification [85]. Epigenetic modifications control biological processes and are specific to the stage of disease development, which is correlated with the regulation of cell differentiation [84, 85].
DNA methylation
The most common epigenetic alteration is methylation, which is a process based on addition of a methyl group to cytosine at C5 position. It takes place especially when it is followed by guanine, in so called CpG dinucleotides that comprise around 1% of the human genome [84]. DNA methylation usually correlates with gene silencing [84, 87]. By now, 132 positions were identified as T1D-methylation variable positions (T1D-MVPs) and they include HLA class II gene (HLA-DQB1) and GAD2 (glutamate decarboxylase 2) encoding GAD65 autoantigen. T1D-MVPs are changes found in monoclonal zygote twins, therefore they cannot be a result of genetic differences. It is known that some T1D-MVPs are found in individuals before T1D diagnosis. This suggests that they arise very early in the process that leads to T1D and are not simply due to post-onset-associated factors [88]. Moreover, methylation sites also comprise HLA-E, HLA-DOB, HLA-DQ2A, INS, IL-2RB and CD226 as well, which may take part in T1D development. Different levels of methylation of INS genes (Ins1 and Ins2) are correlated with T1D progression and they could be considered another marker for this disease. Moreover, it has been proven that during β-cell destruction associated with T1D onset and progression noticeable changes in methylation (presented by those damaged β-cells’ DNA) are present in the insulin genes accountable for regulation of transcription [85].
Histone modifications
Next group of epigenetic factors are histone modifications. Cell’s genetic material is packed into a chromatin structure (linear DNA is wrapped around a histone core). The N-terminal tails of these histones can be covalently modified [84]. The most common types of these modifications are: 1) histone methylation, 2) acetylation (of lysine and arginine), 3) phosphorylation (of serine and threonine), 4) ubiquitination and 5) sumoylation (of lysine) [84, 85]. The level of lysine 9 (belonging to H3 histone protein – H3K9Ac) acetylation demonstrates an increase in the upstream regions of HLA-DRB1 and HLA-DQB1. And those genetic variants are highly common in T1D patients [85].
Imprinting
Imprinting is another very important epigenetic mechanism. It is based on the transcriptional regulation in which only one parental allele of a specific gene is expressed [85]. Phenotype expression of imprinted genes is a consequence of silencing one allele and allowing a normal state of monoallelic gene expression without altering DNA sequence. It is based not only on sequence variation itself but also on methylation and histone modifications [89]. Some of autosomal loci are actively transcribed only on the maternally inherited chromosome and the others are only transcribed on the paternal chromosome [84]. Imprinting takes part in pathogenesis of many types of diabetes, for example transient neonatal diabetes (TNDM) is caused by abnormalities in the imprinted locus on chromosome 6q24. PLAGL1 gene [PLAG1 (pleomorphic adenoma gene 1) like zinc finger 1] resides in this region and it is normally expressed only on the paternal allele. Duplication of the paternal allele and hypomethylation of the maternal allele cause the overexpression of this gene. PLAGL1 regulates the cell cycle arrest and apoptosis. Its overexpression causes apoptosis and loss of β-cell mass. This finally leads to the loss of insulin secretion [85].
Environmental factors
It is widely known that besides typical epigenetic factors associated with autoimmune disorders, we can additionally distinguish environmental factors which are also capable of affecting disease development. Environmental factors include: 1) temperature, 2) climate, 3) increased hygiene and decreased rates of infection in early childhood, 4) vaccinations, 5) antibiotics and drugs, 6) iodine levels, 7) cigarette smoking as well 8) an increasing wealth. On top of that, diet (especially wheat consumption) also has a great impact on the development of T1D. One of the most important environmental factors are infections. It is proved that bacterial infections play a role in development of pancreatic inflammation. Another type of infections are viral infections, for example myocarditis caused by the enteroviruses family [90]. Possible mechanism of action is based on the viral remnants which can be found in pancreatic islets of patients with T1D and are not present in healthy people. Taking antibiotics and various drugs (especially methyl donors) modifies the composition of intestinal microbiota. It has been shown that reduced microbial diversity, especially low on butyrate-producing and mucin-degrading bacteria, takes part in the autoimmunity of islet cells in children [87].
Conclusions
Type 1 diabetes is a chronic autoimmune disease characterized by islets inflammation and destruction of β-cells, which are responsible for insulin production and regulation of glucose homeostasis. It is very important to understand the molecular mechanisms of T1D development based on genetic studies. GWAS analyses led to the discovery of a number of loci modifying the susceptibility to diabetes. Those loci are considered biomarkers of diabetes development (before and after the onset of the disease) in living organisms. Most of T1D-associated loci encode proteins that are involved in the immune response and inflammatory reactions. The biggest group of genes involved in T1D is present in the human leukocyte antigen region (HLA) that contains almost half of genes associated with T1D. Analysis and identification of novel biomarkers is needed for prevention and monitoring of diabetes development. Future genetic studies are crucial to broaden our knowledge of T1D pathogenesis and find a novel effective therapy. Analysis of a genetic background could result in a discovery of new biomarkers of T1D and in a development of a more specific and sensitive diagnostic test.
Acknowledgements
Wszola et al. from Foundation for Research and Science Development, is currently working on research and development of possibilities of using 3D bioprinting method to fabricate fully functional artificial organs – especially bionic pancreas as a STRATEGMED project. Foundation for Research and Science Development is the Leader of the Bionic consortium which consists of teams from: Warsaw University of Technology – team from Materials Science faculty, under the guidance of Prof. Wojciech Swieszkowski; Nencki Institute – under the guidance of Prof. Agnieszka Dobrzyn; Medical University of Warsaw – Biostructures center, under the guidance of Prof. Artur Kaminski; Clinical Hospital of the Child Jesus, under the guidance of Prof. Artur Kwiatkowski – Director Institute of Transplantology. Business and part of biotechnology and engineering solutions in consortium comes from MediSpace Ltd.
Footnotes
The authors declare no conflict of interest.
Funding
This study was financially supported by The National Centre for Research and Development STRATEGMED3/ 305813/2/NCBR/2017.
References
- 1.Jerram ST, Leslie RD (2017): The genetic architecture of type 1 diabetes mellitus. Genes (Basel) 8: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.https://www.who.int/news-room/fact-sheets/detail/diabetes (published 2018).
- 3.Nyaga DM, Vickers MH, Jefferies C, et al. (2018): The genetic architecture of type 1 diabetes mellitus. Mol Cell Endocrinol 477: 70-80. [DOI] [PubMed] [Google Scholar]
- 4.Rich SS (2017): Genetics and its potential to improve type 1 dia-betes care. Curr Opin Endocrinol Diabetes Obes 24: 279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson CC, Rich SS (2018): Genetics of type 1 diabetes. Curr Opin Genet Dev 50: 7-16. [DOI] [PubMed] [Google Scholar]
- 6.Redondo MJ, Steck AK, Pugliese A (2018): Genetics of type 1 diabetes. Pediatr Diabetes 19: 346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delvecchio M, Mozzillo E, Salzano G, et al. (2017): Monogenic diabetes accounts for 6.3% of cases referred to 15 Italian Pediatric Diabetes Centers during 2007 to 2012. J Clin Endocrinol Metab 102: 1826-1834. [DOI] [PubMed] [Google Scholar]
- 8.Pugliese A (2004): Genetics of type 1 diabetes. Endocrinol Metab Clin North Am 33: 1-16. [DOI] [PubMed] [Google Scholar]
- 9.Todd JA (2010): Etiology of type 1 diabetes. Immunity 32: 457-467. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson MA, Macklaren NK (1994): The pathogenesis of insulin-dependent diabetes mellitus. N Engl J Med 331: 1428-1436. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson MA, Eisenbarth GS, Michels AW (2014): Type 1 diabetes. Lancet 383: 69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiffenbauer J (1999): Superantigens and their role in autoimmune disorders. Arch Immunol Ther Exp (Warsz) 1999; 47: 17-24. [PubMed] [Google Scholar]
- 13.Boitard C (1992): The differentiation of the immune system towards anti-islet autoimmunity. Clinical prospects. Diabetologia 35: 1101-1112. [DOI] [PubMed] [Google Scholar]
- 14.American Diabetes Association (2013): Diagnosis and classification of diabetes mellitus, ADA Clinical Practice Recommendations. Diabetes Care 36 Suppl 1: S67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S (2012): Immunotherapies in diabetes mellitus type 1. Med Clin North Am 96: 621-634. [DOI] [PubMed] [Google Scholar]
- 16.Rowe RE, Leech NJ, Nepom GT, McCulloch DK (1994): High genetic risk for IDDM in the pacific northwest: First report from the Washington state diabetes prediction study. Diabetes 43: 87-94. [DOI] [PubMed] [Google Scholar]
- 17.Simmons KM, Michels AW (2015): Type 1 diabetes: a predictable disease. World J Diabetes 6: 380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pugliese A (2013): The multiple origins of type 1 diabetes. Diabet Med 30: 135-146. [DOI] [PubMed] [Google Scholar]
- 19.Cooper JD, Smyth DJ, Smiles AM, et al. (2008): Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet 40: 1399-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Undlien DE, Friede T, Rammensee HG, et al. (1997): HLA-encoded genetic predisposition in IDDM: DR4 subtypes may be associated with different degrees of protection. Diabetes 46: 143-149. [DOI] [PubMed] [Google Scholar]
- 21.Smyth DJ, Plagnol V, Walker NM, et al. (2008): Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 359: 2767-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Permutt MA, Wasson J, Cox N (2005): Genetic epidemiology of diabetes. J Clin Invest 115: 1431-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singal DP, Blajchman MA (1973): Histocompatibility (HLA) antigens, lymphocytotoxic antibodies and tissue antibodies in patients with diabetes mellitus. Diabetes 22: 429-432. [DOI] [PubMed] [Google Scholar]
- 24.Woodrow JC, Cudworth AG (1975): HL-A8 and W15 in diabetes mellitus. Lancet 803. [DOI] [PubMed]
- 25.Bradfield JP, Qu HQ, Wang K, et al. (2011): A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet 7: e1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steck AK, Rewers MJ (2011): Genetics of type 1 diabetes. Clin Chem 57: 176-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies JL, Kawaguchi Y, Bennett ST, et al. (1994): A genome-wide search for human type 1 diabetes susceptibility genes. Nature 371: 130-136. [DOI] [PubMed] [Google Scholar]
- 28.Concannon P, Gogolin-Ewens KJ, Hinds DA, et al. (1998): A second-generation screen of the human genome for susceptibility to insulin-dependent diabetes mellitus. Nat Genet 19: 292-296. [DOI] [PubMed] [Google Scholar]
- 29.Hirschhorn JN, Daly MJ (2005): Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 95-108. [DOI] [PubMed] [Google Scholar]
- 30.Tabor HK, Risch NJ, Myers RM (2002): Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 3: 1-7. [DOI] [PubMed] [Google Scholar]
- 31.Schaid DJ, Chen W, Larson NB (2018): From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet 19: 491-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Wellcome Trust Case Control Consortium (2007): Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rich SS, Concannon P, Erlich H, et al. (2006): The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci 1079: 1-8. [DOI] [PubMed] [Google Scholar]
- 34.Cooper JD, Howson JMM, Smyth D, et al. (2012): Confirmation of novel type 1 diabetes risk loci in families. Diabetologia 55: 996-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp RC, Abdulrahim M, Naser ES, Naser SA (2015): Genetic variations of PTPN2 and PTPN22: Role in the pathogenesis of type 1 diabetes and Crohn’s disease. Front Cell Infect Microbiol 5: 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berchtold LA, Størling ZM, Ortis F, et al. (2011): Huntingtin-interacting protein 14 is a type 1 diabetes candidate protein regulating insulin secretion and β-cell apoptosis. Proc Natl Acad Sci U S A 108: E681-E688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth DJ, Cooper JD, Howson JMM, et al. (2011): FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes 60: 3081-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth DJ, Cooper JD, Bailey R, et al. (2006): A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38: 617-619. [DOI] [PubMed] [Google Scholar]
- 39.Hakonarson H, Grant SFA (2011): Genome-wide association studies (GWAS): Impact on elucidating the aetiology of diabetes. Diabetes Metab Res Rev 27: 685-696. [DOI] [PubMed] [Google Scholar]
- 40.Pociot F, Kaur S, Nielsen LB (2016): Effects of the genome on immune regulation in type 1 diabetes. Pediatr Diabetes 17: 37-42. [DOI] [PubMed] [Google Scholar]
- 41.Espino-Paisán L, Urcelay E, Santiago JL (2009): An insight into the genetics of type 1 Diabetes. Inmunologia 28: 173-181. [Google Scholar]
- 42.Polychronakos C, Li Q (2011): Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet 12: 781-792. [DOI] [PubMed] [Google Scholar]
- 43.Van Belle TL, Coppieters KT, Von Herrath MG (2011): Type 1 diabetes: Etiology, immunology, and therapeutic strategies. Physiol Rev 91: 79-118. [DOI] [PubMed] [Google Scholar]
- 44.Valdes AM, Erlich HA, Noble JA (2005): Human leukocyte antigen class I B and C loci contribute to type 1 diabetes (T1D) susceptibility and age at T1D onset. Hum Immunol 66: 301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Belle TL, Coppieters KT, Von Herrath MG (2011): Type 1 diabetes: Etiology, immunology, and therapeutic strategies. Physiol Rev 91: 79-118. [DOI] [PubMed] [Google Scholar]
- 46.Bennett ST, Lucassen AM, Gough SC, et al. (1995): Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 2: 284-292. [DOI] [PubMed] [Google Scholar]
- 47.De Nanclares GP, Bilbao JR, Calvo B, et al. (2003): 5’-Insulin gene VNTR polymorphism is specific for type 1 diabetes: no association with Celiac or Addison’s disease. Ann N Y Acad Sci 1005: 319-323. doi: 10.1196/annals.1288.050 [DOI] [PubMed] [Google Scholar]
- 48.Pugliese A, Zeller M, Fernandez A, et al. (1997): The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 15: 293-297. [DOI] [PubMed] [Google Scholar]
- 49.Vafiadis P, Bennett ST, Todd JA, et al. (1997): Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 15: 289-292. [DOI] [PubMed] [Google Scholar]
- 50.Bouqbis L, Izaabel H, Akhayat O, et al. (2003): Association of the CTLA4 promoter region (À1661G allele) with type 1 diabetes in the South Moroccan population. Genes Immun 4: 132-137. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Chen S, Gu Y, et al. (2018): CTLA-4 +49 G/A, a functional T1D risk SNP, affects CTLA-4 level in Treg subsets and IA-2A positivity, but not beta-cell function. Sci Rep 8: 10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kavvoura FK, Ioannidis JPA (2005): CTLA-4 gene polymorphisms and susceptibility to type 1 diabetes mellitus: A HuGE review and meta-analysis. Am J Epidemiol 162: 3-16. [DOI] [PubMed] [Google Scholar]
- 53.Ladner MB, Bottini N, Valdes AM, Noble JA (2005): Association of the single nucleotide polymorphism C1858T of the PTPN22 gene with type 1 diabetes. Hum Immunol 66: 60-64. [DOI] [PubMed] [Google Scholar]
- 54.Baxter AG, Jordan MA (2012): From markers to molecular mechanisms: Type 1 diabetes in the post-GWAS era. Rev Diabet Stud 9: 201-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vella A, Cooper JD, Lowe CE, et al. (2005): Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet 76: 773-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goudy K, Aydin D, Barzaghi F, et al. (2013): Human IL2RA null mutation mediates immunodeficiency with lymphoproliferation and autoimmunity. Clin Immunol 146: 248-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu HQ, Montpetit A, Ge B, et al. (2007): Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes 56: 1174-1176. [DOI] [PubMed] [Google Scholar]
- 58.Lowe CE, Cooper JD, Brusko T, et al. (2007): Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet 39: 1074-1082. [DOI] [PubMed] [Google Scholar]
- 59.Maier LM, Lowe CE, Cooper J, et al. (2009): IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet 5: e1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang W, Cui D, Jiang L, et al. (2015): Association of common polymorphisms in the IL2RA gene with type 1 diabetes: Evidence of 32,646 individuals from 10 independent studies. J Cell Mol Med 19: 2481-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knip M, Veijola R, Virtanen SM, et al. (2005): Environmental triggers and determinants of type 1 diabetes. Diabetes 54 (Suppl. 2). [DOI] [PubMed] [Google Scholar]
- 62.Gorman JA, Hundhausen C, Errett JS, et al. (2017): The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol 18: 744-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nejentsev S, Walker N, Riches D, et al. (2009): Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324: 387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S, Wang H, Jin Y, et al. (2009): IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet 18: 358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi J, Fang X, Wan Y, et al. (2015): STAT4 polymorphisms and diabetes risk: A meta-analysis with 18931 patients and 23833 controls. Int J Clin Exp Med 8: 3566-3572. [PMC free article] [PubMed] [Google Scholar]
- 66.Ge Y, Paisie TK, Newman JRB, et al. (2017): UBASH3A mediates risk for type 1 diabetes through inhibition of t-cell receptor-induced NF-κB signaling. Diabetes 66: 2033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Auburger G (2014): 12q24 locus association with type 1 diabetes: SH2B3 or ATXN2? World J Diabetes 5: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori T, Suzuki-Yamazaki N, Takaki S (2018): Lnk/Sh2b3 regulates adipose inflammation and glucose tolerance through group 1 ILCs. Cell Rep 24: 1830-1841. [DOI] [PubMed] [Google Scholar]
- 69. http://www.t1dbase.org/
- 70.Witsø E, Cinek O, Tapia G, et al. (2015): Genetic determinants of enterovirus infections: Polymorphisms in type 1 dia-betes and innate immune genes in the MIDIA study. Viral Immunol 28: 556-563. [DOI] [PubMed] [Google Scholar]
- 71.Cooper JD, Walker NM, Smyth DJ, et al. (2009): Follow-up of 1715 SNPs from the Wellcome Trust Case Control Consortium genome-wide association study in type I diabetes families. Genes Immun 10 (Suppl. 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deane JA, Pisitkun P, Barrett RS, et al. (2007): Control of toll-like receptor 7 expression is essential to restrict auto-immunity and dendritic cell proliferation. Immunity 27: 801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fung EYMG, Smyth DJ, Howson JMM, et al. (2009): Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun 10: 188-191. [DOI] [PubMed] [Google Scholar]
- 74.Catrysse L, Fukaya M, Sze M, et al. (2015): A 20 deficiency sensitizes pancreatic beta cells to cytokine-induced apoptosis in vitro but does not influence type 1 diabetes development in vivo. Cell Death Dis 6: e1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohyama T, Verstreken P, Ly CV, et al. (2007): Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles. J Cell Biol 179: 1481-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakatsu F, Baskin JM, Chung J, et al. (2012): Ptdins4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol 199: 1003-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nogueira TC, Paula FM, Villate O, et al. (2013): GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein bim. PLoS Genet 9: e1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.The ENCODE Project Consortium (2007): Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma L, Bajic VB, Zhang Z (2013): On the classification of long non-coding RNAs. RNA Biol 10: 924-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morán I, Akerman I, Van De Bunt M, et al. (2012): Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 16: 435-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu GC, Pan HF, Leng RX, et al. (2015): Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev 14: 798-805. [DOI] [PubMed] [Google Scholar]
- 82.Mirza AH, Kaur S, Brorsson CA, Pociot F (2014): Effects of GWAS-associated genetic variants on lncRNAs within IBD and T1D candidate loci. PLoS One 9: e105723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akerman I, Tu Z, Beucher A, et al. (2016): Human pancreatic β cell lncRNAs control cell-specific Regulatory Networks. Cell Metab 25: 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Golson ML, Kaestner KH (2017): Epigenetics in formation, function, and failure of the endocrine pancreas. Mol Metab 6: 1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang H, Pollin TI (2018): Epigenetics variation and pathogenesis in diabetes. Curr Diab Rep 18: 1-7. [DOI] [PubMed] [Google Scholar]
- 86.Marchand L, Jalabert A, Meugnier E, et al. (2016): MiRNA-375 a sensor of glucotoxicity is altered in the serum of children with newly diagnosed type 1 diabetes. J Diabetes Res 2016: 1869082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jerram ST, Dang MN, Leslie RD (2017): The role of epigenetics in type 1 diabetes. Curr Diab Rep 17: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rakyan VK, Beyan H, Down TA, et al. (2011): Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet 7: 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitchell BD, Pollin TI (2010): Genomic imprinting in diabetes. Genome Med 2: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coppieters KT, Boettler T, von Herrath M (2012): Virus infections in type 1 diabetes. Cold Spring Harb Perspect Med 2: 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barrett JC, Clayton DG, Concannon P, et al. (2009): Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41: 703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Home - SNP - NCBI. https://www.ncbi.nlm.nih.gov/snp/ (Accessed: December 16, 2019).
- 93.Onengut-Gumuscu S, Chen WM, Burren O, et al. (2015): Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 47: 381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Todd JA, Walker NM, Cooper JD, et al. (2007): Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39: 857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brorsson CA, Pociot F, the Type 1 Diabetes Genetics Consortium (2015): Shared Genetic basis for type 1 diabetes, islet autoantibodies, and autoantibodies associated with other immune-mediated diseases in families with type 1 diabetes. Diabetes Care 38: S8-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Plagnol V, Howson JMM, Smyth DJ, et al. (2011): Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 7: e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McKinnon E, Morahan G, Nolan D, James I, Consortium the T 1 DG (2009): Association of MHC SNP genotype with susceptibility to type 1 diabetes: a modified survival approach. Diabetes Obes Metab 11: 92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brorsson CA, Nielsen LB, Andersen ML, et al. (2016): Genetic risk score modelling for disease progression in new-onset type 1 diabetes patients: Increased genetic load of islet-expressed and cytokine-regulated candidate genes predicts poorer glycemic control. J Diabetes Res 2016: 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swafford ADE, Howson JMM, Davison LJ, et al. (2011): An allele of IKZF1 (Ikaros) conferring susceptibility to childhood acute lymphoblastic leukemia protects against type 1 diabetes. Diabetes 60: 1041-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duarte GCK, Assmann TS, Dieter C, et al. (2017): GLIS3 rs7020673 and rs10758593 polymorphisms interact in the susceptibility for type 1 diabetes mellitus. Acta Diabetol 54: 813-821. [DOI] [PubMed] [Google Scholar]
- 101.Evangelou M, Smyth DJ, Fortune MD, et al. (2014): A method for gene-based pathway analysis using genomewide association study summary statistics reveals nine new type 1 diabetes associations. Genet Epidemiol 38: 661-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xia ZL, Qin QM, Zhao QY (2018): A genetic link between CXCR5 and IL2RA gene polymorphisms and susceptibility to multiple sclerosis. Neurol Res 40: 1040-1047. [DOI] [PubMed] [Google Scholar]
- 103.Reddy MPL, Wang H, Liu S, et al. (2011): Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun 12: 208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Törn C, Hadley D, Lee HS, et al. (2015): Role of type 1 diabetes associated SNPs on risk of autoantibody positivity in the TEDDY Study. Diabetes 64: 1818-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun C, Wei H, Chen X, et al. (2016): ERBB3-rs2292239 as primary type 1 diabetes association locus among non-HLA genes in Chinese. Meta Gene 9: 120-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wallace C, Smyth DJ, Maisuria-Armer M, et al. (2010): The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet 42: 68-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qiu YH, Deng FY, Tang ZX, et al. (2015): Functional relevance for type 1 diabetes mellitus-associated genetic variants by using integrative analyses. Hum Immunol 76: 753-758. [DOI] [PubMed] [Google Scholar]
- 108.Tao JH, Zou YF, Feng XL, et al. (2011): Meta-analysis of TYK2 gene polymorphisms association with susceptibility to autoimmune and inflammatory diseases. Mol Biol Rep 38: 4663-4672. [DOI] [PubMed] [Google Scholar]