Abstract
Background
COVID-19 progresses slowly and negatively affects many people. However, mild to moderate symptoms develop in most infected people, who recover without hospitalization. Therefore, the development of early diagnosis and treatment strategies is essential. One of these methods is proteomic technology based on the blood protein profiling technique. This study aims to classify three COVID-19 positive patient groups (mild, severe, and critical) and a control group based on the blood protein profiling using deep learning (DL), random forest (RF), and gradient boosted trees (GBTs).
Methods
The dataset consists of 93 samples (60 COVID-19 patients, 33 control), and 370 variables obtained from an open-source website. The current dataset contains age, gender, and 368 protein, used to predict the relationship between disease severity and proteins using DL and machine learning approaches (RF, GBTs). An evolutionary algorithm tunes hyperparameters of the models and the predictions are assessed through accuracy, sensitivity, specificity, precision, F1 score, classification error, and kappa performance metrics.
Results
The accuracy of RF (96.21%) was higher as compared to DL (94.73%). However, the ensemble classifier GBTs produced the highest accuracy (96.98%). TGB1BP2 in the cardiovascular II panel and MILR1 in the inflammation panel were the two most important proteins associated with disease severity.
Conclusions
The proposed model (GBTs) achieved the best prediction of disease severity based on the proteins compared to the other algorithms. The results point out that changes in blood proteins associated with the severity of COVID-19 may be used in monitoring and early diagnosis/treatment of the disease.
Key Words: Artificial Intelligence, COVID-19, Random Forest, Deep Learning, Gradient Boosted Trees
1. Introduction
The novel coronavirus disease (COVID-19) has spread rapidly across the globe affecting billions of people's everyday lives. The disease can lead to serious pneumonia, which can lead to death [1]. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak that occurred in December 2019 manifests itself in different situations in infected patients. While it appears as a mild respiratory infection on some infected patients, it may progress to severe pneumonia and acute respiratory distress syndrome (ARDS), resulting in multiple organ failure or even death in some. On the other hand, in some patients, the disease progresses without symptoms. Hence, it is very difficult to determine the percentage of people with COVID-19 severity. However, according to the World Health Organization (WHO), it is estimated that 80% of infections are asymptomatic or mild, 15% are serious infections requiring oxygen support, 5% are critical infections requiring ventilation, and 3% are fatal [2]. Patients classified as COVID-19 clinically severe are diagnosed based on some clinical features such as respiratory rate and mean oxygen saturation. However, when these clinical signs appeared, the patients reached a clinically serious stage. Therefore, patients are either taken to intensive care or can die quickly. Therefore, considering all these negativities, it is very important to detect early which cases may become clinically serious and develop new approaches to prevent deaths from COVID-19. Therefore, many studies have been conducted for early diagnosis and diagnosis. While most of these studies are on the clinical and epidemiological features of COVID-19, one of the most frequently used approaches recently is proteomics technology. Proteomics technology is the study of all proteins in a biological system and is increasingly being used by clinical researchers to identify disease biological markers [3]. Therefore, the detection of molecularly changed proteins in the blood of an infected individual with proteomics technology and the discovery of biomarkers are thought to play an active role in the development of the diagnosis and treatment of COVID-19.
In recent years, machine learning and deep learning-based studies have gained importance in terms of being a decision support mechanism for clinicians for early diagnosis and diagnosis of diseases in the field of health. Therefore, interest in studies involving machine learning algorithms combined with data obtained from methods used in the early diagnosis and diagnosis of COVID-19 has increased considerably. Machine learning is a sub-branch of artificial intelligence, consisting of modeling and algorithms that make inferences from existing data using mathematical and statistical methods and make predictions about the unknown with these inferences [4]. On the other hand, machine learning is an area at the intersection of statistics, artificial intelligence, computers, predictive analysis, and statistical learning about obtaining information from available data [5]. Deep learning, a sub-branch of machine learning, is the name of the system that allows multi-layered neural networks, unlike machine learning [6]. The main factor that distinguishes deep learning from ANN is that deep neural networks consist of many more layers than ANN. It is done by increasing the number of hidden layers to obtain more features from the data to be processed and to make learning better [7].
The relationships between COVID-19 severity and protein profiling technology can be modeled to discover important proteins associated with the pandemic by deep learning and artificial modeling approaches. Therefore, this study aims to classify three COVID-19 positive patient groups (mild, severe, and critical) and a control group based on the blood protein profiling using deep learning and machine learning models (i.e., Random Forest and Gradient Boosted Tree).
2. Material And Methods
2.1. Dataset
The data set used in this study includes age, gender, 368 proteins, obtained from blood protein profiling belong to 93 subjects which are 59 positive COVID-19 cases [mild (n = 26; group 1), severe (n = 9; group 2), critical (n = 24; group 3)] and 28 control groups (control group). In the OLINK proteomics of cardiovascular, immune, inflammation, and neurology panels, each containing 92 proteins, the protein profiles of 87 samples were examined, resulting in 368 protein measurements per subject blood protein profiling. One of the reasons for the data set's missing values was that a sample of the mild symptom group failed the immune and neurology panels' analysis. However, it was not excluded from the analysis for analyzes involving cardiovascular and inflammation panels. Another reason for the missing values in the data set was that in more than 50% of the samples in all four disease groups, thirteen proteins were missing Normalized Protein eXpression (NPX) values or NPX values below the protein-specific detection limit (LOD). Therefore, these 13 proteins were excluded, and 355 proteins remained; 344 were unique in that protein replication in four panels [8].
2.2. Data Preprocessing
Missing values in the data set often complicate the statistical analysis of multivariate data. Therefore, the data set's missing values are complemented by the multiple imputation method using Fully Conditional Specification (FCS), as it is thought that it will negatively affect the model training process [9]. In this method, each variable containing a missing value is determined by a separate model and assigned to that variable with that model [10]. Similarly, since the number of data in each group is unbalanced in the data set, all the classes have been balanced with the "Sample (balance)" operator in Rapidminer Studio. This operator functions the same way as the (absolute) combination of multiplying and sampling works. A fixed number of examples are selected for a given input dataset for each included mark. Current examples are multiplied if insufficient examples are included [11]. Random Forest-Recursive Feature Elimination (RF-RFE) algorithm is used as the variable selection method. In this algorithm, the data set is first trained with a machine learning algorithm, in which variables such as Random Forest (RF), logistic regression, Support Vector Machines (SVM) have certain weights. Then, the variable with the smallest coefficient is removed, and the system is retrained with the remaining variables. This process continues until all features are eliminated, and the variable subset giving the best result is selected. RFE was originally proposed to enable support vector machines to perform feature selection by iteratively training a model, grading features, and then removing the lowest-rated features [12]. However, recently this method has been similarly applied to Random forest (RF), and it is useful in the presence of related features [13], [14], [15].
2.3. Deep Learning
Deep Learning is a machine learning technique developed for machine feature extraction, perception, and learning. It performs its operations using multiple consecutive layers. Each consecutive layer receives the output formed in the previous layer as input [16]. Besides the deep learning algorithm also performs data-based learning, the learning process works with calculations based on network diagrams expressed as a neural network, not a single mathematical model as in standard machine learning algorithms. The deep learning architecture in the study was constituted by using a multi-layer feed-forward neural network with stochastic gradient descent using the backpropagation approach. Epsilon, rho, L1 regularization, L2 regularization, max w2, and dropout hyperparameters were tuned using the Evolutionary optimization algorithm to increase model performance.
2.4. Machine Learning
2.4.1. Random Forest
The random forest (RF) algorithm establishes multiple decision trees on data samples, and one estimate is obtained from each. Afterward, if the problem is a regression problem, the results obtained are averaged, and if the problem is the classification problem, the prediction with the highest number of votes is selected. RF can reduce overfitting, one of the biggest problems of machine learning algorithms. Since this algorithm gives positive results in large data sets, it can work harmoniously in large data sets. The RF provides an advantage by not ignoring outlier observations [17]. The hyperparameters of maximal depth, minimal leaf size, minimal size for the split, and the number of pre pruning alternatives were tuned using the Evolutionary optimization algorithm to increase model performance.
2.4.2. Gradient Boosted Trees (GBTs)
Gradient Boost is a type of ensemble method used in machine learning. The foundations of this algorithm are based on the studies of Friedman et al. [18], [19], [20]. The main disadvantage of decision trees is that it generates a large "bias" in simple trees and a large variance in complex trees. "Bootstrap" is a method of selecting a random number of data from a data set. It is mostly used to reduce the variance of the tree. In this algorithm, a community of stronger learners is produced, usually by compensating for each other's weaknesses, such as decision trees.
According to the Gradient Boosted Tree (GBTs) algorithm, a prediction function is constructed in the first iteration. A loss function is obtained from these differences by calculating the difference between estimates and observations. In the second iteration, the difference between the repetitions and observations is calculated by combining the estimation and loss functions. Thus, the success of the prediction function is tried to be increased by constantly adding on it. The difference between the predictions and the observations obtained is as close to zero as possible [21]. The maximum depth and learning rate hyperparameters were tuned using the Evolutionary optimization algorithm to increase the model performance.
2.5. Performance Evaluation Metrics
A 10-fold cross-validation method was used for the validity of the model. In the 10-fold cross-validation method, all data are divided into ten equal parts. The first part is used as the test set, and the remaining nine parts are used as the training data set, and this process is repeated for each part. In this technique, the general accuracy rate of the model is determined by averaging the accuracy values. Performance metrics for all models are given with accuracy, sensitivity, specificity, precision, F1 score, classification error, and kappa statistics.
2.6. Data Analysis
The compliance of quantitative variables to normal distribution was checked with the Shapiro Wilk test. Quantitative variables fulfilling the normal distribution assumption were summarized with mean and standard deviation and quantitative variables that did not show normal distribution with median and min-max. In statistical analysis, the Kruskal Wallis test was used for variables that did not show normal distribution, and the Conover test was used for pairwise comparisons of variables with differences (p<0.05). One-way analysis of variance (One-way ANOVA) test was used for variables with normal distribution and Tukey test in case of variance homogeneity in multiple comparisons, and Tamhane T2 test when variances were not homogeneous. In this study, in addition to artificial intelligence modeling and basic comparisons, the effect size was calculated to evaluate the effects of each protein on COVID-19 severity and control groups. The effect size is defined as the magnitude of the difference between groups [22]. Generally, the interpretation values in reported literature are small effect between 0.01-0.06, moderate effect between 0.06-0.14, and large effect more than 0.14 [23]. P <0.05 was considered statistically significant. The programming languages of "Statistical Analysis Software" [24], RStudio Version 3.6.2 [25], and RapidMiner Studio Version 9.8 [26] were used in data analysis.
3. Results
3.1. Baseline characteristic of the original data
In the study, 93 subjects are included, which are 34 (36.6%) are female, 59 (63.4%) are male, and the average age is 58.6 (±15.3). Descriptive statistics regarding the COVID-19 positive and control group based on age and gender variables of the data set are given in Table 1 .
Table 1.
escriptive statistics of gender and age variables by COVID-19 positive and control.
| Variable | Control (n=33) |
Mild (n=26) |
Severe (n=9) |
Critical (n=25) |
Effect Size |
p-value | |
|---|---|---|---|---|---|---|---|
| Gender | Female | 16a (48.5%) | 13a (50.0%) | 3a,b(33.3%) | 2b (8.0 %) | 0,61 (Large) | 0.005* |
| Male | 17a (51.5%) | 13a (50.0%) | 6a,b(66.7%) | 23b (92.0)% | |||
| Mean Age ± SD | 61.1a ± 18.2 | 51.3b ± 13.5 | 64.8a ± 12.8 | 60.6a ± 11.3 | 0.09 (Large) | 0.039⁎⁎ | |
a, b: Different characters in each row show a statistically significant difference (p <0.05)
Pearson Chi-square test
Kruskal-Wallis test.
Descriptive statistics of 92 proteins in the cardiovascular II panel according to the COVID-19 positive and control group are given in Table 2 . Additionally, the effect sizes for each protein are estimated in Table 2.
Table 2.
Descriptive statistics of proteins in the cardiovascular II panel by groups.
| Protein Names | Groups |
Effect Size | p-value | |||
|---|---|---|---|---|---|---|
| Control | Mild | Severe | Critical | |||
| Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | |||
| BMP-6 | 4.91a(2.57-6.16) | 4.74a(3.68-5.69) | 5.11a(4.46-5.64) | 5.07a(4.22-6.21) | 0.03 (Large) | 0.121* |
| ADM | 8.19a(6.91-9.07) | 8.47a,b(6.62-9.14) | 8.92b,c(8.22-9.79) | 9.52c(7.83-10.16) | 0.41 (Large) | <0.0001* |
| CD40-L | 6.96a(4.87-8.66) | 4.06b(2.44-7.53) | 5.02b(3.36-6.92) | 3.98b(2.92-7.34) | 0.41 (Large) | <0.0001* |
| PGF | 7.42a(6.92-8.27) | 7.33a(6.1-8.09) | 7.53a,b(7.04-8.25) | 7.92b(6.99-9.66) | 0.22 (Large) | <0.0001* |
| ADAM-TS13 | 5.03 a,b(4.77-5.39) | 5.15a(4.74-5.52) | 4.86a,b(4.68-5.19) | 4.9b(4.27-6.3) | 0.18 (Large) | <0.0001* |
| BOC | 3.79a(2.96-4.53) | 3.76a(2.75-4.36) | 2.99b(2.56-3.44) | 2.73b(2.48-3.59) | 0.49 (Large) | <0.0001* |
| IL-4RA | 2.1a(1.57-2.57) | 2.05a(1.3-3.35) | 2.09a(1.69-4.41) | 2.97b(1.84-5.11) | 0.32 (Large) | <0.0001* |
| IL-1ra | 4.97a(3.9-6.96) | 5.12a(3.97-7.47) | 6.64b(5.45-8.12) | 7.72c(5.99-8.16) | 0.55 (Large) | <0.0001* |
| TNFRSF10A | 2.75a(2.34-3.78) | 2.64a(1.94-3.89) | 3.89b(3.01-4.89) | 4.44b(3.06-6.52) | 0.56 (Large) | <0.0001* |
| STK4 | 6.23a(4.87-6.61) | 2.5b(0.41-5.67) | 3.4b(1.36-4.25) | 3.12b(1.44-5.12) | 0.58 (Large) | <0.0001* |
| PAR-1 | 9.77a(9.21-10.32) | 8.44b(7.72-9.24) | 8.66b(7.79-9.34) | 8.82b(5.73-9.79) | 0.54 (Large) | <0.0001* |
| PRSS27 | 9.32a(8.17-9.77) | 8.95a(7.95-10.36) | 8.21b(7.9-8.53) | 8.04b(7.34-9.03) | 0.42 (Large) | <0.0001* |
| TF | 5.92a(4.92-6.59) | 5.82a(4.63-6.13) | 6.1a,b(5.21-6.94) | 6.26b(5.59-7.51) | 0.19 (Large) | <0.0001* |
| IL1RL2 | 4.51a,b(3.47-4.87) | 4.52a(3.43-5.16) | 4b(2.8-4.96) | 4.4a,b(3.44-5.92) | 0.03 (Large) | 0.103* |
| PDGF subunit B | 11.1a(10.12-11.44) | 9.77b(7.93-10.85) | 10.38b,c(8.78-11.27) | 10.38c(8.01-11.08) | 0.51 (Large) | <0.0001* |
| IL-17D | 2.6a(1.8-4.53) | 2.33a,b(1.56-3.02) | 2.3a,b(1.7-3.33) | 2.08b(1.75-2.83) | 0.07 (Large) | 0.023* |
| LOX-1 | 6.94a(6.31-8.34) | 6.77a(5.2-7.84) | 7.84b(7.15-9.38) | 8.27b(6.45-9.81) | 0.32 (Large) | <0.0001* |
| IL18 | 9.08a,b(8.14-10.62) | 8.7a(7.49-10.27)a | 9.73b,c(8.97-11.09) | 9.85c(8.98-13.73) | 0.41 (Large) | <0.0001* |
| PIgR | 5.76a(5.43-6.21) | 5.69a(5.31-6.01) | 5.67a(5.54-5.76) | 5.66a(5.44-6.23) | 0.02 (Large) | 0.171* |
| RAGE | 13a(12.01-14.07)a | 13.38a(11.32-14.32) | 13.88b(13.55-14.92) | 14.58b(12.6-15.05) | 0.43 (Large) | <0.0001* |
| SOD2 | 10.27a(9.89-10.52) | 10.1b(9.69-10.3) | 10.17a(10.11-10.45) | 10.15a(10.05-10.33) | 0.33 (Large) | <0.0001* |
| FGF-23 | 2.89a(1.89-4) | 2.57a(1.54-5.22) | 2.69a(2.08-3) | 2.95a(1.8-9.17) | <0.001 (Small) | 0.378* |
| SPON2 | 8.17a(7.72-8.63) | 8.39a,b(7.32-8.59) | 8.63c(8.19-8.76) | 8.74c(8.47-8.96) | 0.54 (Large) | <0.0001* |
| GLO1 | 6.54a(5.56-7.55) | 5.23b(4.32-6.63) | 6.67a(5.78-7.42) | 6.01a(5.35-8.88) | 0.38 (Large) | <0.0001* |
| SERPINA12 | 3.01a,b(1.68-4.45) | 3.26a(1.27-6.7) | 1.85a,b(1.24-7.91) | 2.23b(0.59-4.64) | 0.12 (Large) | 0.002* |
| TM | 9.53a,b(9.24-10.15) | 9.55a,b(8.54-10.21) | 9.32a(8.81-10.09) | 9.76b(9.18-11.02) | 0.05 (Large) | 0.045* |
| PRELP | 8.27a(7.86-8.74) | 8.35a(7.63-8.75) | 8.34a(8-8.92) | 8.38a(7.56-9.1) | 0.02 (Large) | 0.215* |
| HO-1 | 11.67a(9.6-12.57) | 11.64a(10.41-12.77) | 12.51b(11.4-12.95) | 12.69b(10.97-13.02) | 0.36 (Large) | <0.0001* |
| XCL1 | 4.63a(3.81-6.76) | 5.07a(3.79-5.66) | 5.39a,b(4.22-5.73) | 5.47b(4.42-7.38) | 0.22 (Large) | <0.0001* |
| CEACAM8 | 4.25a(2.95-5.28) | 4.11a(2.47-5.69) | 4.73a,b(3.89-5.95) | 4.91b(3.62-7.73) | 0.23 (Large) | <0.0001* |
| PTX3 | 3.75a(3.15-5.35) | 3.94a(2.6-5.66) | 5.17b(4.58-6.14) | 5.62b(3.94-6.22) | 0.52 (Large) | <0.0001* |
| PSGL-1 | 4.66a(4.28-5.22) | 4.55a,b(4.08-5.4) | 4.39b(3.84-4.76) | 4.37b,c(3.85-4.9) | 0.19 (Large) | <0.0001* |
| MMP7 | 8.14a(7.07-10.11) | 9.6b(8.49-10.02) | 9.25b,c(8.95-10.15) | 9.91c(8.98-12.18) | 0.48 (Large) | <0.0001* |
| ITGB1BP2 | 8a(6.41-9.32) | 2.53b(1.62-5.53) | 3.71b(1.95-4.15) | 3.09b(1.86-5.21) | 0.60 (Large) | <0.0001* |
| DCN | 4.27a(3.6-5.03) | 4.46a(3.4-4.95) | 4.28a,b(3.99-5.37) | 4.86b(3.93-5.7) | 0.11 (Large) | 0.005* |
| Dkk-1 | 9.74a(7.8-10.47) | 8.29b(7.19-10.09) | 9.05a(8.2-9.76) | 9.28a(7.82-10.52) | 0.37 (Large) | <0.0001* |
| HB-EGF | 3.8a(1.61-4.64) | 0.8b(0.28-1.49) | 0.77b(0.36-1.68) | 1.61c(0.45-2.53) | 0.71 (Large) | <0.0001* |
| BNP | 1.11a(0.56-2.24) | 1.29a,c(0.72-3.29) | 3.38b(1.78-6.24) | 1.77c(0.69-5.87) | 0.21 (Large) | <0.0001* |
| ACE2 | 3.27a(2.5-5.02) | 3.24a(2.57-5.12) | 4.44b(3.19-5.61) | 4.8b(3.08-6.42) | 0.36 (Large) | <0.0001* |
| CTSL1 | 7.21a(6.59-7.73) | 7.44a(6.43-9.71) | 8.75b(8.19-9.74) | 9.53c(8.62-10.24) | 0.61 (Large) | <0.0001* |
| hOSCAR | 10.73a(10.34-11.19) | 10.76a(9.8-11.08) | 11.02b(10.88-11.29) | 11.05b(10.67-11.34) | 0.28 (Large) | <0.0001* |
| TGM2 | 8.85a(5.17-9.59) | 7.79b(6.16-8.92) | 8.22a,b(6.95-9.2) | 8.19a,b(6.53-10.06) | 0.19 (Large) | <0.0001* |
| CA5A | 2.08a(1.35-4.81) | 2.61a(1.39-5.34) | 3.85b(2.63-6) | 4.67b(2.21-7.38) | 0.44 (Large) | <0.0001* |
| PARP-1 | 5.05a(3.83-8.26) | 3.95b(2.88-5.69) | 5.14a,c(4.3-6.97) | 5.83c(4.45-8.92) | 0.03 (Large) | <0.0001* |
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |||
| ANGPT1 | 9.36a ± 0.64 | 8.21b ± 1.04 | 8.69a,b ± 0.82 | 8.30b ± 0.91 | 0.26 (Large) | <0.0001⁎⁎ |
| SLAMF7 | 3.04a ± 0.66 | 3.35a,b ± 0.58 | 3.91b,c ± 0.72 | 4.60c ± 0.77 | 0.48 (Large) | <0.0001⁎⁎ |
| SRC | 7.48a ± 0.34 | 5.54b ± 1.21 | 5.91b ± 0.69 | 5.93b ± 0.98 | 0.47 (Large) | <0.0001⁎⁎ |
| IL6 | 3.16a ± 0.75 | 3.13a ± 1 | 6.56b ± 1.22 | 8.29c ± 1.8 | 0.79 (Large) | <0.0001⁎⁎ |
| IDUA | 5.94a ± 0.47 | 5.59a,b ± 0.42 | 5.25b ± 0.67 | 5.43b,c ± 0.56 | 0.19 (Large) | 0.001⁎⁎ |
| TNFRSF11A | 5.82a ± 0.46 | 5.53a ± 0.41 | 6.07a,b ± 0.6 | 6.52b ± 0.82 | 0.32 (Large) | <0.0001⁎⁎ |
| TRAIL-R2 | 6.10a ± 0.42 | 5.93a ± 0.35 | 7.09b ± 0.59 | 7.97c ± 1.02 | 0.66 (Large) | <0.0001⁎⁎ |
| TIE2 | 7.36a ± 0.25 | 7.34a ± 0.32 | 7.19a ± 0.24 | 7.43a ± 0.25 | 0.06 (Large) | 0.141⁎⁎ |
| IL-27 | 5.78a ± 0.35 | 6.05a ± 0.42 | 7.01b ± 0.44 | 6.89b ± 0.63 | 0.54 (Large) | <0.0001⁎⁎ |
| CXCL1 | 11.78a ± 0.55 | 9.38b ± 0.98 | 10.53c ± 0.93 | 10.67c ± 0.87 | 0.58 (Large) | <0.0001⁎⁎ |
| Gal-9 | 7.99a ± 0.45 | 8.14a ± 0.47 | 9.08b ± 0.29 | 9.31b ± 0.27 | 0.69 (Large) | <0.0001⁎⁎ |
| GIF | 6.98a ± 0.85 | 6.98a ± 0.66 | 6.10b ± 0.87 | 6.60a,b ± 0.84 | 0.12 (Large) | 0.039⁎⁎ |
| SCF | 9.39a ± 0.35 | 9.34a ± 0.37 | 7.85b ± 0.92 | 8.12b ± 0.97 | 0.51 (Large) | <0.0001⁎⁎ |
| FGF-21 | 6.57a ± 1.09 | 6.83a,b ± 1.32 | 6.83a,b ± 2.09 | 7.98b ± 2.65 | 0.10 (Large) | <0.0001⁎⁎ |
| CTRC | 10.17a ± 0.77 | 10.52a ± 0.75 | 9.72a,b ± 0.79 | 9.29b ± 1.37 | 0.21 (Large) | 0.279⁎⁎ |
| GH | 8.20a,b ± 2.02 | 6.94a ± 2.18 | 9.26b ± 0.84 | 8.28a,b ± 1.42 | 0.14 (Large) | <0.0001⁎⁎ |
| FS | 10.76a ± 0.55 | 10.74a ± 0.52 | 10.79a ± 0.48 | 11.05a ± 0.68 | 0.05 (Large) | 0.008⁎⁎ |
| CD84 | 5.84a ± 0.43 | 4.59b ± 0.37 | 4.58b ± 0.41 | 4.68b ± 0.42 | 0.67 (Large) | 0.296⁎⁎ |
| PAPPA | 3.19a ± 0.6 | 2.77a ± 0.71 | 2.77a ± 0.59 | 3.01a ± 0.67 | 0.07 (Large) | <0.0001⁎⁎ |
| REN | 6.65a ± 0.67 | 6.26a ± 0.55 | 5.99a ± 0.76 | 7.30b ± 0.89 | 0.30 (Large) | 0.12⁎⁎ |
| DECR1 | 7.92a ± 1.06 | 3.22b ± 0.74 | 3.89b,c ± 0.34 | 4.39c ± 0.77 | 0.85 (Large) | <0.0001⁎⁎ |
| MERTK | 6.31a ± 0.4 | 6.48a,b ± 0.49 | 6.83b,c ± 0.6 | 7.16c ± 0.32 | 0.40 (Large) | <0.0001⁎⁎ |
| KIM1 | 8.22a ± 0.89 | 8.14a ± 0.78 | 9.13a,b ± 0.58 | 9.42b ± 1.29 | 0.27 (Large) | <0.0001⁎⁎ |
| THBS2 | 5.50a ± 0.2a | 5.54a ± 0.15a | 5.68a,b ± 0.18a.b | 5.75b ± 0.2b | 0.25 (Large) | <0.0001⁎⁎ |
| VSIG2 | 3.87a ± 0.59 | 3.69a ± 0.42 | 3.54a ± 0.61 | 4.00a ± 0.78 | 0.06 (Large) | <0.0001⁎⁎ |
| AMBP | 7.60a ± 0.23 | 7.60a ± 0.21 | 7.52a ± 0.28 | 7.57a ± 0.25 | 0.01 (Moderate) | 0.249⁎⁎ |
| IL16 | 6.73a ± 0.62 | 6.55a ± 0.61 | 7.01a,b ± 0.6 | 7.21b ± 0.67 | 0.16 (Large) | 0.008⁎⁎ |
| SORT1 | 8.96a ± 0.38 | 8.36b ± 0.37 | 8.65a,b ± 0.34 | 8.75a ± 0.23 | 0.34 (Large) | <0.0001⁎⁎ |
| CCL17 | 10.95a ± 0.88 | 8.47b ± 0.98 | 8.16b ± 0.87 | 8.23b ± 1.77 | 0.52 (Large) | <0.0001⁎⁎ |
| CCL3 | 6.66a,b ± 0.84 | 6.06a ± 0.63 | 7.30b,c ± 0.94 | 7.65c ± 0.89 | 0.39 (Large) | <0.0001⁎⁎ |
| IgG Fc receptor II-b | 3.03a ± 0.89 | 2.96a ± 0.7 | 3.51a ± 0.54 | 3.34a ± 0.67 | 0.07 (Large) | 0.08⁎⁎ |
| LPL | 9.53a ± 0.47 | 10.08b ± 0.37 | 9.47a ± 0.46 | 9.42a ± 0.47 | 0.30 (Large) | <0.0001⁎⁎ |
| PRSS8 | 8.96a ± 0.39 | 8.78a ± 0.44 | 9.08a ± 0.25 | 9.05a ± 0.35 | 0.09 (Large) | 0.103⁎⁎ |
| AGRP | 5.19a ± 0.36 | 5.44a,b ± 0.59 | 5.74b,c ± 0.57 | 6.12c ± 0.57 | 0.35 (Large) | <0.0001⁎⁎ |
| GDF-2 | 9.17a ± 0.65 | 8.80a ± 0.51 | 7.54b ± 0.67 | 7.60b ± 0.87 | 0.52 (Large) | <0.0001⁎⁎ |
| FABP2 | 8.04a ± 0.99 | 8.02a ± 0.86 | 7.29a,b ± 1.07 | 7.09b ± 1.35 | 0.15 (Large) | 0.009⁎⁎ |
| THPO | 4.50a ± 0.54 | 3.65b ± 0.44 | 3.70b,c ± 0.54 | 4.20a,c ± 0.66 | 0.32 (Large) | <0.0001⁎⁎ |
| MARCO | 6.83a ± 0.25 | 6.92a ± 0.2 | 6.89a ± 0.18 | 6.91a ± 0.28 | 0.03 (Large) | 0.389⁎⁎ |
| GT | 2.32a ± 0.63 | 2.59a ± 0.62 | 2.09a ± 0.94 | 2.43a ± 1.08 | 0.03 (Large) | 0.259⁎⁎ |
| MMP12 | 7.13a ± 0.86 | 6.89a ± 0.5 | 6.25a ± 0.83 | 6.51a ± 1.24 | 0.11 (Large) | 0.038⁎⁎ |
| PD-L2 | 3.49a ± 0.32 | 3.44a ± 0.42 | 3.51a ± 0.5 | 3.65a ± 0.42 | 0.04 (Large) | 0.248⁎⁎ |
| TNFRSF13B | 9.64 ± 0.42a | 9.73 ± 0.45a.b | 10.22 ± 0.6b.c | 10.26 ± 0.55 | 0.26 (Large) | <0.0001⁎⁎ |
| LEP | 6.03a ± 1.56 | 5.91a ± 1.33 | 5.56a ± 1.7 | 6.22a ± 0.8 | 0.02 (Large) | 0.817⁎⁎ |
| HSP 27 | 10.33a ± 0.31 | 9.57b ± 0.77 | 10.10a,b ± 0.32 | 10.25a ± 0.39 | 0.29 (Large) | <0.0001⁎⁎ |
| CD4 | 4.46a ± 0.34 | 4.78b ± 0.38 | 5.05b,c ± 0.34 | 5.27c ± 0.54 | 0.38 (Large) | <0.0001⁎⁎ |
| NEMO | 8.08a ± 0.8 | 4.55b ± 0.84 | 5.27b,c ± 0.65 | 5.52c ± 0.95 | 0.76 (Large) | <0.0001⁎⁎ |
| VEGFD | 7.79a ± 0.4 | 7.76a ± 0.28 | 7.80a,b ± 0.24 | 7.42b ± 0.53 | 0.15 (Large) | 0.011⁎⁎ |
| HAOX1 | 5.03a ± 1.26 | 5.72a ± 1.22 | 6.33a,b ± 1.6 | 7.62b ± 1.35 | 0.39 (Large) | <0.0001⁎⁎ |
a, b,c: Different characters in each row show a statistically significant difference (p <0.05)
Kruskal-Wallis test
One-way analysis of variance.
According to Table 2, the difference between groups in terms of protein in the cardiovascular panel except for BMP-6, IL1RL2, PIgR, FGF-23, PRELP, TIE2, CTRC, CD84, REN, AMBP, IgG Fc receptor II-b, PRSS8, MARCO, GT, PD-L2, and LEP proteins is statistically significant (p<0.05). According to the effect size findings, the two proteins most prominently affecting the severity of COVID-19 and control groups in the cardiovascular II panel are DECR-1 (0.85) and IL-6 (0.79) proteins. Similarly, the descriptive statistics of 92 proteins in the immune response panel according to the COVID-19 positive and control group are given in Table 3 .
Table 3.
Descriptive statistics of proteins in the immune response panel by groups.
| Protein Names | Groups |
Effect Size | p-value | |||
|---|---|---|---|---|---|---|
| Control | Mild | Severe | Critical | |||
| Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | |||
| PPP1R9B | 6.7a(5.3-7.29) | 1.22b(0.74-3.72) | 1.83b(0.63-2.45) | 1.77b(0.73-3.62) | 0.63 (Large) | <0.0001* |
| GLB1 | 3.8a(1.61-4.64) | 0.8b(0.28-1.49) | 0.77b(0.36-1.68) | 1.61c(0.45-2.53) | 0.71 (Large) | <0.0001* |
| PSIP1 | 4.48a(2.72-6.71) | 2.16b(1.54-4.56) | 3.87a(3.16-5.43) | 4.43a(3.47-7.4) | 0.49 (Large) | <0.0001* |
| ZBTB16 | 4.09a(2.53-8.12) | 0.4b(-0.07-2.25) | 1.2b,c(0.42-2.69) | 1.45c(0.54-3.28) | 0.71 (Large) | <0.0001* |
| IRAK4 | 6.74a(5-7.89) | 1.56b(0.85-3.69) | 2.35b(1.29-3.51) | 2.04b(1.12-4.29) | 0.64 (Large) | <0.0001* |
| HCLS1 | 8.19a(6.97-8.74) | 3.67b(2.46-6) | 5.37c(4.15-6.71) | 5.33c(4.08-7.51) | 0.69 (Large) | <0.0001* |
| CNTNAP2 | 1.97a(1.39-3.32) | 1.46b(0.65-1.99) | 1.39b(0.69-1.76) | 1.09b(0.56-2.3) | 0.37 (Large) | <0.0001* |
| CLEC4G | 3.34a(2.58-4.47) | 3.4a(2.85-4.53) | 3.88a,b(3.08-4.86) | 4.32b(3.37-6.35) | 0.36 (Large) | <0.0001* |
| IRF9 | 3.11a(2.09-4.31) | 1.33b(0.85-1.87) | 1.74b,c(1.15-2.85) | 2.21c(1.23-5.42) | 0.58 (Large) | <0.0001* |
| EDAR | 4.22a(2.3-5.71) | 1.72b(1.11-3.92) | 1.36b(0.9-1.7) | 1.47b(0.92-2.68) | 0.63 (Large) | <0.0001* |
| IL6 | 2.21a(1.47-3.79) | 1.9a(1.16-4.87) | 5.02b(4.25-8.14) | 7.13c(2.91-10.47) | 0.64 (Large) | <0.0001* |
| DGKZ | 1.44a(0.58-2.65) | 0.39b(-0.1-0.68) | 0.6b(0.29-0.89) | 0.57b(0.02-3.48) | 0.54 (Large) | <0.0001* |
| CLEC4C | 3.88a,b(2.58-4.88) | 3.98a(3.3-5.44) | 3.51b,c(2.41-4.31) | 3.05c(2.07-4.27) | 0.25 (Large) | <0.0001* |
| IRAK1 | 4.82a(3.32-6.07) | 1.6b(1.13-2.97) | 2.19b,c(1.68-2.81) | 2.39c(1.9-4.37) | 0.72 (Large) | <0.0001* |
| CLEC4A | 4.19a(3.05-4.52) | 3.84b(2.85-4.24) | 2.98c(2.14-3.61) | 2.94c(2.31-4) | 0.60 (Large) | <0.0001* |
| PRDX1 | 5.32a(3.86-6.62) | 1.24b(0.71-6.25) | 2.14b,c(1.28-4.48) | 2.81c(1.83-5.08) | 0.67 (Large) | <0.0001* |
| PRDX3 | 2.4a(-0.07-4.38) | 0.63b(-0.98-0.43) | 0.26b,c(-0.65-0.91) | 0.25c(-0.32-1.47) | 0.70 (Large) | <0.0001* |
| FGF2 | 3.64a(2.03-5.74) | 0.24b(-0.13-1.31) | 0.98b(0.15-1.83) | 0.44b(-0.07-1.94) | 0.64 (Large) | <0.0001* |
| PRDX5 | 7.65a(7.06-8.44) | 3.27b(2.53-6.61) | 4.55c(3.79-6.31) | 5.19c(4.04-7.05) | 0.70 (Large) | <0.0001* |
| TRIM5 | 3.83a(2.42-5.49) | 1.2b(0.92-2.47) | 1.66b,c(1.43-3.49) | 2.29c(1.57-4.61) | 0.67 (Large) | <0.0001* |
| DCTN1 | 7.42a(5.65-8.63) | 2.81b(1.54-4.47) | 3.37b,c(2.46-3.9) | 3.6c(2.6-7.18) | 0.63 (Large) | <0.0001* |
| ITGA6 | 5.1a(1.43-6.34) | 0.88b(0.03-3.98) | 0.56b(0.12-1.17) | 0.69b(-0.11-2.09) | 0.59 (Large) | <0.0001* |
| CDSN | 3.16a(2.28-3.96) | 2.6a(1.47-6.23) | 2.29a(1.89-3.65) | 2.59a(1.54-4.55) | 0.13 (Large) | 0.002* |
| FXYD5 | 2.25a(0.77-4.13) | 0.02b(-0.51-0.85) | 0.32b,c(0.03-1.25) | 0.56c(-0.7-3.06) | 0.64 (Large) | <0.0001* |
| TRAF2 | 3.19a(2.16-5.3) | 1.31b(0.86-4.42) | 1.81b(1.08-2.55) | 1.73b(1.09-3.7) | 0.54 (Large) | <0.0001* |
| LILRB4 | 3.05a(2.09-4.62) | 3.42a(2.65-5.38) | 4.86b(4.19-6.22) | 5.47c(4.62-7.38) | 0.64 (Large) | <0.0001* |
| NTF4 | 1.89a(1.31-2.81) | 1.91a(1.4-3.54) | 1.54a(1.33-1.95) | 1.58a(0.95-2.53) | 0.09 (Large) | 0.011* |
| KRT19 | 2.17a(1.17-4.07) | 1.84a(0.08-4.19) | 4.84b(2.69-7.31) | 5.57b(3.11-7.72) | 0.63 (Large) | <0.0001* |
| HNMT | 9.12a(7.7-10.2) | 8.96a(6.91-10.66) | 9.55a,b(8.9-11.63) | 9.91b(9.13-14.96) | 0.31 (Large) | <0.0001* |
| CCL11 | 7.23a(6.32-8.56) | 7.41a(5.32-8.33) | 6.69a(6.21-7.59) | 7.11a(6.23-8.04) | 0.06 (Large) | 0.036* |
| EGLN1 | 1.77a(1.03-2.59) | 1.16b(0.86-1.98) | 1.69a(0.76-2.61) | 1.88a(1.19-4.68) | 0.32 (Large) | <0.0001* |
| NFATC3 | 0.94a(0.43-2.78) | 0.57b(0.12-2.1) | 0.81a,b(0.32-2.1) | 1.38a(0.53-2.45) | 0.26 (Large) | <0.0001* |
| EIF5A | 0.25a(-0.17-1.95) | 0.03b(-0.53-0.45) | 0.01b,c(-0.3-0.39) | 0.24a,c(-0.63-1.4) | 0.20 (Large) | <0.0001* |
| EIF4G1 | 7.51a(6.32-8.13) | 2.57b(1.51-5.26) | 4.07c(2.79-4.86) | 4.41c(3.12-6.49) | 0.72 (Large) | <0.0001* |
| CD28 | 1.61a(1.22-4.38) | 1.65a(0.94-3.75) | 1.37a(0.98-1.77) | 1.57a(0.92-2.3) | 0.03 (Large) | 0.130* |
| PTH1R | 3.9a(3.28-5.38) | 4.09a(3.47-4.92) | 3.6a(3.45-4.4) | 3.85a(3.26-4.78) | 0.08 (Large) | 0.019* |
| BIRC2 | 2.13a(1.16-4.28) | 0.31b(-0.01-0.86) | 0.59b,c(0.3-1.01) | 0.71c(0.18-1.58) | 0.70 (Large) | <0.0001* |
| HSD11B1 | 3.1a,b(2.55-3.8) | 3.17a(2.64-4.36) | 2.59b(2.12-3.74) | 2.89b,c(1.96-4.06) | 0.11 (Large) | 0.005* |
| NF2 | 4.01a(2.22-5.11) | 1.72b(-2.48-0.4) | 1.31b(-1.98–0.62) | 1.19b(-2.32-1.56) | 0.63 (Large) | <0.0001* |
| SH2B3 | 6.72a(5.05-8.86) | 2.7b(1.53-6.8) | 3.23b(2.46-3.65) | 2.79b(1.67-5) | 0.59 (Large) | <0.0001* |
| FCRL3 | 1.08a(0.48-1.91) | 0.96a(0.46-1.74) | 0.79a(0.63-2.11) | 0.92a(0.55-1.88) | 0.01 (Moderate) | 0.293* |
| CKAP4 | 4.74a(3.79-5.3) | 4.71a(3.65-5.47) | 5.82b(5.26-7.72) | 6.7c(5.16-9.94) | 0.64 (Large) | <0.0001* |
| JUN | 0.55a(-0.4-5.39) | 0.23a(-0.76-2.76) | 0.2a(-0.57-1.3) | 0.59a(-0.39-3.84) | 0.03 (Large) | 0.121* |
| HEXIM1 | 8.48a(5.94-9.71) | 3.86b(2.64-5.69) | 5.07c(4.51-6.96) | 5.67c(4.68-7.8) | 0.78 (Large) | <0.0001* |
| CLEC4D | 3.36a(2.49-4.54) | 3.49a(2.02-4.47) | 3.82a,b(3.48-5.27) | 4.42b(3.09-7.93) | 0.29 (Large) | <0.0001* |
| PRKCQ | 1.86a(0.8-5.11) | 0.4b(-0.3-1.2) | 0.56b(-0.09-0.93) | 0.57b(0.04-4.82) | 0.57 (Large) | <0.0001* |
| CXADR | 2.22a(1.57-3.45) | 2.18a(1.29-3.3) | 2.5a,b(1.6-3.67) | 2.77b(1.95-4.84) | 0.19 (Large) | <0.0001* |
| IL10 | 3.51a(2.86-6.58) | 3.7a(2.84-7.15) | 5.1b(4.24-6.77) | 5.92b(4.35-7.35) | 0.53 (Large) | <0.0001* |
| SRPK2 | 5.57a(2.85-7.63) | 0.19b(-0.15-1.87) | 1.01b,c(0.41-2.37) | 1.55c(0.74-4.52) | 0.76 (Large) | <0.0001* |
| KLRD1 | 6.44a(5.61-8.36) | 6.62a,b(5.71-7.56) | 7.15a,b(5.98-7.59) | 7.15b(5.83-8.87) | 0.13 (Large) | 0.002* |
| BACH1 | 2.79a(1.46-5.36) | 0.73b(0.22-3.18) | 1.49b,c(1.17-2.98) | 2.12c(1.33-3.79) | 0.61 (Large) | <0.0001* |
| PIK3AP1 | 4.88a(3.49-7.4) | 2.51b(1.32-3.67) | 3.05b,c(2.28-4.72) | 3.18c(1.94-5.86) | 0.54 (Large) | <0.0001* |
| SPRY2 | 7.1a(4.38-9.22) | 1.9b(0.76-7.81) | 2.4b(1.16-2.71) | 2.2b(1.18-5.02) | 0.56 (Large) | <0.0001* |
| STC1 | 5.55a(4.39-6.49) | 7.01b(5.55-7.72) | 7.29b,c(6.65-7.54) | 7.52c(6.43-7.72) | 0.66 (Large) | <0.0001* |
| ARNT | 0.84a,b(0.21-2.32) | 0.36a(-0.3-1.63) | 0.63a,b(0.39-3.91) | 0.83b(-0.07-5.86) | 0.17 (Large) | <0.0001* |
| FAM3B | 4.88a(3.96-5.64) | 4.87a(3.16-5.58) | 4.31b(3.21-4.87) | 4.45a,b(3.34-6.53) | 0.13 (Large) | 0.002* |
| DFFA | 7.31a(5.91-8.94) | 3.8b(2.91-5.09) | 4.61c(4.44-5.93) | 5.24c(4.26-8.38) | 0.73 (Large) | <0.0001* |
| DAPP1 | 9.03a(7.08-9.75) | 2.33b(1.29-5.55) | 2.87b(0.8-3.57) | 2.4b(1.45-5.48) | 0.60 (Large) | <0.0001* |
| PADI2 | 1.41a,c(0.74-2.77) | 0.59b(0.25-3.05) | 0.79a,b(0.52-1.59) | 1.36c(0.51-4.41) | 0.28 (Large) | <0.0001* |
| CLEC7A | 3.33a(-0.07-4.19) | 3.19a(2.1-3.66) | 3.36a(2.88-4.28) | 3.35a(-0.16-6.42) | 0.01 (Moderate) | 0.265* |
| IL12RB1 | 2a(1.57-5.67) | 2.25a(1.73-2.98) | 2.65a,b(2.34-3.33) | 3.05b(2.1-5.01) | 0.46 (Large) | <0.0001* |
| TANK | 3.71a(1.94-5.94) | 1.06b(0.31-2.2) | 1.5b(0.81-2.56) | 1.44b(0.09-4.18) | 0.57 (Large) | <0.0001* |
| KPNA1 | 1.18a(0.05-2.16) | 0.83b(-1.59-0.16) | 0.57b,c(-1.19-1.41) | 0.45c(-1.17-3.03) | 0.57 (Large) | <0.0001* |
| LAG3 | 2.39a(1.79-4.03) | 2.77b(2.2-4.4) | 3.16b,c(2.68-4.04) | 3.28c(2.32-4.64) | 0.34 (Large) | <0.0001* |
| IL5 | 0.63a(0.18-3.5) | 0.55a(0.11-6.91) | 0.9a(0.2-2.99) | 0.66a(0.07-4.38) | 0.01 (Moderate) | 0.259* |
| CD83 | 2.91a(2.41-3.61) | 2.91a(2.17-3.51) | 2.69a(2.36-3.36) | 2.91a(2.18-4.72) | <0.0001 (Small) | 0.397* |
| ITGB6 | 3.07a,b(2.21-3.72) | 2.99a,b(1.85-4.06) | 2.85a(2.2-3.02)a | 3.24b(2.32-5.26)b | 0.08 (Large) | 0.018* |
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |||
| TPSAB1 | 4.81a ± 0.6 | 4.51a ± 0.53 | 4.70a ± 0.59 | 4.46a ± 0.62 | 0.06 (Large) | 0.121⁎⁎ |
| DPP10 | 1.66a ± 0.43 | 1.32b ± 0.35 | 1.02b ± 0.33 | 1.44a,b ± 0.53 | 0.18 (Large) | 0.002⁎⁎ |
| GALNT3 | 2.63a ± 0.46 | 2.99a ± 0.58 | 3.00a ± 0.45 | 3.59b ± 0.56 | 0.35 (Large) | <0.0001⁎⁎ |
| TRIM21 | 5.93a ± 0.63 | 2.52b ± 0.79 | 3.40c ± 0.98 | 4.19c ± 0.93 | 0.75 (Large) | <0.0001⁎⁎ |
| ITM2A | 2.36a ± 0.42 | 2.28a ± 0.95 | 1.84a ± 0.83 | 2.59a ± 1.29 | 0.05 (Large) | 0.202⁎⁎ |
| MILR1 | 2.82a ± 0.46 | 3.16a ± 0.43 | 3.28a,b ± 0.61 | 3.68b ± 0.52 | 0.33 (Large) | <0.0001⁎⁎ |
| LY75 | 3.11a ± 0.35 | 2.86a,c ± 0.45 | 2.42b ± 0.35 | 2.66b,c ± 0.36 | 0.27 (Large) | <0.0001⁎⁎ |
| PLXNA4 | 5.69a ± 0.91 | 3.48b ± 0.82 | 4.14b ± 0.79 | 3.62b ± 0.98b | 0.56 (Large) | <0.0001⁎⁎ |
| MGMT | 7.33a ± 0.59 | 3.34b ± 1.06 | 3.96b,c ± 0.98 | 4.33c ± 0.97 | 0.79 (Large) | <0.0001⁎⁎ |
| TREM1 | 2.36a ± 0.57 | 2.33a ± 0.51 | 3.11b ± 0.49 | 3.20b ± 0.67 | 0.34 (Large) | <0.0001⁎⁎ |
| SH2D1A | 3.23a ± 1.05 | 1.66b ± 0.51 | 1.99b ± 0.5 | 2.15b ± 0.62 | 0.43 (Large) | <0.0001⁎⁎ |
| ICA1 | 3.74a ± 0.91 | 0.78b ± 0.31 | 1.20b,c ± 0.38 | 1.27c ± 0.49 | 0.82 (Large) | <0.0001⁎⁎ |
| DCBLD2 | 7.84a ± 0.36 | 7.96a ± 0.35 | 7.94a ± 0.36 | 7.89a ± 0.44 | 0.02 (Large) | 0.580⁎⁎ |
| FCRL6 | 3.07a ± 0.55 | 3.62b ± 0.65 | 2.97a,b ± 0.56 | 3.46a,b ± 0.77 | 0.14 (Large) | 0.014⁎⁎ |
| NCR1 | 3.48a,b ± 0.41 | 3.27a ± 0.36 | 3.61a,b ± 0.47 | 3.83b ± 0.78 | 0.14 (Large) | 0.004⁎⁎ |
| CXCL12 | 1.55a ± 0.25 | 1.35b ± 0.18 | 1.65a ± 0.27 | 1.72a ± 0.24 | 0.29 (Large) | <0.0001⁎⁎ |
| AREG | 3.39 ± 0.49a | 3.21 ± 0.63a | 4.99 ± 0.77b | 5.78 ± 1.03c | 0.71 (Large) | <0.0001⁎⁎ |
| IFNLR1 | 2.43a ± 0.37 | 2.58a ± 0.28 | 3.02b ± 0.39 | 3.12b ± 0.44 | 0.40 (Large) | <0.0001⁎⁎ |
| SIT1 | 1.79a ± 0.33 | 2.22a ± 0.65 | 3.24b ± 0.99 | 2.93b ± 1.07 | 0.34 (Large) | <0.0001⁎⁎ |
| MASP1 | 2.27a ± 0.33 | 1.96b ± 0.33 | 1.28c ± 0.2 | 1.42c ± 0.28 | 0.62 (Large) | <0.0001⁎⁎ |
| LAMP3 | 4.24a ± 0.61 | 4.68a ± 0.91 | 5.53b ± 0.66 | 6.07b ± 0.47 | 0.56 (Large) | <0.0001⁎⁎ |
| CLEC6A | 1.94a ± 0.56 | 2.26a ± 0.51 | 3.01b ± 0.61 | 3.26b ± 0.65 | 0.49 (Large) | <0.0001⁎⁎ |
| DDX58 | 4.34a ± 0.75 | 2.81b ± 0.82 | 4.09a ± 0.98 | 4.94a ± 1.3 | 0.43 (Large) | <0.0001⁎⁎ |
| ITGA11 | 3.21a ± 0.36 | 2.92a ± 0.42 | 2.12b ± 0.34 | 2.18b ± 0.6 | 0.52 (Large) | <0.0001⁎⁎ |
| BTN3A2 | 3.21a,b ± 0.53 | 2.83a ± 0.39 | 3.39b ± 0.59 | 4.24c ± 0.62 | 0.53 (Large) | <0.0001⁎⁎ |
a, b,c: Different characters in each row show a statistically significant difference (p <0.05)
Kruskal-Wallis test
One-way analysis of variance.
Considering Table 3, except for CD28, FCRL3, JUN, CLEC7A, IL5, CD83, TPSAB1, ITM2A, DCBLD2 proteins, the difference in protein between groups in the immune response panel and IL12 was statistically significant ( p<0.05). Considering the effect size findings, the two proteins that most remarkably affect the COVID-19 severity and control groups in the immune response panel are ICA1 (0.82) and MGMT (0.79) proteins. Descriptive statistics of 92 proteins in the inflammation panel according to the COVID-19 positive and control group are given in Table 4 .
Table 4.
Descriptive statistics of proteins in the inflammation panel by groups.
| Protein Names | Groups |
Effect Size | p-value | |||
|---|---|---|---|---|---|---|
| Control | Mild | Severe | Critical | |||
| Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | |||
| IL8 | 5.46a(4.6-7.13) | 4.34b(3.29-6.57) | 6.11a(5.12-6.9) | 7.36c(5.96-9.01) | 0.64 (Large) | <0.0001* |
| CD8A | 9.28a(7.84-10.83) | 9.9b(8.75-11.66) | 9.58a,b(8.27-11.22) | 9.91b(8.68-11.79) | 0.09 (Large) | 0.011 |
| MCP-3 | 1.77a(1.03-4.31) | 1.66a(0.85-4.33) | 4.65b(2.25-6.67) | 5.87c(3.83-8.06) | 0.64 (Large) | <0.0001* |
| GDNF | 2a(1.09-2.7) | 2.03a,b(1.29-3.29) | 1.93a,b(1.61-3.23) | 2.29b(1.74-3.29) | 0.09 (Large) | 0.013 |
| CD244 | 5.95a(5.4-7.28) | 5.96a(5.08-6.63) | 5.55b(4.96-6.11) | 5.54b(5.06-6.26) | 0.23 (Large) | <0.0001* |
| IL7 | 3.77a(2.39-5.61) | 1.83b(1.25-3.46) | 2.62b,c(1.46-3.35) | 2.8c(1.34-4.35) | 0.52 (Large) | <0.0001* |
| uPA | 9.58a(9.12-10.47) | 9.74a(8.53-10.61) | 9.69a(9.34-10.45) | 10.12b(9.57-11.17) | 0.20 (Large) | <0.0001* |
| IL6 | 2.5a(1.42-4.27) | 1.96a(1.08-5.28) | 5.42b(4.58-8.32) | 7.42c(3.21-10.62) | 0.64 (Large) | <0.0001* |
| IL-17C | 2.12a(1.22-4.96) | 2.18a(1.16-4.03) | 2.53a,b(2.15-5.09) | 3.39b(2.02-5.6) | 0.34 (Large) | <0.0001* |
| IL-17A | 1.52a,b(0.75-2.93) | 1.38a(0.68-2.87) | 1.8b5(1.33-2.92) | 1.72a,b(0.91-2.56) | 0.08 (Large) | 0.015 |
| AXIN1 | 7.42a(5.71-10.85) | 2.39b(1.15-4.52) | 3.25b(1.86-3.55) | 2.75b(1.93-4.95) | 0.63 (Large) | <0.0001* |
| TRAIL | 7.54a(6.88-8.1) | 7.87a(6.99-8.82) | 7.18a,b(7.01-7.95) | 6.95b(6.03-8.27) | 0.30 (Large) | <0.0001* |
| IL-20RA | 0.87a(0.51-1.35) | 0.79a(0.25-3.01) | 1.02a(0.5-1.82) | 0.97a(0.54-2.47) | 0.07 (Large) | 0.031 |
| CXCL9 | 7.21a(5.64-10.71) | 7.02a(5.82-8.45) | 8.17b(7.96-10.67) | 8.9b(7.47-11.01) | 0.37 (Large) | <0.0001* |
| CST5 | 6.64a(5.83-8.33) | 6.33a(5.63-7.68) | 6.34a(5.54-7.04) | 6.08a(5.51-8.58) | 0.03 (Large) | 0.116 |
| IL-2RB | 0.71a(0.1-1.43) | 0.57b(-0.09-1.02) | 0.54a,b(0.25-0.87) | 0.71a(0.17-1.74) | 0.10 (Large) | 0.007 |
| IL-1 alpha | 0.02a(-1.34-1.75) | -0.74b(-1.21-3.14) | -0.58a,b(-1.12–0.18) | -0.84b(-1.14-0.2) | 0.23 (Large) | <0.0001* |
| IL2 | 0.7a(0.44-1.02) | 0.55a(0.24-1.08) | 0.94b(0.58-1.37) | 0.6a(0.22-1.08) | 0.17 (Large) | <0.0001* |
| TSLP | 1.11a(0.16-1.83) | 1a(0.47-2.66) | 0.95a(0.6-6.43) | 1.15a(0.19-4.12) | 0.28 (Small) | 0.931 |
| CCL4 | 6.09a(5.28-8.11) | 5.59b(4.09-6.85) | 6.44a(5.46-8.1) | 6.41a(5.01-8.26) | 0.19 (Large) | <0.0001* |
| IL18 | 9.34a,b(8.24-10.83) | 8.89a(7.75-10.45) | 9.88b,c(9.27-11.26) | 10.06c(9.17-14.12) | 0.41 (Large) | <0.0001* |
| TGF-alpha | 2.54a(2.09-3.29) | 2.96a(1.59-3.55) | 3.39b(2.91-4.94) | 4.19c(2.62-6.04) | 0.59 (Large) | <0.0001* |
| CCL11 | 7.59a(6.63-9.25) | 7.79a(5.7-8.72) | 7.17a(6.57-7.96) | 7.48a(6.68-8.57) | 0.06 (Large) | 0.046 |
| TNFSF14 | 6.15a(4.57-7.45) | 4.2b(2.91-5.41) | 6.08a(5.24-7.06) | 6.42a(4.37-7.22) | 0.52 (Large) | <0.0001* |
| FGF-23 | 2.18a(1.48-3.34) | 1.79a(0.83-4.18) | 2.05a(1.35-2.46) | 2.17a(1.08-8.45) | 0.01 (Moderate) | 0.285 |
| IL-10RA | 0.89a(0.55-2.49) | 0.82a(0.58-3.03) | 0.96a(0.7-2.44) | 1.01a(0.46-2.7) | 0.04 (Large) | 0.073 |
| FGF-5 | 1.03a(0.8-1.64) | 1.03a(0.6-1.39) | 1.16a(0.86-1.45) | 1.04a(0.56-1.6) | 0.008 (Small) | 0.517 |
| LIF-R | 4.09a(3.55-4.55) | 4.24a(3.05-4.94) | 4.33a,b(3.93-5.29) | 4.66b(4.13-5.1) | 0.35 (Large) | <0.0001* |
| CCL19 | 9.39(8.05-11.76)a | 9.41(8.16-11.8)a | 9.98(9.31-11.93)a | 11.15(9.87-12.56)b | 0.46 (Large) | <0.0001* |
| IL-15RA | 1.24a(0.89-1.87) | 1.36a(0.58-1.75) | 1.6a,b7(1.14-2.25) | 1.63b(1.02-3.15) | 0.14 (Large) | 0.002 |
| IL-22 RA1 | 1.66a(0.91-3.67) | 1.49a(0.65-2.43) | 1.73a(0.84-2.72) | 1.55a(0.61-2.44) | 0.01 (Moderate) | 0.284 |
| Beta-NGF | 0.02a(-0.75-0.64) | -0.12b(-0.41–0.01) | -0.11a,b(-0.3-0.17) | -0.08a,b(-0.27-0.12) | 0.25 (Large) | <0.0001* |
| CXCL5 | 13.2a9(11.96-13.91) | 10.16b(7.59-12.72) | 11.08b(7.34-12.58) | 10.31b(6.78-12.77) | 0.58 (Large) | <0.0001* |
| TRANCE | 4.55a(3.71-5.59) | 4.82a(3.2-6.49) | 2.99b(2.5-4.33) | 2.66b(1.46-5.15) | 0.54 (Large) | <0.0001* |
| HGF | 8.57a(7.71-10.01) | 8.11a(6.28-8.94) | 9.42b(8.85-10.62) | 10.19c(8.73-12.55) | 0.61 (Large) | <0.0001* |
| IL-24 | 0.92a(-0.07-2.11) | 0.74a(-0.1-2.63) | 1.61b(0.98-2.55) | 1.95b(0.46-4.28) | 0.31 (Large) | <0.0001* |
| IL13 | 0.71a(0.23-3.82) | 0.62a(0.03-1.73) | 0.52a(0.2-0.83) | 0.7a(0.24-4.28) | 0.03 (Large) | 0.116 |
| ARTN | 0.88a(0.23-1.44) | 0.84a,b(0.46-2.24) | 0.94a,b(0.32-1.21) | 1.07b(0.59-2.95) | 0.04 (Large) | 0.106 |
| IL10 | 3.29a(2.55-6.25) | 3.55a(2.74-7.07) | 4.93b(4.01-6.37) | 5.66b(4.09-7.13) | 0.53 (Large) | <0.0001* |
| TNF | 2.74a(1.71-4.27) | 2.82a(2-3.8) | 3.58b(3.25-4.62) | 4.05b(3.09-5.33) | 0.51 (Large) | <0.0001* |
| CCL23 | 10.33a(9.05-11.25) | 10.37a(9.43-10.99) | 11.47b(9.19-12.09) | 11.76b(9.92-12.68) | 0.45 (Large) | <0.0001* |
| CD5 | 5.4a(4.28-6.35) | 5.38a(4.46-6.37) | 5.28a(4.41-6.02) | 5.09a(4.58-6.22) | 0.005 (Small) | 0.469 |
| CXCL6 | 10.45a(8.98-11.75) | 7.45b(4.32-9.61) | 8.63b,c(7.1-9.48) | 8.38c(7.37-10.59) | 0.60 (Large) | <0.0001* |
| CXCL10 | 9.85a(8.61-12.55) | 9.64a(8.48-13.4) | 12.52b(11.58-13.64) | 13.49b(10.41-13.75) | 0.56 (Large) | <0.0001* |
| IL-20 | 0.62a(0.43-1.11) | 0.46b(-0.03-0.61) | 0.71a(0.45-1.02) | 0.55a(0.38-0.78) | 0.31 (Large) | <0.0001* |
| SIRT2 | 8.66a(7.03-9.76) | 3b(2.27-5.58) | 4.07c(3.18-5.74) | 4.48c(3.51-8.07) | 0.70 (Large) | <0.0001* |
| DNER | 8.74a(8.23-9.38) | 8.68a(7.51-9.04) | 8.14b(7.72-8.81) | 8.09b(7.54-8.57) | 0.43 (Large) | <0.0001* |
| EN-RAGE | 2.3a(1.56-4.9) | 2.3a(1.35-3.89) | 4.68b(3.13-5.89) | 5.11b(3.37-7.51) | 0.61 (Large) | <0.0001* |
| CD40 | 11.81a(10.92-13.1) | 10.95b(9.84-11.51) | 11.58a(10.98-12.45) | 11.9a(11.14-14.07) | 0.48 (Large) | <0.0001* |
| IL33 | 0.77a(0.49-1.16) | 0.62a(0.27-1.26) | 1.12b(0.69-1.56) | 1.13b(0.64-2.06) | 0.36 (Large) | <0.0001* |
| IFN-gamma | 6.89a(5.29-11.09) | 6.82a(5.42-11.77) | 8.88b(7.7-11.8) | 10.45b(6.12-13.42) | 0.43 (Large) | <0.0001* |
| IL4 | -0.1a(-1.21-2.78) | -0.41b(-1.56-0.44) | -0.15a,b(-0.8-0.24) | -0.09a,b(-0.83-1.13) | 0.06 (Large) | 0.041 |
| LIF | -0.02a(-0.39-1.8) | -0.12a(-0.42-0.28) | 0.26a,b(-0.09-0.64) | 0.51b(-0.13-2.75) | 0.42 (Large) | <0.0001* |
| NRTN | 0.55a(0.17-1.06) | 0.37b(0.16-0.7) | 0.58a,b(0.37-0.87) | 0.57a(0.2-1.33) | 0.12 (Large) | 0.003 |
| MCP-2 | 9.65a(7.47-11.47) | 9.21a(8.22-11.96) | 9.22a,b(8.44-12.73) | 10.84b(8.92-12.19) | 0.15 (Large) | 0.001 |
| CASP-8 | 6.7a(5.69-7.7) | 2.41b(1.62-4.24) | 3.36b,c(2.82-4.8) | 3.86c(2.89-7.28) | 0.70 (Large) | <0.0001* |
| CCL20 | 6.96a(6.39-12.02) | 7.47a(5.44-9.36) | 7.9a(7.1-10.35) | 9.82b(7.02-12.25) | 0.33 (Large) | <0.0001* |
| ST1A1 | 6.47a(5.55-7.18) | 1.21b(0.6-3.48) | 1.88b(0.76-2.65) | 1.58b(0.98-4.49) | 0.64 (Large) | <0.0001* |
| STAMBP | 8.21a(6.87-9.92) | 4.19b(3.29-5.83) | 4.82c(4.51-5.86) | 4.93c(3.77-7.12) | 0.66 (Large) | <0.0001* |
| IL5 | 0.77a(0.14-3.82) | 0.48a(0.16-7.15) | 0.98a(0.35-3.32) | 0.78a(0.22-3.9) | 0.09 (Large) | 0.011 |
| ADA | 6.08a(5.31-7.43) | 5.33b(4.59-6.36) | 5.84a,b(5.66-6.66) | 6.11a(5.09-7.5) | 0.21 (Large) | <0.0001* |
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |||
| VEGFA | 11.41a ± 0.59 | 10.77b ± 0.53 | 11.86a,c ± 0.27 | 12.11c ± 0.55 | 0.50 (Large) | <0.0001⁎⁎ |
| CDCP1 | 2.74a ± 0.69 | 2.69a ± 0.65 | 3.63b ± 0.35 | 4.31b ± 0.91 | 0.50 (Large) | <0.0001⁎⁎ |
| OPG | 10.14a,b ± 0.53 | 10.00a ± 0.38 | 10.54b ± 0.41 | 11.08c ± 0.57 | 0.46 (Large) | <0.0001⁎⁎ |
| LAP TGF-beta-1 | 7.59a ± 0.74 | 6.03b ± 0.5 | 6.67c ± 0.62 | 6.65c ± 0.48 | 0.53 (Large) | <0.0001⁎⁎ |
| MCP-1 | 11.28a ± 0.46 | 11.34a ± 0.71 | 12.16b ± 0.61 | 13.30c ± 0.92 | 0.62 (Large) | <0.0001⁎⁎ |
| CXCL11 | 9.19a ± 0.98 | 8.13b ± 0.98 | 10.38c ± 0.67 | 10.53c ± 0.63 | 0.57 (Large) | <0.0001⁎⁎ |
| OSM | 4.17a ± 0.77 | 4.06a ± 0.88 | 6.30b ± 1.05 | 6.38b ± 1.05 | 0.60 (Large) | <0.0001⁎⁎ |
| CXCL1 | 10.92a ± 0.57 | 8.70b ± 0.98 | 9.77c ± 0.91 | 9.89c ± 0.83 | 0.55 (Large) | <0.0001⁎⁎ |
| CD6 | 6.07a,b ± 0.64 | 6.35a ± 0.72 | 5.85a,b ± 0.73 | 5.69b ± 0.57 | 0.14 (Large) | 0.005⁎⁎ |
| SCF | 9.34a ± 0.35 | 9.26a ± 0.37 | 7.80b ± 0.92 | 8.05b ± 0.94 | 0.51 (Large) | <0.0001⁎⁎ |
| SLAMF1 | 2.37a ± 0.45 | 2.40a ± 0.31 | 3.06b ± 0.55 | 3.00b ± 0.59 | 0.31 (Large) | <0.0001⁎⁎ |
| MCP-4 | 15.62a ± 0.72 | 14.05b ± 0.76 | 14.28b ± 0.69 | 14.50b ± 0.8 | 0.44 (Large) | <0.0001⁎⁎ |
| MMP-1 | 15.08a ± 0.88 | 13.92b ± 0.99 | 15.33a ± 0.97 | 14.79a ± 0.87 | 0.25 (Large) | <0.0001⁎⁎ |
| FGF-21 | 5.30a ± 1.02 | 5.49a ± 1.24 | 5.55a ± 1.85 | 6.49a ± 2.44 | 0.08 (Large) | 0.325⁎⁎ |
| IL-10RB | 6.22a ± 0.31 | 6.07a ± 0.4 | 6.29a ± 0.31 | 6.28a ± 0.45 | 0.05 (Large) | 0.357⁎⁎ |
| IL-18R1 | 8.38a ± 0.41 | 8.39a ± 0.61 | 9.27b ± 0.53 | 9.75b ± 0.38 | 0.63 (Large) | <0.0001⁎⁎ |
| PD-L1 | 5.51a ± 0.5 | 5.48a ± 0.58 | 6.51b ± 0.38 | 6.93b ± 0.54 | 0.61 (Large) | <0.0001⁎⁎ |
| IL-12B | 6.50a ± 0.75 | 6.56a ± 0.64 | 6.74a ± 0.51 | 6.81a ± 0.72 | 0.04 (Large) | 0.405⁎⁎ |
| MMP-10 | 9.20a,b ± 0.68 | 9.03a ± 0.34 | 9.04a,b ± 0.65 | 9.61b ± 0.6 | 0.15 (Large) | 0.006⁎⁎ |
| CCL3 | 5.81a ± 0.64 | 5.38a ± 0.56 | 6.71b ± 0.72 | 7.24b ± 0.83 | 0.56 (Large) | <0.0001⁎⁎ |
| Flt3L | 9.31a ± 0.45 | 9.10a,b ± 0.49 | 8.77a,b ± 0.55 | 8.89b ± 0.68 | 0.11 (Large) | 0.030⁎⁎ |
| 4E-BP1 | 10.45a ± 0.88 | 7.91b ± 0.96 | 9.25c ± 0.98 | 9.07c ± 0.78 | 0.57 (Large) | <0.0001⁎⁎ |
| CCL28 | 2.65a ± 0.65 | 2.36a,b ± 0.53 | 2.38a,b ± 0.43 | 2.21b ± 0.46 | 0.10 (Large) | 0.093⁎⁎ |
| FGF-19 | 8.79a ± 0.8 | 8.63a ± 0.91 | 7.55b ± 1.13 | 8.42a,b ± 0.99 | 0.14 (Large) | 0.023⁎⁎ |
| CCL25 | 6.22a ± 0.62 | 6.26a ± 0.62 | 6.10a ± 0.67 | 6.43a ± 0.43 | 0.03 (Large) | 0.451⁎⁎ |
| CX3CL1 | 4.10a ± 0.4 | 4.10a ± 0.55 | 4.46a,b ± 0.72 | 4.91b ± 0.55 | 0.32 (Large) | <0.0001⁎⁎ |
| TNFRSF9 | 6.77a ± 0.55 | 6.64a ± 0.39 | 6.50a ± 0.47 | 6.65a ± 0.88 | 0.02 (Large) | 0.806⁎⁎ |
| NT-3 | 2.61a ± 0.45 | 2.04b ± 0.5 | 1.92b ± 0.5 | 1.77b ± 0.51 | 0.34 (Large) | <0.0001⁎⁎ |
| TWEAK | 9.10a ± 0.37 | 8.77b ± 0.44 | 8.24c ± 0.24 | 8.08c ± 0.51 | 0.51 (Large) | <0.0001⁎⁎ |
| TNFB | 4.57a,b ± 0.55 | 4.89a ± 0.37 | 4.37b ± 0.32 | 4.35b,c ± 0.49 | 0.19 (Large) | 0.001⁎⁎ |
| CSF-1 | 10.13a ± 0.21 | 10.14a ± 0.37 | 10.67b ± 0.18 | 10.78b ± 0.14 | 0.59 (Large) | <0.0001⁎⁎ |
a, b,c: Different characters in each row show a statistically significant difference (p <0.05)
Kruskal-Wallis test
One-way analysis of variance.
When Table 4 is examined, except for CST5, TSLP, FGF-23, IL-10RA, FGF-5, IL-22 RA1, IL13, ARTN, CD5, FGF-21, IL-10RB, IL-12B, CCL28, CCL25, and TNFRSF9 proteins, the difference in protein between the groups in the inflammation panel, and IL12 is statistically significant (p <0.05). When the effect size values are examined, the two proteins that most markedly affect the COVID-19 severity and control groups in the inflammation panel are SIRT2 (0.70) and CASP-8 (0.70) proteins. Finally, descriptive statistics of 92 proteins in the neurology panel according to the COVID-19 positive and control group are given in Table 5 .
Table 5.
Descriptive statistics of proteins in the neurology panel by groups.
| Protein Names | Groups |
Effect Size | p-value | |||
|---|---|---|---|---|---|---|
| Control | Mild | Severe | Critical | |||
| Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | Median (Min-Max) | |||
| NMNAT1 | 3.01a(1.85-7.05) | 2.87a(1.7-5.38) | 3.81a,b(2.05-4.72) | 4.5b(3.86-8.47) | 0.34 (Large) | <0.0001* |
| NRP2 | 8.23a,b(7.94-8.44) | 8.21a(7.93-8.36) | 8.26a,b(7.91-8.33) | 8.33b(7.93-8.54) | 0.09 (Large) | 0.012* |
| MAPT | 0.09a,b(-0.49-0.88) | 0.19a(-0.6-0.73) | 0.29b(-0.25-0.77) | 0.09b,c(-0.44-1.33) | 0.16 (Large) | 0.001* |
| CADM3 | 3.66a(2.57-4.31) | 3.26a(1.72-3.89) | 2.62a(2.45-4.36) | 2.92a(2.07-5.46) | 0.12 (Large) | 0.004* |
| GDNF | 1.92a(1.09-2.55) | 1.88a,b(1.38-3.24) | 2.09a,b(1.4-3.03) | 2.17b(1.6-3.23) | 0.07 (Large) | 0.022* |
| UNC5C | 4.62a(3.91-5.3) | 4.39a(2.9-4.95) | 4.44a(3.78-5.15) | 4.61a(3.91-5.59) | 0.02 (Large) | 0.193* |
| VWC2 | 5.55a(4.57-7.29) | 5.41a(4.41-6.5) | 5.88a(5.13-6.47) | 5.46a(4.17-6.6) | 0.02 (Large) | 0.193* |
| Siglec-9 | 4.92a(4.36-5.48) | 4.86a(4.37-5.42) | 5.06a,b(4.45-5.4) | 5.29b(4.28-5.94) | 0.16 (Large) | 0.001* |
| CLM-6 | 5.86a(5.34-6.61) | 5.87a(5.23-6.57) | 6.1a,b(5.8-6.82) | 6.42b(5.9-7.63) | 0.31 (Large) | <0.0001* |
| NBL1 | 5.07a(4.72-5.3) | 4.88b(4.44-5.03) | 4.93a,b(4.67-5.14) | 4.92b(4.72-5.17) | 0.22 (Large) | <0.0001* |
| EFNA4 | 2.9a(2.43-3.61) | 2.8b(1.99-3.3) | 3.04a,b(2.78-3.83) | 3.38b(2.61-5.99) | 0.29 (Large) | <0.0001* |
| SCARB2 | 4.5a(3.94-5.32) | 4.63a,b(3.77-6.02) | 5.14b,c(4.53-7.29) | 5.72c(4.55-7.97) | 0.43 (Large) | <0.0001* |
| ROBO2 | 5.58a(5.1-6.53) | 5.7a(3.99-6.18) | 4.58b(4.16-4.95) | 4.6b(3.88-5.55) | 0.54 (Large) | <0.0001* |
| CRTAM | 5.04a(4.32-7.05) | 5.47a(4.48-6.52) | 5.23a(4.67-6.57) | 5.5a(4.01-6.43) | 0.03 (Large) | 0.131* |
| RGMA | 10.96a(10.34-11.45) | 11a(9.68-11.39) | 10.4b(9.71-10.67) | 10.14b(9.28-11.16) | 0.40 (Large) | <0.0001* |
| MSR1 | 6.55a(4.91-7.61) | 6.62a(5.31-7.54) | 6.72a,b(6.35-7.6) | 7.08b(6.14-7.75) | 0.18 (Large) | <0.0001* |
| Alpha-2-MRAP | 9.75a(8.34-10.68) | 7.9b(6.8-8.4) | 8.18b,c(7.95-8.95) | 8.74c(8.07-10.48) | 0.63 (Large) | <0.0001* |
| sFRP-3 | 5.51a,b(4.74-6.01) | 5.33a(4.24-5.8) | 5.41a,b(4.9-6.19) | 5.93b(3.93-6.33) | 0.16 (Large) | 0.001* |
| EPHB6 | 4.04a(3.35-4.69) | 3.85a(2.43-4.43) | 3.57a(3.31-4.4) | 3.81a(3.17-4.93) | 0.06 (Large) | 0.041* |
| CNTN5 | 4.9a(4.23-5.8) | 4.74a(3.6-5.67) | 3.88b(2.78-4.62) | 3.44b(2.98-4.95) | 0.52 (Large) | <0.0001* |
| MATN3 | 8.94a(7.91-10.34) | 8.84a(7.56-9.82) | 9.66b(8.95-13.73) | 10.58b(9.67-12.18) | 0.59 (Large) | <0.0001* |
| RSPO1 | 2.79a(2.34-4.02) | 2.51b(1.49-3.24) | 2.98a,b,c(2.31-4.76) | 3.31c(2.42-5.11) | 0.30 (Large) | <0.0001* |
| GAL-8 | 8.6a(6.87-9.84) | 5.49b(4.7-6.53) | 6.38c(5.65-7.04) | 6.6c(6.12-7.86) | 0.76 (Large) | <0.0001* |
| LAYN | 5.45a(4.53-6.3) | 5.22a(4.17-5.94) | 5.32a,b(4.49-6.48) | 5.65b(5.09-8.93) | 0.09 (Large) | 0.010* |
| NEP | 2.17a(1.5-4.21) | 2.31a(1.31-3.92) | 3.26a(1.39-5.65) | 3.1a(1.27-5.3) | 0.04 (Large) | 0.097* |
| THY 1 | 10.06a(9.49-10.53) | 9.86b(8.63-10.29) | 9.65b(8.96-10.25) | 9.86a,b(9.09-10.88) | 0.16 (Large) | 0.001* |
| TMPRSS5 | 2.79a(1.93-3.6) | 2.7a(2.1-3.57) | 2.2b(1.32-2.52) | 2.19b(1.6-3.02) | 0.35 (Large) | <0.0001* |
| GM-CSF-R-alpha | 5.66a(3.3-6.84) | 5.84a(3.96-6.71) | 4.96a(4.52-6.05) | 5.91a(4.12-6.32) | 0.01 (Moderate) | 0.309* |
| Beta-NGF | 1.2a(0.86-2.64) | 1.17a(0.8-1.86) | 1.41a,b(1.12-2.36) | 1.69b(1.27-2.66) | 0.44 (Large) | <0.0001* |
| CD200 | 6.41a(5.67-7.28) | 6.5a(5.19-6.84) | 5.8b(4.89-6.64) | 6.03a,b(5.21-7.05) | 0.15 (Large) | 0.001* |
| G-CSF | 3.01a(2.14-4.17) | 2.71a(1.86-4.74) | 2.62a(1.81-4.94) | 3.74b(1.52-6.1) | 0.26 (Large) | <0.0001* |
| DRAXIN | 3.16a(2.5-4.67) | 3.24a(1.84-4.45) | 3.09a(2.63-4.44) | 4.02b(2.73-6.97) | 0.10 (Large) | 0.008* |
| PVR | 8.14a(7.31-8.61) | 8.17a(6.6-9.09) | 8.77b(8.1-9.26) | 9.07b(8.56-9.47) | 0.56 (Large) | <0.0001* |
| TNFRSF12A | 4.97a(4.11-5.91) | 4.82a(3.12-6.03) | 4.71a(4.51-6.2) | 5.67b(4.34-7.51) | 0.25 (Large) | <0.0001* |
| SKR3 | 6.9a(6.37-7.6) | 6.79a(5.96-7.44) | 6.98a,b(6.49-8.16) | 7.24b(6.48-9.92) | 0.14 (Large) | 0.001* |
| FLRT2 | 2.35a(1.76-2.94) | 2.34a,b(1.44-2.68) | 2.08b(1.86-2.18) | 2.32a,b(1.65-3.03) | 0.10 (Large) | 0.007* |
| MDGA1 | 5.04a(4.23-6.04) | 3.39b(1.98-5.03) | 3.82b(2.3-4.37) | 4.3c(3.37-5.87) | 0.19 (Large) | <0.0001* |
| CDH6 | 4.78a(3.93-5.23) | 4.61b(3.48-5.02) | 3.73c(3.09-4.51) | 3.89c(2.57-5.11) | 0.44 (Large) | <0.0001* |
| DDR1 | 7.36a(6.93-7.81) | 7.28a(6.3-7.65) | 6.9b(6.17-7.35) | 7.26a(6.61-7.9) | 0.12 (Large) | 0.003* |
| JAM-B | 8.06a(7.53-8.75) | 7.73a,b(6.79-8.38) | 7.27b(6.97-8.07) | 7.51b,c(6.68-9.81) | 0.20 (Large) | <0.0001* |
| NAAA | 3.58a(2.66-4.46) | 3.25a(2.79-4.02) | 3.05a(1.89-4.87) | 3.07a(1.92-5.3) | 0.10 (Large) | 0.006* |
| N2DL-2 | 3.06a(2.31-4.19) | 2.89a(1.98-3.38) | 3.51a(2.86-4.23) | 4.17b(2.94-7.95) | 0.42 (Large) | <0.0001* |
| PLXNB1 | 1.99a(1.43-2.65) | 1.6a(1.23-4.49) | 2.01a,b(1.78-2.74) | 2.62(1.79-3.65) | 0.38 (Large) | <0.0001* |
| Dkk-4 | 3.7a(2.57-5.05) | 3.35a(2.24-4.32) | 3.42a(2.98-4.55) | 3.39a(2.72-5.88) | 0.01 (Moderate) | 0.304* |
| EDA2R | 4.7a,b(0.79-6.12) | 4.23a(2.84-4.79) | 4.7a,b(3.88-5.74) | 5.13b(3.55-7.91) | 0.15 (Large) | 0.001* |
| LAT | 10.04a(8.89-10.56) | 5.03b(3.31-7.04) | 6.4c(5.33-7.52) | 6.47c(4.11-8.3) | 0.71 (Large) | <0.0001* |
| NTRK3 | 7.54a(6.81-8.27) | 7.36a(6.22-7.72) | 6.93b(6.38-7.08) | 6.66b(5.18-7.55) | 0.49 (Large) | <0.0001* |
| LAIR-2 | 5.05a(0.66-8.4) | 4.77a(3.28-7.55) | 5.1a(4.27-8.78) | 5.37a(4.38-8.87) | 0.01 (Moderate) | 0.320* |
| Nr-CAM | 9.66a(9.26-10.05) | 9.57b(8.75-9.83) | 9.21b(8.96-9.52) | 9.42b(9.13-9.8) | 0.23 (Large) | <0.0001* |
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |||
| EZR | 3.92a ± 0.3 | 3.75a ± 0.37 | 4.45b ± 0.35 | 4.84c ± 0.37 | 0.64 (Large) | <0.0001⁎⁎ |
| SMOC2 | 7.96a ± 0.43 | 7.69a,b ± 0.53 | 7.40b ± 0.46 | 7.56a,b ± 0.66 | 0.12 (Large) | 0.006⁎⁎ |
| NCAN | 8.51a ± 0.33 | 8.60a ± 0.31 | 8.28a ± 0.45 | 8.48a ± 0.41 | 0.06 (Large) | 0.147⁎⁎ |
| PRTG | 6.63a ± 0.24 | 6.55a ± 0.35 | 6.20b ± 0.16 | 6.31b ± 0.31 | 0.23 (Large) | <0.0001⁎⁎ |
| PLXNB3 | 5.49a ± 0.47 | 3.91b ± 0.52 | 3.81b ± 0.19 | 3.99b ± 0.42 | 0.73 (Large) | <0.0001⁎⁎ |
| CPA2 | 10.18a ± 0.56 | 9.97a ± 0.83 | 9.05b ± 0.61 | 9.03b ± 0.93 | 0.32 (Large) | <0.0001⁎⁎ |
| CD38 | 5.57a ± 0.56 | 5.76a,b ± 0.41 | 6.19b,c ± 0.44 | 6.66c ± 0.54 | 0.46 (Large) | <0.0001⁎⁎ |
| SMPD1 | 4.39a ± 0.43 | 4.58a ± 0.39 | 5.04b ± 0.54 | 5.27b ± 0.35 | 0.46 (Large) | <0.0001⁎⁎ |
| RGMB | 6.02a ± 0.33 | 5.84a,c ± 0.4 | 5.41b ± 0.42 | 5.65b,c ± 0.43 | 0.21 (Large) | <0.0001⁎⁎ |
| SIGLEC1 | 5.81a ± 0.51 | 6.25a,b ± 0.84 | 6.89b,c ± 0.76 | 7.34c ± 0.41 | 0.51 (Large) | <0.0001⁎⁎ |
| ADAM 22 | 4.48a ± 0.41 | 4.27a ± 0.43 | 3.67b ± 0.37 | 3.85b ± 0.62 | 0.29 (Large) | <0.0001⁎⁎ |
| CLEC1B | 12.34a ± 0.55 | 9.52b ± 0.78 | 10.92c ± 0.8 | 11.19c ± 0.9 | 0.69 (Large) | <0.0001⁎⁎ |
| ADAM 23 | 4.26a ± 0.45 | 3.97a,b ± 0.7 | 3.59b ± 0.53 | 3.54b,c ± 0.53 | 0.23 (Large) | <0.0001⁎⁎ |
| HAGH | 7.84a ± 0.61 | 5.48b ± 1.04 | 6.56c ± 1.31 | 6.11b,c ± 1.24 | 0.49 (Large) | <0.0001⁎⁎ |
| LXN | 2.79a ± 0.5 | 1.63b ± 0.18 | 1.84b ± 0.4 | 1.75b ± 0.34 | 0.66 (Large) | <0.0001⁎⁎ |
| BCAN | 4.50a ± 0.42 | 4.22a ± 0.40 | 3.69b ± 0.34 | 3.71b ± 0.59 | 0.36 (Large) | <0.0001⁎⁎ |
| GDF-8 | 3.66a ± 0.56 | 3.64a ± 0.79 | 2.41b ± 0.34 | 2.53b ± 1.07 | 0.35 (Large) | <0.0001⁎⁎ |
| WFIKKN1 | 3.52a ± 0.36 | 3.69a ± 0.47 | 3.07b ± 0.18 | 3.10b ± 0.52 | 0.27 (Large) | <0.0001⁎⁎ |
| CDH3 | 7.19a ± 0.27 | 7.29a ± 0.37 | 7.06a ± 0.43 | 7.36a ± 0.57 | 0.05 (Large) | 0.217⁎⁎ |
| GFR-alpha-1 | 6.79a ± 0.36 | 6.95a ± 0.46 | 7.78b ± 0.54 | 8.18b ± 0.59 | 0.62 (Large) | <0.0001⁎⁎ |
| SCARA5 | 8.72a ± 0.3 | 8.52a,b ± 0.38 | 8.20b ± 0.34 | 8.35b,c ± 0.38 | 0.21 (Large) | <0.0001⁎⁎ |
| NTRK2 | 6.64a ± 0.22 | 6.32b ± 0.33 | 5.76c ± 0.27 | 5.66c ± 0.44 | 0.62 (Large) | <0.0001⁎⁎ |
| GZMA | 5.85a ± 0.44 | 6.28b ± 0.46 | 6.51b,c ± 0.46 | 6.71c ± 0.47 | 0.37 (Large) | <0.0001⁎⁎ |
| SCARF2 | 6.39a ± 0.49 | 6.04b ± 0.43 | 5.63b ± 0.56 | 5.74b ± 0.48 | 0.28 (Large) | <0.0001⁎⁎ |
| GDNFR-alpha-3 | 5.14a ± 0.35 | 4.96a ± 0.43 | 4.10b ± 0.31 | 4.29b ± 0.51 | 0.49 (Large) | <0.0001⁎⁎ |
| CPM | 6.80a ± 0.19 | 6.66a ± 0.21 | 6.14b ± 0.32 | 6.15b ± 0.46 | 0.48 (Large) | <0.0001⁎⁎ |
| CLEC10A | 5.20a ± 0.36 | 5.14a,b ± 0.7 | 4.58b,c ± 0.5 | 4.47c ± 0.56 | 0.28 (Large) | <0.0001⁎⁎ |
| GCP5 | 5.26a ± 0.73 | 4.74b ± 0.63 | 4.12b ± 0.75 | 4.23b ± 0.57 | 0.32 (Large) | <0.0001⁎⁎ |
| BMP-4 | 4.33a ± 0.53 | 4.99b ± 0.62 | 4.60a,b ± 0.5 | 4.18a ± 0.57 | 0.26 (Large) | <0.0001⁎⁎ |
| FcRL2 | 4.89a ± 0.54 | 5.01a,b ± 0.56 | 5.11a,b ± 0.48 | 5.45b ± 0.65 | 0.14 (Large) | 0.009⁎⁎ |
| IL-5R-alpha | 4.02a ± 0.77 | 3.70a ± 0.55 | 5.15b ± 0.7 | 5.14b ± 0.94 | 0.42 (Large) | <0.0001⁎⁎ |
| PDGF-R-alpha | 5.25a ± 0.31 | 5.28a ± 0.36 | 5.42a,b ± 0.46 | 5.77b ± 0.39 | 0.28 (Large) | <0.0001⁎⁎ |
| CTSC | 5.03a ± 0.41 | 3.59b ± 0.74 | 3.65b ± 0.63 | 4.33c ± 0.6 | 0.52 (Large) | <0.0001⁎⁎ |
| CTSS | 5.61a ± 0.24 | 5.62a ± 0.24 | 5.95b ± 0.4 | 6.22b ± 0.23 | 0.53 (Large) | <0.0001⁎⁎ |
| N-CDase | 3.72a ± 0.61 | 3.61a,b ± 0.69 | 3.00b ± 0.48 | 3.31a,b ± 0.82 | 0.11 (Large) | 0.014⁎⁎ |
| TNFRSF21 | 8.28a ± 0.24 | 8.22a ± 0.29 | 8.01a ± 0.34 | 8.10a ± 0.32 | 0.09 (Large) | 0.054⁎⁎ |
| CLM-1 | 6.08a ± 0.98 | 6.28a,b ± 0.82 | 7.19b,c ± 0.63 | 7.56c ± 0.84 | 0.36 (Large) | <0.0001⁎⁎ |
| SPOCK1 | 2.38a ± 0.27 | 2.42a ± 0.27 | 2.55a,b ± 0.33 | 2.68b ± 0.35 | 0.16 (Large) | 0.009⁎⁎ |
| IL12 | 8.30a ± 0.77 | 8.35a ± 0.69 | 8.54a ± 0.55 | 8.53a ± 0.69 | 0.02 (Large) | 0.579⁎⁎ |
| MANF | 8.98a ± 0.07 | 6.85b ± 1.14 | 7.36b ± 0.95 | 7.50b ± 0.98 | 0.52 (Large) | <0.0001⁎⁎ |
| TN-R | 4.27a ± 0.36 | 4.36a ± 0.55 | 4.00a,b ± 0.38 | 3.86b ± 0.53 | 0.17 (Large) | 0.003⁎⁎ |
| CD200R1 | 4.28a,b ± 0.33 | 4.37a ± 0.33 | 4.04a,b ± 0.35 | 4.11b ± 0.32 | 0.13 (Large) | 0.008⁎⁎ |
| KYNU | 9.04a ± 0.74 | 8.35b ± 0.6 | 9.37a,c ± 0.47 | 9.62c ± 0.58 | 0.39 (Large) | <0.0001⁎⁎ |
a, b, c: Different characters in each row show a statistically significant difference (p <0.05)
Kruskal-Wallis test
One-way analysis of variance.
According to Table 5, the difference between groups in terms of protein in the neurology panel except for proteins UNC5C, VWC2, CRTAM, NEP, GM-CSF-R-alpha, Dkk-4, LAIR-2, NCAN, CDH3, TNFRSF21, and IL12 is statistically significant (p<0.05). As the effect sizes are appreciated, the two proteins that most markedly affect the COVID-19 severity and control groups in the neurology panel are PLXNB3 (0.73) and GAL-8 (0.70).
3.2. Results of Preprocessing data
In the data set, a total of 7 missing values were determined in other variables except for gender and age variables. As a result of the assignment made using the Fully Conditional Specification method instead of the determined missing values, the missing values in the data set were completed. The class imbalance problem is solved using the "Sample (balance)" operator on the data set that does not contain any missing value. As a result, the data set became balanced, with 33 people in each group. As a result of the feature selection with the Recursive Feature Elimination method to increase the model performance, the number of variables in the data set decreased to 138 proteins.
3.3. Artificial intelligence models and Performance Evaluation
In this study, artificial intelligence models (deep learning (DL), Random Forest (RF), Gradient Boosted Trees (GBTs) are constructed to classify three COVID-19 positive patient groups (mild, severe, and critical) and a control group based on the blood protein profiling. The hyperparameters for the models are tuned by the Evolutionary optimization method, which uses an evolutionary approach. As a result, the hyperparameters for the deep learning model were optimized as 1.0e-8 for epsilon, 0.99 for rho, 0.0 for L1, 0.17 for L2, and 10.0 for maxw2. Hyperparameters for the random forest model were tuned as 35.0 for maximal depth, 77.0 for minimal leaf size, 55.0 for minimal size for the split, and 28.0 for the number of pre pruning alternatives. Finally, the hyperparameters for the GBTs model have been optimized as 62.0 for maximal depth, 0.94 for learning rate, 20.0 for the number of bins, and 1.0e-5 for min split improvement.
According to the model performance metric results in Table 6 , the GBTs classification algorithm gave the most successful result. The accuracy rate based on Random Forest (96.21%) was more successful than the accuracy rate based on classic machine learning (DL 94.73%). In kappa statistic, which measures the reliability of the statistical fit, the GBTs, RF, and DL approaches represent a perfect fit with the values of 0.96, 0.95, and 0.93.
Table 6.
Performance metrics of the models.
| Models | Groups | Sensitivity (%) | Specificity (%) | Precision (%) | F1 score (%) | Accuracy (%) | Class. Error | Kappa |
|---|---|---|---|---|---|---|---|---|
| Gradient Boosted Trees (GBTs) | Mild | 96.97 | 97.98 | 94.12 | 95.52 | 96.98 | 3.02 | 0.96 |
| Severe | 1.00 | 98.99 | 97.06 | 98.51 | ||||
| Critical | 90.91 | 98.99 | 96.77 | 93.75 | ||||
| Control | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Random Forest (RF) | Mild | 96.97 | 98.99 | 96.97 | 96.97 | 96.21 | 3.79 | 0.95 |
| Severe | 1.00 | 97.98 | 94.29 | 97.06 | ||||
| Critical | 87.88 | 98.99 | 96.67 | 92.06 | ||||
| Control | 1.00 | 98.99 | 97.06 | 98.51 | ||||
| Deep Learning (DL) | Mild | 96.97 | 96.97 | 91.43 | 94.12 | 94.73 | 5.27 | 0.93 |
| Severe | 1.00 | 97.98 | 94.29 | 97.06 | ||||
| Critical | 87.88 | 97.98 | 93.55 | 90.62 | ||||
| Control | 93.94 | 1.00 | 1.00 | 96.88 |
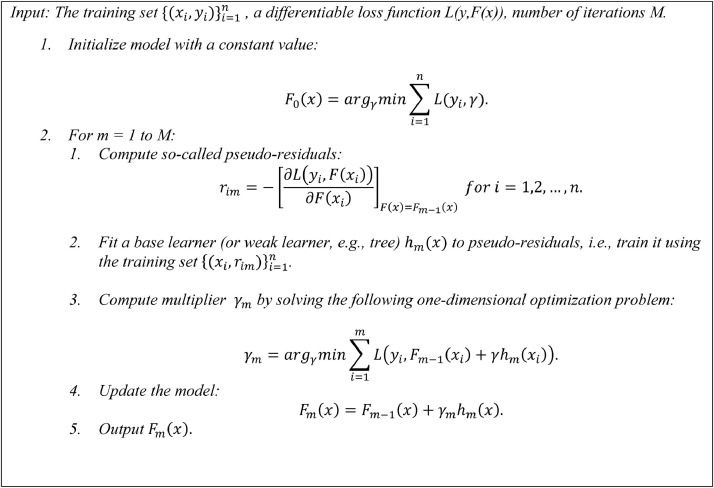
Fig. 1 displays the pseudo-codes of the GBTs algorithm, which produces the best prediction in classifying the severity of COVID19 disease based on the proteomics data.
Fig. 2.
The graphic of variable importance values of the top ten proteins for the GBTs algorithms.
Fig. 1.
The pseudo-codes of the GBTs algorithm.
Input: The training set , a differentiable loss function L(y,F(x)), number of iterations M.
-
1Initialize model with a constant value:
-
2
For m = 1 to M:
-
3Compute so-called pseudo-residuals:
-
4
Fit a base learner (or weak learner, e.g., tree) hm(x) to pseudo-residuals, i.e., train it using the training set.
-
5Compute multiplier γm by solving the following one-dimensional optimization problem:
-
6Update the model:
-
7
Output Fm(x).
Figure 1. The pseudo-codes of the GBTs algorithm
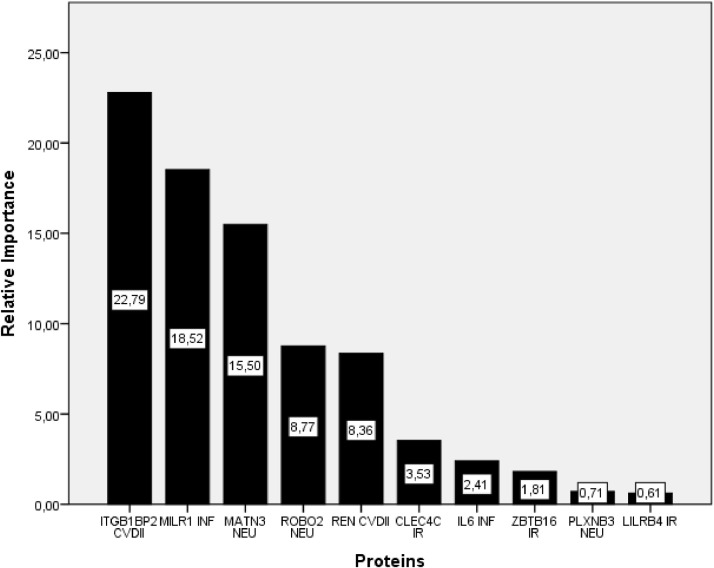
Table 7 and Figure 1 depict the importance levels of the top ten proteins in COVID-19 positive and control individuals on the severity of the disease in the GBTs modeling.
Table 7.
Variable importance values of the top ten proteins for the GBTs algorithms.
| Protein Names | Relative Importance | Percentage |
|---|---|---|
| ITGB1BP2 CVDII | 22.79 | 0.26 |
| MILR1 INF | 18.52 | 0.21 |
| MATN3 NEU | 15.50 | 0.18 |
| ROBO2 NEU | 8.77 | 0.10 |
| REN CVDII | 8.36 | 0.09 |
| CLEC4C IR | 3.53 | 0.04 |
| IL6 INF | 2.41 | 0.03 |
| ZBTB16 IR | 1.81 | 0.02 |
| PLXNB3 NEU | 0.71 | 0.01 |
| LILRB4 IR | 0.61 | 0.01 |
CVDII: Cardiovascular II Panel; IR: Immune Response Panel; INF: Inflammation Panel; NEU: Neurology Panel
ITGB1BP2 CVDII (22.79-0,26%), MILR1 INF (18,52-0,21%), MATN3 NEU (15,50-0,18%) and ROBO2 NEU (8,77-0,10%) provided the highest importance, while the lowest importance values were for PLXNB3 NEU (0,71-0,01%) and LILRB4 IR (0,61-0,01%) from the GBTs technique.
4. Discussion
COVID-19 is widespread, with high morbidity and high mortality in chronically ill patients all over the world. According to recent studies, older patients are more likely to become infected with COVID-19, particularly those with underlying diseases. The seriousness of cases puts great pressure on the deficiency of services for intensive care. Unfortunately, basic clinical characteristics of COVID-19 at distinct serious stages remain unknown to date. Several attempts are being made to develop automated systems to enable the early identification of the disease by medical experts based on medical images or -omics technologies. Prediction models that combine factors or characteristics to predict the likelihood of people being infected are helping clinicians cope with the outbreak of COVID-19 [27].
During the severe acute respiratory syndrome-new coronavirus-2 pandemic, the insufficiency of laboratory diagnostic tools and the take a long time led clinicians to more rapid diagnosis methods. Although COVID-19 can be effectively diagnosed at an early stage by the approaches based on proteomic analysis, the detection of serious COVID-19 patients before the manifestation of severe symptoms to reduce mortality is equally important. In this study, three positive (mild, extreme, and critical) COVID-19 patient groups and a control group may be separated based on deep learning and multiple machine learning models (i.e., Random Forest, Gradient Boosted Tree) related to blood protein profiling. According to the experimental results from the current study, it can be concluded that the models based on blood proteins generate promising prediction results in classifying COVID-19 (mild/severe/critical) severity levels and the control group. When the prediction results of the algorithms are compared according to the performance metrics (i.e., accuracy, sensitivity, specificity, precision, F1, kappa, and classification error), the GBTs algorithm slightly outperforms deep learning and random forest techniques on the classification problem under question. The ten top proteins, ITGB1BP2, MILR1, MATN3, ROBO2, REN, CLEC4C, IL6, ZBTB16, PLXNB3, and LILRB4, calculated from the best performing GBTs algorithm, can be used as biomarkers in the COVID-19 severity classification. A similar paper has been reported that six proteins (IL6, CKAP4, Gal-9, IL-1ra, LILRB4, and PD-L1) are associated with the severity of the COVID-19 disease, and complex variations in blood proteins associated with the severity of the disease may be used as early biomarkers to screen the severity of the disease in COVID-19 and act as future therapeutic targets [8]. On the other hand, when the effect sizes for all proteins are examined, the five proteins with the highest values are DECR-1 (0.85), ICA1 (0.82), IL-6 (0.79), MGMT (0.79), and PLXNB3 (0.73), respectively. Besides, the findings of the proposed GBTs model indicate that the two proteins (i.e., IL6 and LILRB4) are significantly related to COVID-19 severity, as reported by the previous work [8].
Different studies on the proteomics profiling of the COVID-19 pandemic have been reported to identify the differences in proteins. A novel paper [28] has performed RNA-seq and high-resolution mass spectrometry on 128 blood samples from COVID-19-positive and COVID-19-negative patients with various disease severities and outcomes and mapped 219 molecular features with high significance for the status and severity of COVID-19. Finally, the related study presents a web-based platform to be interactively explored and demonstrated through a machine learning approach (ExtraTrees classifier) to COVID-19 severity prediction [28]. Another research has profiled host responses to COVID-19 by studying plasma proteomics in a population of patients with COVID-19, including non-survivors and survivors emerging from moderate or extreme symptoms, and revealed numerous plasma protein alterations associated with COVID-19. To classify 11 proteins as biomarkers and a range of biomarker combinations validated by an independent cohort and precisely differentiated and projected COVID-19 outcomes, we developed a pipeline based on machine-learning (penalized logistic regression) [29]. A recent study has been described the development of a proteomic risk score (PRS) based on 20 blood proteomic biomarkers linked to progression to severe COVID-19 and established that using a machine learning model (Light Gradient Boosting Machine), a core group of gut microbiota could reliably predict the blood proteomic biomarkers of COVID-19 [30]. The study conducted by Gomila et al. [31] used matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI TOF MS) to analyze the mass spectra profiles of the sera from 80 COVID-19 patients, clinically classified as mild (33), severe (26), critical (21), and 20 healthy controls and they found a clear variability of the serum peptidome profile depending on COVID-19 severity. The two support vector machines discrete severe (severe and critical) and non-severe (mild) patients with 90% precision in the study of the resulting matrix of peak intensity and estimated correctly the non-negative outcome of the severe patients in 85% of the cases and the negative in 38% of the cases. Yet, the current work additionally encapsulates the use of a deep learning approach in the proteomics analysis of the COVID-19 severity, which is an important difference from the studies given earlier.
To sum up, the proposed model (Gradient Boosted Tree) achieved the best prediction of disease severity based on the proteins compared to the other algorithms. The results point out that changes in blood proteins associated with the severity of the disease may be used in monitoring the severity of COVID-19 disease and in early diagnosis and treatment.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study does not require ethical approval because the open-source data set is used. Also, there is no conflict of interest among the authors. Any institution or organization did not financially support this research.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cmpb.2021.105996.
Appendix. Supplementary materials
Reference
- 1.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation. Computers in Biology and Medicine. 2020;126 doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization W.H. World Health Organization; 2020. Coronavirus Disease 2019 (COVID-19) [PubMed] [Google Scholar]
- 3.Legg K., McKiernan H., Reisdorph N. Molecular Diagnostics, Elsevier; 2017. Application of Proteomics to Medical Diagnostics; pp. 233–248. [Google Scholar]
- 4.Hastie T., Tibshirani R., Friedman J. Springer Science & Business Media; 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- 5.Müller A.C., Guido S. Introduction to machine learning with Python: a guide for data scientists. " O'Reilly Media, Inc. 2016 [Google Scholar]
- 6.Goodfellow I., Bengio Y., Courville A., Bengio Y. MIT press Cambridge; 2016. Deep learning. [Google Scholar]
- 7.Bengio Y., LeCun Y. Scaling learning algorithms towards AI. Large-scale kernel machines. 2007;34:1–41. [Google Scholar]
- 8.Patel H., Ashton N.J., Dobson R.J., Anderson L.-m., Yilmaz A., Blennow K., Gisslen M., Zetterberg H. Proteomic blood profiling in mild, severe and critical COVID-19 patients. medRxiv. 2020 doi: 10.1038/s41598-021-85877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown M.L., Kros J.F. Data mining and the impact of missing data. Industrial Management & Data Systems. 2003 [Google Scholar]
- 10.Van Buuren S., Brand J.P., Groothuis-Oudshoorn C.G., Rubin D.B. Fully conditional specification in multivariate imputation. Journal of statistical computation and simulation. 2006;76:1049–1064. [Google Scholar]
- 11.Akthar F., Hahne C. RapidMiner 5, Operator Reference. www. rapid-i. com. 2012 [Google Scholar]
- 12.Guyon I., Weston J., Barnhill S., Vapnik V. Gene selection for cancer classification using support vector machines. Machine learning. 2002;46:389–422. [Google Scholar]
- 13.Svetnik V., Liaw A., Tong C., Wang T. International Workshop on Multiple Classifier Systems. Springer; 2004. Application of Breiman's random forest to modeling structure-activity relationships of pharmaceutical molecules; pp. 334–343. [Google Scholar]
- 14.Jiang H., Deng Y., Chen H.-S., Tao L., Sha Q., Chen J., Tsai C.-J., Zhang S. Joint analysis of two microarray gene-expression data sets to select lung adenocarcinoma marker genes. BMC bioinformatics. 2004;5:81. doi: 10.1186/1471-2105-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregorutti B., Michel B., Saint-Pierre P. Correlation and variable importance in random forests. Statistics and Computing. 2017;27:659–678. [Google Scholar]
- 16.LeCun Y., Bengio Y., Hinton G. Deep learning. nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 17.Biau G., Scornet E. A random forest guided tour. Test. 2016;25:197–227. [Google Scholar]
- 18.Friedman J.H. 1999 Reitz Lecture. The Annals of Statistics. 2001;29:1189–1232. [Google Scholar]
- 19.Friedman J., Hastie T., Tibshirani R. Additive logistic regression: a statistical view of boosting (with discussion and a rejoinder by the authors) The annals of statistics. 2000;28:337–407. [Google Scholar]
- 20.Freund Y., Schapire R.E. A decision-theoretic generalization of on-line learning and an application to boosting. Journal of computer and system sciences. 1997;55:119–139. [Google Scholar]
- 21.Krauss C., Do X.A., Huck N. Deep neural networks, gradient-boosted trees, random forests: Statistical arbitrage on the S&P 500. European Journal of Operational Research. 2017;259:689–702. [Google Scholar]
- 22.Sullivan G.M., Feinn R. Using effect size—or why the P value is not enough. Journal of graduate medical education. 2012;4:279–282. doi: 10.4300/JGME-D-12-00156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen J. Academic press; 2013. Statistical power analysis for the behavioral sciences. [Google Scholar]
- 24.Yaşar Ş., Arslan A., Colak C., Yoloğlu S. A Developed Interactive Web Application for Statistical Analysis: Statistical Analysis Software. Middle Black Sea Journal of Health Science. 2020;6:227–239. [Google Scholar]
- 25.Verzani J. Getting started with RStudio. " O'Reilly Media, Inc.". 2011 [Google Scholar]
- 26.Hofmann M., Klinkenberg R. CRC Press; 2016. RapidMiner: Data mining use cases and business analytics applications. [Google Scholar]
- 27.López-Úbeda P., Díaz-Galiano M.C., Martín-Noguerol T., Luna A., Ureña-López L.A., Martín-Valdivia M.T. COVID-19 detection in radiological text reports integrating entity recognition. Computers in Biology and Medicine. 2020;127 doi: 10.1016/j.compbiomed.2020.104066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.K.A. Overmyer, E. Shishkova, I.J. Miller, J. Balnis, M.N. Bernstein, T.M. Peters-Clarke, J.G. Meyer, Q. Quan, L.K. Muehlbauer, E.A.J.C.s. Trujillo, Large-scale Multi-omic Analysis of COVID-19 Severity, (2020). [DOI] [PMC free article] [PubMed]
- 29.Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., Zou X., Yang Q., Yuan Y., Bie Y. Immunity; 2020. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.W. Gou, Y. Fu, L. Yue, G.-d. Chen, X. Cai, M. Shuai, F. Xu, X. Yi, H. Chen, Y. Zhu, Gut Microbiota May Underlie the Predisposition of Healthy Individuals to COVID-19-Sensitive Proteomic Biomarkers, (2020).
- 31.Gomila R.M., Martorell G., Fraile-Ribot P.A., Domenech-Sanchez A., Oliver A., Garcia-Gasalla M., Alberti S. Rapid classification and prediction of COVID-19 severity by MALDI-TOF mass spectrometry analysis of serum peptidome. medRxiv. 2020 doi: 10.1093/ofid/ofab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.