Abstract
Objective
Electronic health records (EHR) are increasingly being recognized as a major source of data reusable for medical research and quality monitoring, although patient identification and assessment of symptoms (characterization) remain challenging, especially in complex diseases such as systemic lupus erythematosus (SLE). Current coding systems are unable to assess information recorded in the physician’s free‐text notes. This study shows that text mining can be used as a reliable alternative.
Methods
In a multidisciplinary research team of data scientists and medical experts, a text mining algorithm on 4607 patient records was developed to assess the diagnosis of 14 different immune‐mediated inflammatory diseases and the presence of 18 different symptoms in the EHR. The text mining algorithm included key words in the EHR, while mining the context for exclusion phrases. The accuracy of the text mining algorithm was assessed by manually checking the EHR of 100 random patients suspected of having SLE for diagnoses and symptoms and comparing the outcome with the outcome of the text mining algorithm.
Results
After evaluation of 100 patient records, the text mining algorithm had a sensitivity of 96.4% and a specificity of 93.3% in assessing the presence of SLE. The algorithm detected potentially life‐threatening symptoms (nephritis, pleuritis) with good sensitivity (80%‐82%) and high specificity (97%‐97%).
Conclusion
We present a text mining algorithm that can accurately identify and characterize patients with SLE using routinely collected data from the EHR. Our study shows that using text mining, data from the EHR can be reused in research and quality control.
INTRODUCTION
Electronic health records (EHR) are increasingly being recognized as a major source of data reusable for medical research and quality monitoring. In the EHR, clinical information for the patient’s diagnostic and therapeutic trajectory is collected. This includes logistic information, such as time and date of appointments, as well as results from laboratory tests, lists of medication, and, arguably most important, the physicians’ notes of the patients’ visits. Some of the data are recorded by using the structured International Classification of Diseases (ICD) codes, yet these are too rigid to reflect clinically relevant subtleties, are often unreliable, and have large interobserver variability (1, 2, 3). However, crucial information about the diagnosis and symptoms is often recorded in the physician’s unstructured free‐text notes.
For physicians, the main advantage of using free text is that it offers the opportunity for nuance and expression (4). This nuance and expression is important in diseases with different phenotypes and courses of disease, such as autoimmune diseases. For research and quality control, however, accumulation of relevant information in free text in the EHR presents a challenge. Manual extraction of information from free text is time consuming and is therefore not practical in studies with large amounts of patients. In addition, manual extraction is prone to errors and limits reproducibility (5, 6). Moreover, a new or altered research question will result in having to review all data again. Because of this, other methods of retrieving these data are currently being developed.
Text mining methods are increasingly being used to collect information from free text. With text mining, it is possible to scrutinize free text for key words or phrases regarding variables of interest, which can be captured in a structured way. However, recent studies show that identification of patients with a certain diagnosis improves when free text is also used, as opposed to coding systems alone (7, 8). Text mining can also be used to characterize patients by evaluating the presence of risk factors or multiple manifestations of disease, whereas coding systems are mainly used for diagnoses and a small selection of symptoms (9, 10, 11). This can be especially helpful in studying the heterogeneous clinical course of patients with immune‐mediated inflammatory diseases (IMIDs).
One IMID in which this characterization is important is systemic lupus erythematosus (SLE). SLE is a systemic IMID with a large but varied set of associated symptoms, ranging from relatively mild dermatological manifestations to possibly life‐threatening nephritis. SLE can be difficult to diagnose because it shares multiple symptoms and often overlaps with other IMIDs, such as antiphospholipid syndrome (APS). Serological examination can be used to aid in the diagnostic process. Autoantibodies against double‐stranded DNA (anti‐dsDNA) are highly specific for SLE and are only examined in practice whenever there is clinical suspicion of SLE because they are not associated with any other disease, although it is known that they are also present in a small percentage of the healthy population (12).
Reliable identification of the patients is a necessity for clinical research and quality control when reusing data from the EHR. Furthermore, for diseases in which there is large heterogeneity in the clinical course, patient characterization can be of great added value. In this study, we present a rule‐based text mining algorithm capable of reliably assessing the diagnosis and clinical manifestations of a cohort of patients suspected of having SLE.
PATIENTS AND METHODS
Study population
This study was performed in the University Medical Center Utrecht (UMC Utrecht), a tertiary care center in the Netherlands. To ensure a balanced cohort of patients with and without the diagnosis of SLE, all patients who were tested for anti‐dsDNA between February 2014 and July 2017 were included in a cohort, irrespective of the result of the test. Anti‐dsDNA testing is an important component of the diagnostic process for all patients who are suspected of having SLE and is also part of the annual laboratory follow‐up for all patients with SLE, according to the hospital protocol. The date of each anti‐dsDNA test in this cohort was regarded as an individual time point for which the diagnosis and symptoms were assessed in this study. For some patients, samples were collected at multiple time points. Informed consent was not collected in this study; the requirement for obtaining informed consent was waived by the Biobank Research Ethics Committee because of the large number of patients, many of whom are no longer managed at UMC Utrecht.
EHRs
UMC Utrecht uses ChipSoft HiX as its electronic health record system. Within this software system, all relevant medical data can be accessed, reviewed, and recorded by the physician. This includes, among other things, laboratory data, medication data, physician’s notes by all of the physicians treating the patient in the hospital (including physicians from different departments), and letters to the patient’s general practitioner (GP) (which were also used in the current study). Medication data and laboratory data are recorded in a structured format and are therefore easily usable in research. Although the physician’s notes and letters to the GP usually follow a certain format, they consist mostly of free text and are therefore considered unstructured.
Because drawing of blood did not always happen on the same day as the visit to the clinician, clinical data for up to 14 days after sample collection were used. Data were extracted from the EHR through the Utrecht Patient Oriented Database (UPOD). The structure and function of UPOD is described elsewhere (13). In brief, UPOD is an infrastructure of relational databases comprising data on patient characteristics, hospital discharge diagnoses, medical procedures, medication orders, and laboratory tests for all patients treated at UMC Utrecht since 2004. This study was in accordance with the guidelines approved by the medical ethical committee and was approved by the biobank committee of UMC Utrecht. All collected data were pseudonymized for privacy reasons.
Text mining
To search the EHR for relevant data, a rule‐based text mining algorithm for the Dutch language was developed in R. The algorithm looks for specific key words that indicate that something (eg, a diagnosis) is present in the text. Key words were constructed for SLE as well as several other IMIDs that can cause symptoms similar to those of SLE and could therefore explain the anti‐dsDNA test requested by the physician. These key words were used to identify patients with a clinical diagnosis of an IMID, as recorded in the EHR, rather than to identify patients fulfilling classification criteria for a specific IMID.
Key words for diagnoses and symptoms
Key words were formulated for SLE as well as several other autoimmune diseases that can overlap with or mimic symptoms of SLE. The list of symptoms for which key words were made was based on the individual clinical components of the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) (14). Hematological and serological components of the SLEDAI can be extracted from laboratory results directly; therefore, there is no need to use text mining for these manifestations. Ambiguous terms or abbreviations that can indicate multiple things (eg, “DM” can indicate the diagnosis of either dermatomyositis or diabetes mellitus) were excluded from the list of key words. When seen in the context of the total report in the EHR, the true meaning of such an ambiguous term is usually relatively easy to interpret for a person but not for a text mining algorithm. The complete list of diagnoses and symptoms for which key words were formulated is available in Supplementary Tables S1a and S1b.
Key words were formulated on the basis of terms used in literature, terms used in clinical practice, and known synonyms using regular expressions. Results of the most recent version of the algorithm were compared to manual review in an iterative process. Discordances were noted and used to improve the algorithm. More than 100 patient records were manually reviewed in five iterations to improve performance. To facilitate incorporation of expert knowledge within the algorithm, the process of constructing key words was supervised by a specialized clinical immunologist (ML).
Context mining of negative and neutral records
In the EHR, key words can be present in either a positive, negative, or neutral context. In a positive context, the key words indicate that the disease or symptom is present and should be recorded as such. However, when mentioned in a negative context (eg, “diagnosis X is unlikely”), this instance should function as an indication that a diagnosis is less likely. Seventy variations of negative context were constructed. By using both the positive and negative instances of the key words, a calculation was made to assess the likelihood of actual presence (see below). This calculation does not include neutral records (eg, records mentioned in the differential diagnosis, because a diagnosis mentioned in the differential diagnosis is neither definitively present nor absent and therefore does not influence the likelihood of the definitive diagnosis).
Confidence
To assess the likeliness of the presence of each individual diagnosis or symptom, we calculated a confidence score:
Established diagnoses are often repeated in the EHR; therefore, the number of times that a disease that is present in the EHR is mentioned should be relatively high, resulting in a high level of confidence. New patients will have fewer records of their diagnoses, but because the total number of documents included is small as well, the resulting confidence level will still be high. To limit the number of false‐positives, negative records were assigned more weight than positive records because positive records might occasionally be found in the EHR of nonpatients, whereas negative records in the EHR of patients are scarce.
On the basis of iterative manual checking for diagnoses given by the text mining program during the development of the algorithm, we identified a cutoff of a confidence level of 14% as appropriate to limit the number of false‐positives and false‐negatives. If there was no single diagnosis with a confidence level of 14% or higher for a sample, this was defined as a “no diagnosis” category. Patients with more than one diagnosis with a confidence level of 14% or higher were recorded as having multiple diagnoses; for example, if there was a confidence level of 60% for both SLE and APS, this was recorded as the patient having both diagnoses.
For symptoms, a similar approach was used. However, unlike the diagnosis, the presence of symptoms can be different at each visit to the physician and are therefore mentioned less often in the total number of documents, resulting in lower confidence levels. This lower level of confidence mainly affects non–life‐threatening symptoms because critical symptoms, such as nephritis or pericarditis, are commonly repeated in the patient history if ever present. Similar to the diagnosis, negative records of symptoms lowered the confidence level and neutral records were ignored. With the goal of creating an overview of the symptoms in the patient’s history, we chose to set the level for inclusion of symptoms to a confidence level of 1%, meaning there had to be more positive records than negative records. This approach means that assigned symptoms should be interpreted as having been present at least once and not necessarily at the time of record.
Assessment of accuracy of the text mining algorithm
To assess the accuracy of the final version of the text mining algorithm, 100 patients from the cohort were randomly selected, independent of the records used to determine the cutoff for the level of confidence, and the diagnoses were manually gathered from the EHR by a clinical immunologist with expertise in the field of SLE (ML). The clinical immunologist was blinded for the outcome of the text mining algorithm during this process. The diagnoses reported by the clinical immunologist were used as the gold standard to compare to the diagnoses reported by the text mining algorithm and used to calculate sensitivity and specificity. A similar approach was used to determine the accuracy of the text mining algorithm for symptoms. Because of the low prevalence of several symptoms, we only analyzed the accuracy for symptoms with a prevalence of at least 10% in the 100 randomly selected patients.
RESULTS
Patients and demographics
Between 2014 and 2017, 2038 patients were tested for the presence of anti‐dsDNA a total of 4607 times. The mean age of all patients at the time of their first visit was 44.9 years, and 63.4% of all patients were female.
Diagnoses
The text mining algorithm assigned the diagnosis SLE to 484 patients (23.7%) at the first record, making it the most common diagnosis (Figure 1, Supplementary Table S2). One thousand two hundred two patients (59.0%) were assigned no diagnosis at the first record. Twenty‐six patients were assigned SLE after the first record, bringing the total number of patients with SLE during the entire follow‐up to 510 (25%). The diagnosis of SLE was assigned in 2726 records (59%).
FIGURE 1.
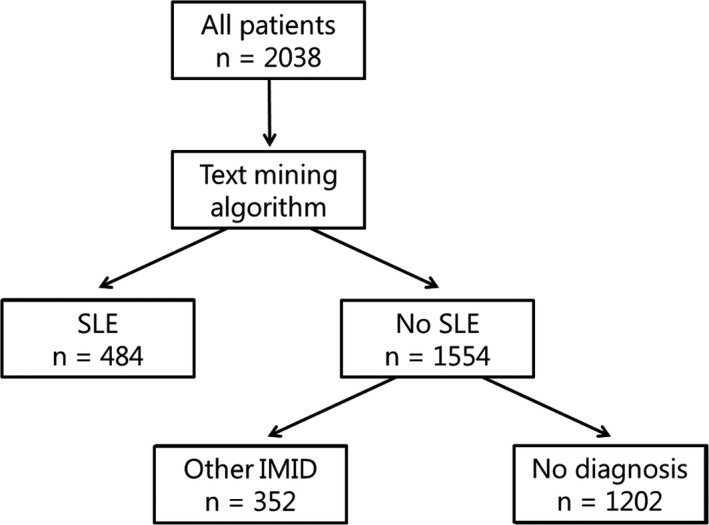
Flowchart of identification of patients with systemic lupus erythematosus (SLE) at first record. The first records of all 2038 patients in the cohort were evaluated by the text mining algorithm, which assigned the diagnosis SLE to 484 patients; 352 patients were assigned another immune‐mediated inflammatory disease (IMID): antiphospholipid syndrome, cutaneous lupus erythematosus, giant cell arteritis, granulomatosis with polyangiitis, idiopathic inflammatory myopathy, juvenile idiopathic arthritis, lupus‐like disease (also known as incomplete SLE), mixed connective tissue disease, polymyalgia rheumatica, primary Sjögren syndrome, rheumatoid arthritis, systemic sclerosis, or undifferentiated connective tissue disease.
Overlap of multiple diagnoses was common in our cohort. Overlap of APS and SLE was the most common (in 64 patients at the time of the first sample). Our algorithm reported both a diagnosis of cutaneous lupus erythematosus (CLE) and SLE in 59 patients. Lupus‐like disease (LLD) is also called incomplete SLE, and some patients with LLD will eventually develop SLE. A combination of both LLD and SLE was recorded in 28 patients.
Symptoms
The prevalence of all symptoms is presented in Table 1. Fatigue was the most common symptom in patients both with and without SLE. Nephritis was reported in 33.3% of all patients with SLE.
Table 1.
Prevalence of symptoms
| Feature | Prevalence, n (%) |
|---|---|
| Total number of patients | 2038 |
| Symptoms | |
| Alopecia | 292 (14.3) |
| Arthritis | 328 (16.1) |
| Arthralgia | 322 (15.8) |
| Cranial nerve disorder | 141 (6.9) |
| Digital ulceration | 14 (0.7) |
| Fatigue | 1030 (50.5) |
| Mucosal ulcers | 323 (15.6) |
| Myositis | 55 (2.7) |
| Nephritis | 203 (10.0) |
| Rash | 310 (15.2) |
| Pericarditis | 134 (6.6) |
| Pleurisy | 117 (5.7) |
| Psychosis | 59 (2.9) |
| Raynaud | 432 (21.2) |
| Seizure | 139 (6.8) |
| Stroke | 317 (15.6) |
| Vasculitis | 270 (13.2) |
| Visual disturbance | 20 (1.0) |
Prevalence of clinical symptoms in all patients at the time of the first record is presented. Because laboratory parameters can be assessed by using structured data, and no text mining is required to assess these data, these parameters are not shown. “Rash” includes photosensitivity and malar rash.
Performance of the algorithm
The results of the comparison between the outcome of the text mining algorithm and the judgement of the clinical immunologist are shown in Table 2. In 71 of the 100 randomly selected patients, the diagnoses of the text mining algorithm and of the clinical immunologist matched completely. In nine cases, the text mining algorithm recorded overlap of SLE with CLE and/or LLD, whereas the clinical immunologist recorded only SLE to be present. Seven patients were assigned another diagnosis overlapping with SLE by the text mining algorithm, for example, overlap of SLE with APS, whereas the clinical immunologist only reported the diagnosis SLE. The algorithm misdiagnosed five patients, whom were determined to have no diagnosis according to the clinical immunologist. Conversely, the algorithm missed a diagnosis in eight cases.
Table 2.
Performance of the text mining algorithm for diagnosis
| Outcome | n |
|---|---|
| Complete match | 71 |
| Algorithm reported overlapping diagnosis of LLD/CLE in context of SLE, whereas immunologist did not report overlap | 9 |
| Algorithm missed an overlapping diagnosis | 4 a |
| Algorithm assigned an extra overlapping diagnosis | 4 a |
| Diagnosis wrongly assigned by algorithm | 5 |
| Diagnosis missed by algorithm | 8 |
Comparison of 100 randomly selected patients: diagnosis assessed by text mining algorithm compared to assessment of a clinical immunologist.
Abbreviations: CLE, cutaneous lupus erythematosus; LLD, lupus‐like disease; SLE, systemic lupus erythematosus.
In one case, the text mining algorithm missed one overlapping diagnosis and misdiagnosed another overlapping diagnosis. This case is represented in both rows.
The clinical immunologist assigned SLE as a diagnosis to 55 of the 100 patients. Fifty‐three of these fifty‐five were correctly recorded by the text mining algorithm, and three false‐positives were recorded, resulting in a sensitivity and specificity of 96.4% and 93.3%, respectively, with a positive predictive value (PPV) of 94.6%. The algorithm correctly reported 22 of the 26 patients with no assigned diagnosis and mistakenly reported that eight patients did not have an IMID, resulting in a sensitivity and specificity of 84.6% and 89.2%, respectively, for identifying patients without an IMID diagnosis (Table 3).
Table 3.
Performance of the text mining algorithm for diagnosis of SLE
| Diagnosis | n (according to manual review) | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| SLE | 53 | 0.96 | 0.93 | 0.95 | 0.95 |
| APS | 11 | 0.72 | 1 | 1 | 0.97 |
| No diagnosis | 26 | 0.85 | 0.89 | 0.73 | 0.94 |
Sensitivity, specificity, PPV, and NPV for all diagnoses with a prevalence of >10% in the 100 randomly selected patients (n = 90). Ten patients had a diagnosis other than SLE or APS.
Abbreviations: APS, antiphospholipid syndrome; NPV, negative predictive value; PPV, positive predictive value; SLE, systemic lupus erythematosus.
Ten of the eighteen studied symptoms were present in at least 10% of the 100 patients. The sensitivity of the text mining algorithm was variable for different symptoms, ranging from poor (0.38 for mucosal ulcerations) to good (0.84 for fatigue). Specificity generally was very good, with a specificity of more than 0.90 for most symptoms (Table 4). The algorithm detected the possibly life‐threatening symptoms, pleuritis and nephritis, with good sensitivity and very high specificity. Two‐by‐two tables for all symptoms are available in Supplementary Table S3.
Table 4.
Performance of the text mining algorithm for symptoms
| Symptom | n (according to manual review) | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| Alopecia | 26 | 0.65 | 0.95 | 0.81 | 0.89 |
| Arthritis | 41 | 0.41 | 0.92 | 0.69 | 0.77 |
| Arthralgia | 45 | 0.56 | 0.93 | 0.86 | 0.72 |
| Fatigue | 67 | 0.84 | 0.64 | 0.82 | 0.66 |
| Mucosal ulcers | 21 | 0.38 | 0.81 | 0.35 | 0.83 |
| Nephritis | 39 | 0.82 | 0.97 | 0.94 | 0.89 |
| Rash | 49 | 0.51 | 0.90 | 0.66 | 0.83 |
| Pleuritis | 10 | 0.80 | 0.97 | 0.73 | 0.98 |
| Raynaud | 26 | 0.69 | 0.88 | 0.67 | 0.89 |
| Vasculitis | 17 | 0.53 | 0.94 | 0.64 | 0.91 |
Sensitivity, specificity, PPV, and NPV for all symptoms with a prevalence of >10% in the 100 randomly selected patients.
Abbreviations: NPV, negative predictive value; PPV, positive predictive value.
DISCUSSION
Patient identification is a vital part of research and quality control when reusing data from the EHR. Because the reliability of traditional coding systems is limited for these purposes, we investigated an alternative method of patient identification as well as patient characterization. In this study, we developed a rule‐based text mining algorithm to assess the presence of a diagnosis of autoimmune diseases in the EHR as well as to assess SLE‐related disease manifestations.
Our text mining algorithm has good accuracy for the diagnosis of SLE, as well as high sensitivity and specificity, when compared to the assessment of a medical specialist. Moreover, it outperforms other algorithms for identifying patients with SLE presented in previous studies. For example, Jorge et al (15) and Barnado et al (16) used algorithms processing a combination of structured parameters, such as number of corresponding ICD codes, positive laboratory test results, and use of medication (PPV 92%, sensitivity 47% and PPV 91%, sensitivity 40%, respectively). Jorge et al (15) also developed an algorithm that included both structured parameters and natural language processing, but this algorithm performed slightly worse than the algorithm using only structured parameters (PPV 90%, sensitivity 41%). Murray et al (17) used a combination of these structured parameters and also included the use of a single search term for lupus in the EHR notes but reached similar results to the studies that did not include this single free‐text search term (PPV 85 and 98%, sensitivity 97 and 84% ). A limitation of these machine learning models is that they have limited portability, restricting their widespread use, although recent improvements in this regard have been made (15). A possible explanation for why our algorithm outperforms these models is that our algorithm does not include ICD codes and does include context mining regarding text processing. Furthermore, our text mining algorithm does not consider our primary inclusion criterion, anti‐dsDNA, whereas these studies selected patients on the basis of SLE‐associated ICD codes, which were subsequently assessed by the corresponding algorithms, possibly introducing a bias.
It is noteworthy that our algorithm assesses the clinical diagnosis recorded in the EHR rather than the diagnosis according to classification criteria. Although classification criteria provide a point of reference to clinicians as to which patients should be given a clinical diagnosis, the main use of classification criteria is to define a homogeneous group of patients for research (18). In our study, we chose to focus on the clinical diagnosis.
Ideally we would have also compared the outcome of the algorithm to the recorded ICD coding. Unfortunately in the Netherlands, the ICD coding is not recorded directly into the EHR, but rather it is a derivative of the financial coding system, called DBC (a diagnosis‐treatment combination), which uses different coding from the ICD coding. For registration purposes, an ICD code is later linked to all different DBC codes. This indirect coding introduces two translational steps with loss of information and chances for mistakes. Furthermore, these coding systems are not always suited for specific diagnoses (eg, CLE is often coded as inflammatory dermatosis, which covers several other diseases as well). We compared the outcome of the 100 manually reviewed patients to the recorded DBC code. For SLE, this resulted in similar percentages of sensitivity and specificity; however, for most other diseases, the DBC coding was often inaccurate or incomplete in cases of overlapping diagnoses. For example, a DBC code that could be related to the diagnosis APS was only recorded in 3 of 11 patients in the 100 patients reviewed.
To our knowledge, only one other study investigated assessment of symptoms by text mining in SLE. This study by Gianfrancesco et al (19) focused solely on lupus nephritis. Our study shows that patient characterization and assessment of a wide range of symptoms is also possible with the use of text mining in a heterogeneous disease such as SLE. The sensitivity of our algorithm varied for different symptoms, which implies that the key words used for some symptoms could be improved. However, sensitivity for possible life‐threatening symptoms was good, possibly because of more frequent repetition in the medical history in the EHR. Our algorithm performed slightly better than a text mining algorithm used by Gianfrancesco et al (19) (sensitivity 79%, specificity 86%) to detect lupus nephritis in the EHR. It should be noted that the gold standard used to compare to a text mining algorithm, manual review of EHR records, is prone to human errors. For example, in several cases in which there were discrepancies between the outcomes produced by the algorithm and manual review, further investigation revealed that a symptom was overlooked by manual review and the algorithm was correct. Yet for this article, only the initial assessment by the clinical immunologist was used for analysis.
Because of the low prevalence of certain symptoms, we were not able to adequately test the performance of the algorithm for these symptoms, which therefore were excluded from analysis. We were also not able to reliably assess some clinical features that usually are recorded only once in the physician’s notes. Smoking status, family history, and use of alcohol and drugs are usually only extensively discussed and recorded during the first visit of the patient to the physician. Because they are only mentioned in one visit, this results in low levels of confidence and also requires that the first visit of the patient be included in the data supplied to the text mining program. Improvements in identifying smokers with the use of text mining recently have been made in our hospital, although this method was not included in this study (20).
With increasingly smart and flexible text mining programs being developed, we predict that the use of clinical data from the EHR in clinical research and quality control will increase in the future. In computer science, the phrase “garbage in, garbage out” is often used to indicate the necessity of quality input data to produce a quality outcome. Although administration is often seen as a burden by health care professionals, their role in this process is vital. Researchers and health care professionals should work together to find a way to record accurate data that are reusable in research, while limiting the associated burden of administration on health care professionals (21).
Text mining algorithms have great flexibility and adaptability, allowing for use in different research projects. Although the text mining algorithm presented in this study is a relatively simple and transparent rule‐based algorithm and can still be improved on, it can relatively easily be adapted to better suit research questions from other cohort studies, in contrast to machine learning algorithms. Further research is needed to evaluate the portability of our algorithm to other hospitals because this would, for example, pave the way for a national registry for rare diseases with routinely collected clinical data. Moreover, we plan to investigate whether the performance of the algorithm can be improved when our method is combined with machine learning methods using more than only free‐text data, similar to those used in previous studies (15, 16, 17).
There are several hurdles to be overcome before an algorithm like this can be implemented on a large scale. It is likely that the algorithm would have to be amended for use in hospitals using another EHR program because the structure of the necessary data might be different, which would likely also influence the level of confidence used for a cutoff. Furthermore, our algorithm was developed in a Dutch‐speaking environment; language‐specific adaptations are required for implementation in other countries.
Our study presents a method to assess the diagnosis and clinical manifestations of patients with SLE recorded in the EHR for large groups of patients with great accuracy. Our algorithm can both be used for patient identification and characterization of patients with SLE. Our study demonstrates that text mining can be used for performing large‐scale research and quality control with clinical data from the EHR in heterogeneous diseases such as SLE.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Brunekreef had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Otten, van Laar, Limper.
Acquisition of data
Brunekreef, van den Bosch, Hoefer, Limper, Haitjema.
Analysis and interpretation of data
Brunekreef, Otten, van den Bosch, Hoefer, van Laar, Limper, Haitjema.
ROLE OF THE STUDY SPONSOR
Thermo Fisher Scientific had no role in the study design or in the collection, analysis, or interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript for publication. Publication of this article was not contingent upon approval by Thermo Fisher Scientific.
Supporting information
Table S1‐S2
Table S3
Supported by Thermo Fisher Scientific.
No potential conflicts of interest relevant to this article were reported.
References
- 1. Stausberg J, Lehmann N, Kaczmarek D, Stein M. Reliability of diagnoses coding with ICD‐10. Int J Med Inform 2008;77:50–7. [DOI] [PubMed] [Google Scholar]
- 2. Moores KG, Sathe NA. A systematic review of validated methods for identifying systemic lupus erythematosus (SLE) using administrative or claims data. Vaccine 2013;31 Suppl 10:K62–73. [DOI] [PubMed] [Google Scholar]
- 3. Otsa K, Talli S, Harding P, Parsik E, Esko M, Teepere A, et al. Administrative database as a source for assessment of systemic lupus erythematosus prevalence: Estonian experience. BMC Rheumatol 2019;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lovis C, Baud RH, Planche P. Power of expression in the electronic patient record: structured data or narrative text? [Comparative Study]. Int J Med Inform 2000;58–59:101–10. [DOI] [PubMed] [Google Scholar]
- 5. Klein DO, Rennenberg R, Gans R, Enting R, Koopmans R, Prins MH. Limited external reproducibility restricts the use of medical record review for benchmarking. BMJ Open Qual 2019;8:e000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanskamp‐Sebregts M, Zegers M, Vincent C, van Gurp PJ, de Vet HC, Wollersheim H. Measurement of patient safety: a systematic review of the reliability and validity of adverse event detection with record review. BMJ Open 2016;6:e011078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ford E, Carroll JA, Smith HE, Scott D, Cassell JA. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc 2016;23:1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ford E, Nicholson A, Koeling R, Tate A, Carroll J, Axelrod A, et al. Optimising the use of electronic health records to estimate the incidence of rheumatoid arthritis in primary care: what information is hidden in free text? [Original research paper]. BMC Med Res Methodol 2013;13:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jonnalagadda SR, Adupa AK, Garg RP, Corona‐Cox J, Shah SJ. Text mining of the electronic health record: an information extraction approach for automated identification and aubphenotyping of HFpEF patients for clinical trials. J Cardiovasc Transl Res 2017;10:313–21. [DOI] [PubMed] [Google Scholar]
- 10. Jonnagaddala J, Liaw ST, Ray P, Kumar M, Chang NW, Dai HJ. Coronary artery disease risk assessment from unstructured electronic health records using text mining. J Biomed Inform 2015;58 Suppl:S203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karystianis G, Nevado AJ, Kim CH, Dehghan A, Keane JA, Nenadic G. Automatic mining of symptom severity from psychiatric evaluation notes. Int J Methods Psychiatr Res 2018;27:e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kavanaugh AF, Solomon DH. Guidelines for immunologic laboratory testing in the rheumatic diseases: anti‐DNA antibody tests. Arthritis Rheum 2002;47:546–55. [DOI] [PubMed] [Google Scholar]
- 13. Ten Berg MJ, Huisman A, van den Bemt PM, Schobben AF, Egberts AC, van Solinge WW. Linking laboratory and medication data: new opportunities for pharmacoepidemiological research. Clin Chem Lab Med 2007;45:13–9. [DOI] [PubMed] [Google Scholar]
- 14. Gladman DD, Ibañez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol 2002;29:288–91. [PubMed] [Google Scholar]
- 15. Jorge A, Castro VM, Barnado A, Gainer V, Hong C, Cai T, et al. Identifying lupus patients in electronic health records: development and validation of machine learning algorithms and application of rule‐based algorithms. Semin Arthritis Rheum 2019;49:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barnado A, Casey C, Carroll RJ, Wheless L, Denny JC, Crofford LJ. Developing electronic health record algorithms that accurately identify patients with systemic lupus erythematosus. Arthritis Care Res (Hoboken) 2017;69:687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murray SG, Avati A, Schmajuk G, Yazdany J. Automated and flexible identification of complex disease: building a model for systemic lupus erythematosus using noisy labeling. J Am Med Inform Assoc 2019;26:61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bertsias GK, Pamfil C, Fanouriakis A, Boumpas DT. Diagnostic criteria for systemic lupus erythematosus: has the time come? [Review]. Nat Rev Rheumatol 2013;9:687–94. [DOI] [PubMed] [Google Scholar]
- 19. Gianfrancesco MA, Tamang S, Schmajuk G, Yazdany J. Application of text mining methods to identify lupus nephritis from electronic health records [abstract]. Lupus Sci Med 2019;6 Suppl 1:A142. [Google Scholar]
- 20. Groenhof TK, Koers LR, Blasse E, de Groot M, Grobbee DE, Bots ML, et al. Data mining information from electronic health recordsproduced high yield and accuracy for current smoking status. J Clin Epidemiol 2020;118:100–6. [DOI] [PubMed] [Google Scholar]
- 21. Bezemer T, de Groot MC, Blasse E, ten Berg MJ, Kappen TH, Bredenoord AL, et al. A human(e) factor in clinical decision support systems. J Med Internet Res 2019;21:e11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S2
Table S3


