Abstract
Background:
Although previous experiments have implicated sphingosine-1-phosphate (S1P) as a links between immune reactions and cancer progression, the exact mechanism of this interaction has not comprehensively studied in clinical human samples. This study sought to evaluate the S1P regulation by sphingosine kinase 1 (SPHK1), an S1P-producing enzyme, in the immunity/immuno-reactivity of clinical human breast cancer surgical specimens.
Methods:
S1P levels were examined in tumor, peri-tumoral and normal human breast samples by mass spectrometry. Genomics Data Commons data portal of The Cancer Genome Atlas cohort was used to assess the expression of S1P-related and immune-related genes.
Results:
S1P levels were significantly higher in tumor samples compared to peri-tumoral (P < 0.05) or normal human breast samples (P < 0.001). SPHK1 gene expression was elevated in tumoral samples compared to normal breast samples (P < 0.01). Furthermore, the elevated expression of SPHK1 in breast cancer tissue was associated with an increased expression of the different kinds of immune-related genes, such as CD68, CD163, CD4, and FOXP3 (forkhead box P3), in HER2-negative breast cancer. Network analysis showed the central role of SPHK1 in the interaction of S1P signaling and expression of immune cell-related proteins.
Conclusion:
We demonstrated that S1P is mainly produced by tumor tissue, rather than peri-tumoral tissue, in breast cancer patients. Our data revealed the involvement of S1P signaling in the regulation of immune-related genes, suggesting the links between S1P and complicated immune-cancer interactions in breast cancer patients.
Keywords: Breast cancer, Immune cell-related gene, Tumor immune microenvironment, Sphingosine kinase 1, Sphingolipid, Sphingosine-1-phosphate
Introduction
Breast cancer accounts for the top oncologic diagnosis and, after lung cancer, the top oncologic risk for mortality in U.S. women.1 Therefore, focusing on the biological cellular mechanisms of interaction between breast cancer tissue and its stromal microenvironment has become a critical area of interest for investigational efforts to augment patient long-term survivorship. Stromal cells that surround a tumor are increasingly recognized as being important for cancer progression due to their ability to establish a tumor microenvironment (TME). TME components of relevance for cancer progression include cancer cells, vessels (lymphatic, arterial and venous), lymphoid and other immune-active cells, signaling compounds and other molecular factors, as well as interstitial fluid.2–4 Due to its role in cancer progression, TME has now been recognized as one of the important strategic targets for cancer treatment.5,6
Sphingosine-1-phosphate (S1P) is an important bioactive lipid mediator which is actively involved in the sequential molecular and cellular milestones of cancer progression, invasion and metastasis.4,7–11 S1P is generated intracellularly by the phosphorylation of sphingosine by two producing enzymes, sphingosine kinase 1 and 2 (SPHK1 and SPHK2), and transporters then transport S1P into the extracellular space. The mechanism by which extracellular S1P exerts its influence on other cells is via a family of five G protein-coupled receptors (S1PR1–5) in an autocrine, and/or paracrine manner, coined “inside-out” signaling of S1P.10–16 S1P bioactive concentration regulation takes place via an equilibrium between phosphorylation enzymatic activity versus S1P specific phosphorylase/S1P specific lyase activity.7,16 Although an increasing number of non-human experiments have revealed the relevance of S1P to cancer progression, there are only limited patient clinical data reported.17 Elevated transcription of SPHK1 was previously reported in clinical human breast cancer samples which correlated with a worse clinical outcomes and lack of a clinical response to cytotoxic systemic therapy.18–20 Moreover, we have recently showed that elevated concentrations of S1P in cancer samples correlated with lymphatic nodal metastatic disease in the clinical setting.21
S1P is also involved in the immune response.11,16 Differential S1P levels in different organs is a factor for S1PR1-dependent T cell emigration out of secondary lymphoid organs to the systemic circulation, both lymphatic and hematologic.22,23 This lymphocytic phenomenon in both T cells and B cells was found in association with S1PR1 and in the endothelium with protein spinster homolog 2 (SPNS2).23 Although previous in vivo studies indicated that S1P links immune reactions and cancer progression, the exact mechanism of this immune-oncologic interaction has not been fully elucidated in human samples.
Here, we determined the concentrations of the bioactive sphingolipids, sphingosine (Sph), S1P, dihydro-Sph (DHSph), and dihydro-S1P (DHS1P), in surgical specimens from patient breast cancer using highly sensitive liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). We also analyzed gene expression of S1P-related enzymes using the The Cancer Genome Atlas (TCGA) cohort. Furthermore, we utilized the TCGA database and investigated the association between S1P-related genes and immune-related gene expression.
Material and Methods
Patient tissue samples
From November 2015 to February 2016, 37 clinical samples from breast cancer surgical specimens at Niigata University Medical and Dental Hospital. The Institutional Review Board of Niigata University approved this study and the methods. After obtaining informed consent, breast specimens were collected from 20 breast cancer cases with malignant tumors greater than 1.5 cm in size. Breast cancer, peri-tumoral, and normal breast samples were collected immediately after surgery and preserved in liquid nitrogen. Peri-tumoral samples were defined as being within 1 cm from the gross tumoral edge. Normal breast samples were from a breast quadrant distinct from the malignancy, free of neoplasm. All samples were stored at −80°C.
Quantification of Sphingolipids by LC-ESI-MS/MS
Sphingolipid quantification was obtained by sample lipid extracts undergoing quantification by LC-ESI-MS/MS (4000 QTRAP, ABI) at the Virginia Commonwealth University Lipidomics Core as described previously.13,21,24–26 Internal standards were purchased from Avanti Polar Lipids (Alabaster, AL), and 500 pmol added to each sample in 20 µl of an ethanol:methanol:water (7:2:1) mixture. The HPLC-grade solvents were obtained from VWR (West Chester, PA).
Gene expression analysis of S1P-related enzymes using the TCGA cohort
S1P gene expression enzyme-encoding was evaluated by retrieving RNA sequencing and gene expression quantification data from breast tumor tissue (n = 1047) and paired normal breast tissue (n = 112) using the Genomics Data Commons data portal. Gene expression levels were derived using normalization methods provided in the DESeq2 package27 and log2 transformed. The differences in expression of S1P-related genes between normal and their paired cancer tissues are assessed. We divided the 1047 tumor samples into high or low groups based on SPHK1 expression level applying optimal cutoff point of SPHK1. We then retrieved the data from tumor breast tissues which HER2 status is negative (n = 540) or positive (n = 157). Differential expression of S1P-related and immune-related genes between HER2-negative and SPHK1-high breast tissue (n = 452), and HER2-negative and SPHK1-low breast tissue (n = 88) was measured. The S1P-related and immune-related genes were extracted in a molecular interaction network, based on the methodology described in the following paragraph.
Pathologic Examination
All immunohistochemical staining in this study was performed and evaluated as previously described.21 Briefly, paraffin-embedded blocks from each resected specimens were used for histology and immunohistochemistry. Serial 4 mm sections were cut and were used for staining with hematoxylin and eosin, anti-phosphorylated SPHK1, anti-CD4, anti-CD8, anti-CD68 and anti-CD163 immuno-histochemistry. Slides containing cancer tissue were deparaffinized in xylene and were rehydrated in graded alcohols to water. For phosphorylated SPHK1 (pSPHK1), CD4 staining, antigen retrieval was performed by pressure cooking the cancer tissues in target retrieval solution, pH 9 (Agilent Technologies, Inc., Santa Clara, CA, USA), at 120 degree Celsius for 10 min. For CD8, CD68 and CD163 staining, Antigen Retrieval Citra Solution, pH 6 (BioGenex, San Ramon, CA, USA), was used, and tissues were pressure cooked at the same temperature and time as aforementioned. Endogenous peroxidase was blocked using 0.3% H2O2 for 30 min. After blocking nonspecific background, the sections were incubated over-night with the primary antibody (pSPHK1 polyclonal antibody; 1:100 dilution; ECM Biosciences LLC, Versailles, KY, USA;CD4 monoclonal antibody; 1:50 dilution; NICHIREI BIOSCIENCE INC., Chuo-ku, Tokyo, Japan; CD8 monoclonal antibody; 1:50 dilution; NICHIREI BIOSCIENCE INC., Chuo-ku, Tokyo, Japan; CD68 monoclonal antibody; 1:100 dilution; Agilent Technologies, Inc., Santa Clara, CA, USA; CD163 monoclonal antibody; 1:100 dilution; Leica Biosystems Newcastle Ltd, Benton Lane, Newcastle upon Tyne, UK) at 4 degree Celsius. Then the sections were incubated with biotinylated secondary anti-rabbit antibody at room temperature for 30 min for pSPHK1 staining and secondary anti-mouse antibody at room temperature for 30 min for CD4, CD8, CD68 and CD163 staining. After 30 min with streptavidin-peroxidase complex, sections were stained with diaminobenzidine substrate and were counterstained with hematoxylin. pSPHK1 expression was defined as the presence of cytosolic staining with the anti-pSPHK1 antibody according to our previous report.21 Normal rabbit/mouse immuno-globulin was substituted as the primary antibodies in the negative control.
Network for S1P-related and immune-related proteins
To construct a molecular interaction network, the protein-protein association data of the STRING database28 were imported to Cytoscape29 with stringApp plugin software. For the human protein-related association network, S1P-related and immune-related proteins were uploaded to the STRING database and the association network was constructed with a cut-off edge confidence score of 0.15. Using the network information downloaded from the database, the molecular interaction network figure was generated with Cytoscape.
Statistical Analysis
Statistical comparison included Friedman test followed by Dunn’s test for tumoral samples, peri-tumoral samples, and normal breast samples. Paired t-tests statistical comparison was used to evaluate differential expression differences between tumoral samples and normal samples from the TCGA data. Unpaired t-tests statistical comparison was utilized to evaluate differential expression in SPHK1-high and SPHK1-low breast tissue. The expression level correlation matrix analysis of interested genes in tumoral and normal samples were evaluated by Pearson correlation and genes were grouped by the hierarchical clustering method. Statistical evaluations were performed using the GraphPad Prism (GraphPad software Inc, California, USA) and R/Bioconductor packages.30 All analyses were two-sided and significance was set as P values < 0.05.
Results
S1P and other sphingolipid levels in clinical normal, peri-tumoral and tumoral samples
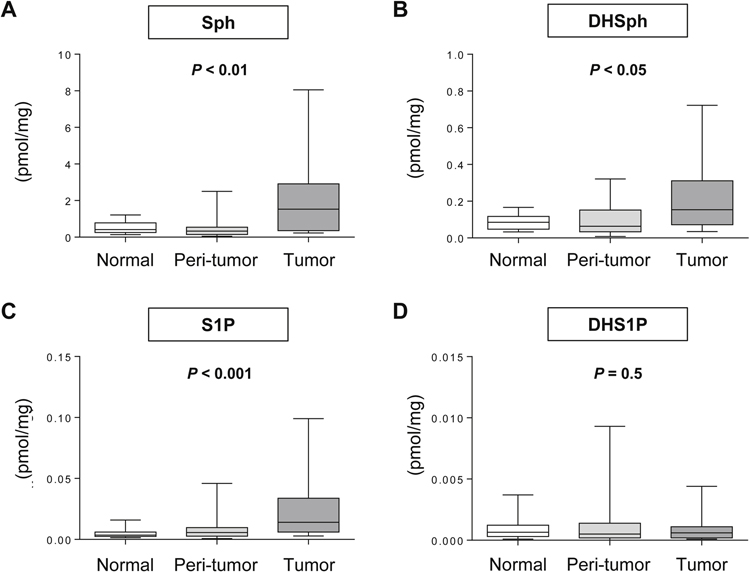
We examined S1P and other sphingolipid levels in 20 clinical breast cancer samples (Fig. 1). The levels of the sphingolipids; Sph, DHSph, S1P, and DHS1P, were successfully determined in tumoral, peri-tumoral, and normal samples from the 20 patients. Pathological results of the 20 patients are shown in Supplementary Table 1. As expected, a Friedman test revealed that S1P levels were significantly different depending on the location (P < 0.001; Fig. 1C). Dunn’s multiple comparison demonstrated higher concentrations of S1P in tumors than normal samples and peri-tumoral samples (P < 0.05, P < 0.001). Similarly, Sph and DHSph levels were significantly different depending on the location (P < 0.01; Fig. 1A, P < 0.05; Fig. 1B). Sph levels in tumors were significantly higher than in normal samples and peri-tumoral samples (P < 0.05, P < 0.001).
Fig. 1 – Levels of S1P and other sphingolipids in different locations of breast tissue.
The levels of Sph (A), DHSph (B), S1P (C) and DHS1P (D) were determined by LC-ESI-MS/MS in tumor, peri-tumor, and normal breast tissue from surgical specimens. Each box plot represents the minimum, mean, max and 25th and 75th percentiles of the indicated sphingolipid (mmol/mg).
S1P-related gene expression in tumoral and normal samples
S1P-related gene expression in tumoral and normal samples were evaluated in the TCGA database (Fig. 2). Of note, both SPHK1 and SPHK2 gene expression concentrations in tumoral samples were evaluated compared to normal samples. Interestingly, expression of some of the S1P-related genes; S1PR3, ATP-binding cassette C1 (ABCC1), S1P lyase 1 (SGPL1), and ORMDL sphingolipid biosynthesis regulator 2 (ORMDL2), were elevated in the tumoral samples compared to normal samples. On the other hand, there was significantly decreased expression of the S1P-related genes; S1PR1, S1PR2, ATP binding cassette subfamily G member 2 (ABCG2), SPNS2, S1P phosphatase 1 (SGPP1) and ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3), in tumoral versus normal samples.
Fig. 2 – Expression of S1P-related genes in breast cancer and normal breast tissue.
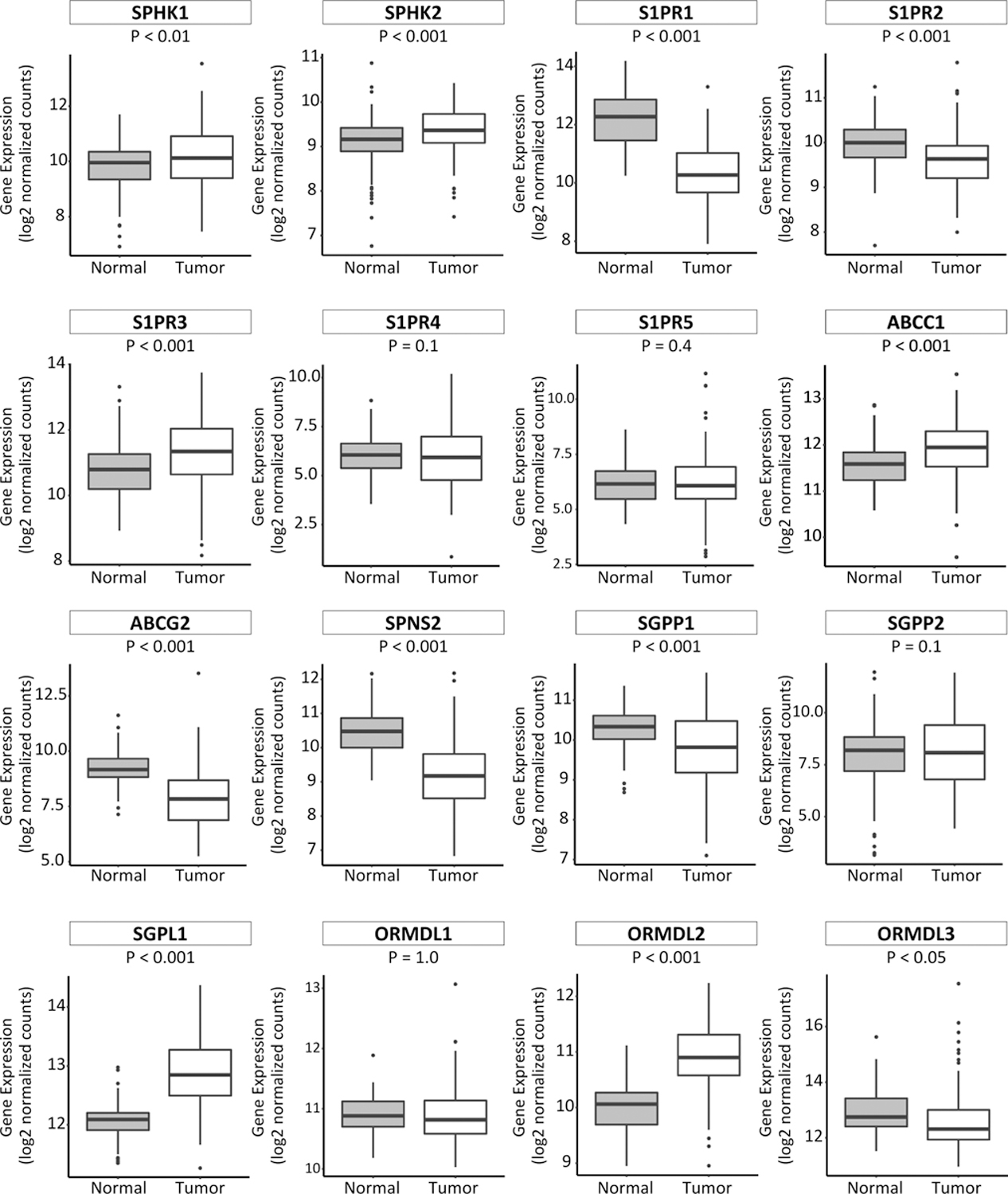
Expression levels of S1P-related genes, as determined by RNA sequencing, in breast cancer 112 normal and their paired tumor breast tissues, were obtained from the TCGA database. In each boxplot the top and bottom represent the 25th and 75th percentile, respectively, and the band near the middle of the box is the median gene expression level.
The association of SPHK1 and S1P-related gene expression in tumoral samples
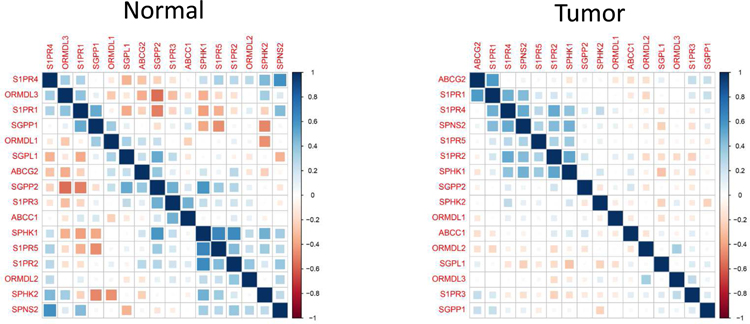
We next examined how the S1P-related genes associated to each other utilizing using correlation matrix analysis for TCGA cohort (Fig. 3). The results showed that there are different patterns of the interactions among the genes in the normal and tumor tissue. Although there are various interactions each other among the genes in the normal tissue, only SPHK1 and limited number of genes showed significant interactions in the tumor (Fig. 3). Therefore, we next evaluated the correlation between the tumoral expression of SPHK1 and other S1P-related genes (Fig. 4). We examined the expression of S1P-related genes in patients with high-SPHK1 expression and those with low-SPHK1 expression in HER2-negative breast cancer patients, since we have previously demonstrated that the S1P axis is a critical player especially in HER2-negative breast cancer.21
Fig. 3 – The correlation matrix analysis for S1P-related genes in normal and tumor tissue.
Levels of correlation of each gene are shown. Blue squares indicated positive correlation and red squares indicated inverse correlation.
Fig. 4 – Expression of S1P-related genes in breast cancer tissue with high-or low-SPHK1 expression in the HER2-negative group.
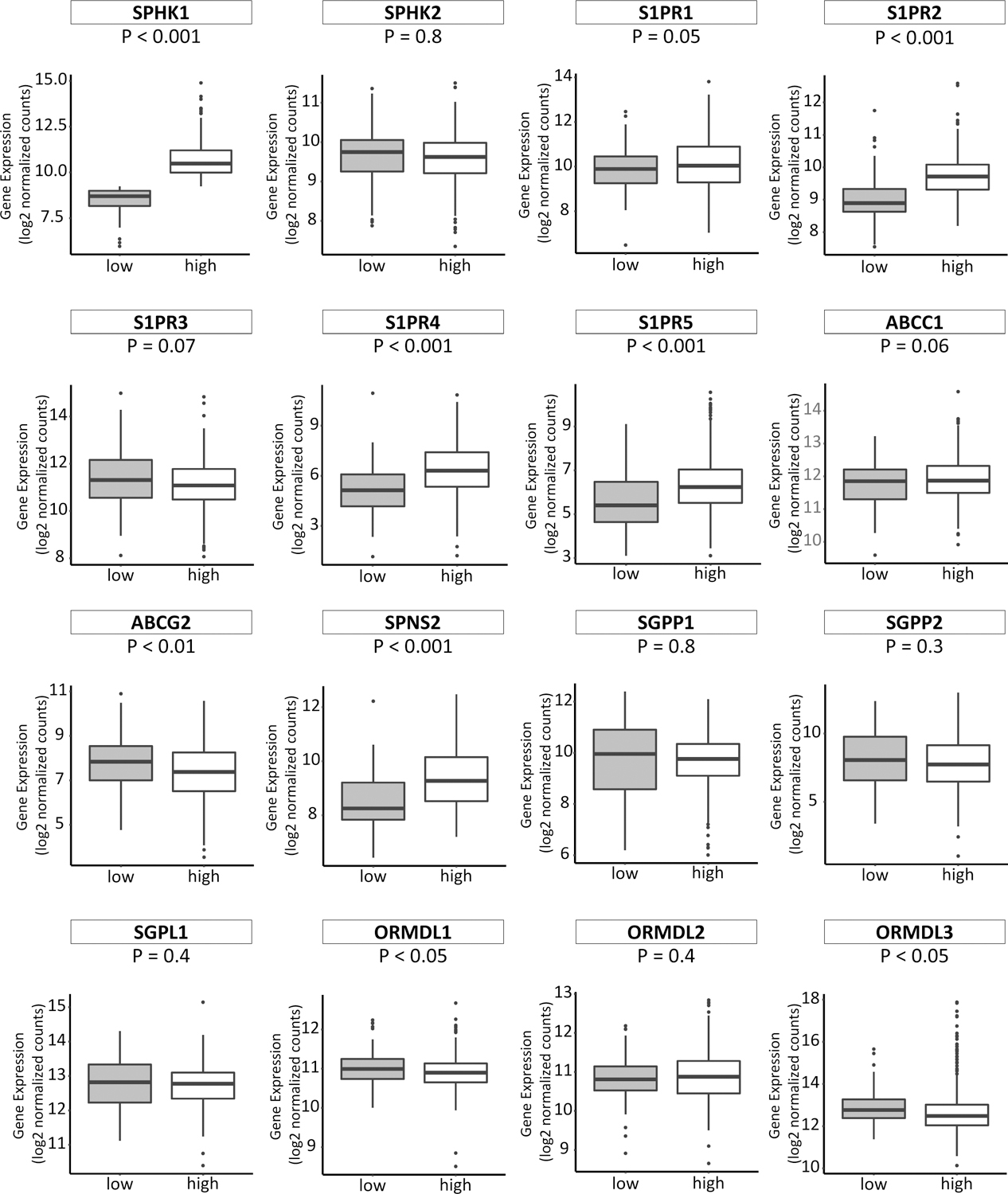
In each boxplot the top and bottom represent the 25th and 75th percentile, respectively, and the band near the middle of the box is the median gene expression level. low: low-SPHK1 expression; high: high-SPHK1 expression.
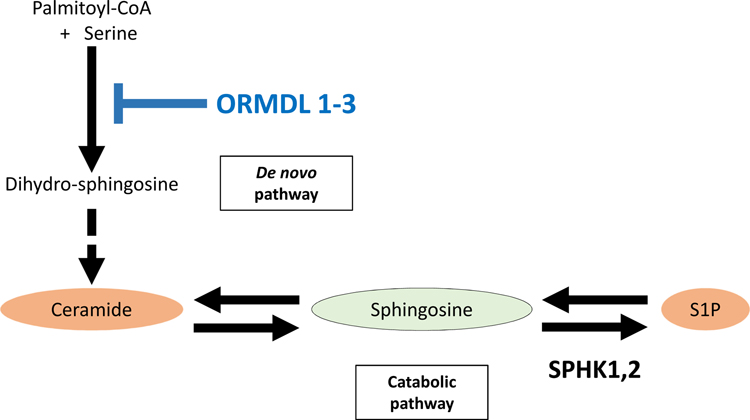
The expression of some of the S1P-related genes; S1PR2, S1PR4, S1PR5 and SPNS2, were significantly increased in HER2 negative breast cancer cases with high-SPHK1 versus low-SPHK1 (Fig. 4). According to the previous reports, it has been suggested that ORMDL sphingolipid biosynthesis regulator 1 (ORMDL1) and ORMDL3 may be a master regulator of sphingolipids, which suppress S1P production (Fig. 5).31,32 Interestingly, we observed that the expression of ORMDL1 and ORMDL3 were significantly decreased in HER2 negative breast cancer cases with high-SPHK1 versus low-SPHK1 as expected (Fig. 4).
Fig. 5 – Regulation by ORMDL 1–3 in sphingolipid metabolism.
ORMDL1–3 is a master regulator of sphingolipid metabolism, which suppress production of dihydro-sphingosine resulting in the suppression of S1P production.
Immune-related gene expression in association with SPHK1
We evaluated the correlation of differential genetic expression in SPHK1 in of immune-related genes in HER2-negative breast cancer tissue by utilizing the TCGA database (Fig. 6). We found a significant increase in the expression of the immune-related genes, including CD68, CD163, C-C motif chemokine ligand 1 (CCL1), tumor necrosis factor (TNF), IL6, IL1B, IL12A, IL12B, transforming growth factor beta 1 (TGFB1), C-C motif chemokine ligand 2 (CCL2), and IL10, as macrophage markers in SPHK1-high HER2-negative breast cancer tissues than in SPHK1-low. T cell markers including CD4, CD8A, CD8B, are increased in SPHK1-high HER2-negative breast cancer tissues. Regulatory T cell markers including programmed cell death 1 (PDCD1), CD274, CD80, cytotoxic T-lymphocyte associated protein 4 (CTLA4) and forkhead box P3 (FOXP3), are also increased in SPHK1-high HER2-negative breast cancer tissues. Moreover, adipocyte-related genes, such as homeobox C9 (HOXC9), peroxisome proliferator activated receptor gamma (PPARG), PPARG coactivator 1 alpha (PPARGC1A) and leptin (LEP), were also increased in SPHK1-high HER2-negative breast cancer tissue.
Fig. 6 – Expression of immune-related genes in breast cancer tumor tissue with high or low-SPHK1 expression in the HER2-negative group.
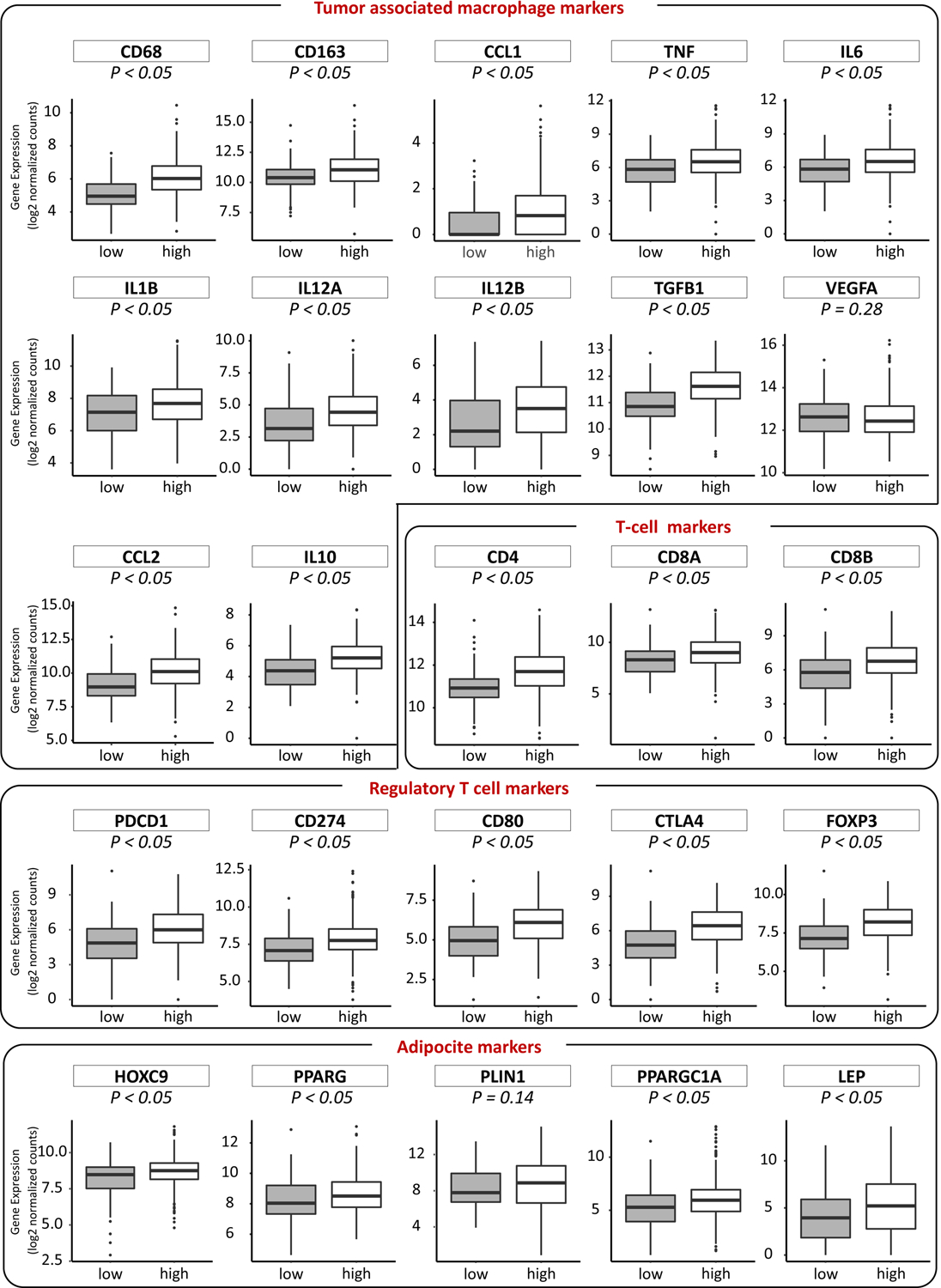
In each boxplot the top and bottom represent the 25th and 75th percentile, respectively, and the band near the middle of the box is the median gene expression level. low: low-SPHK1 expression; high: high-SPHK1 expression.
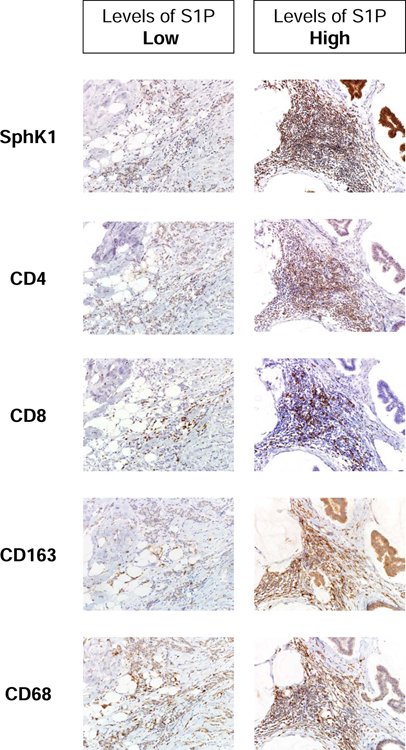
We investigated the association between levels of S1P and Infiltration of inflammatory cells, such as CD4 or CD8-positive T cells, and CD68 or CD163-positive monocytes/macrophages (Fig. 7). The expression of active form of pSPHK1 is positive in the S1P concentration high specimen as we previously reported.21 We examined the immune cell-related protein expression of high S1P concentration and pSPHK1 expression specimen and low S1P concentration and pSPHK1 expression specimen. Expressions of CD4, CD8, CD68 and CD163 is high in the high S1P concentration and pSPHK1 expression specimen. On the other hand, those were low in the low S1P concentration and pSPHK1 expression specimen. Consistent with the result of gene expression analysis, immunohistochemistry of surgical specimens revealed Infiltration of inflammatory cells, such as CD4 or CD8-positive T cells, and CD68 or CD163-positive monocytes/macrophages.
Fig. 7 – Immunohistochemical expression of immune cells in the surgical specimens.
Immunohistochemical expression of active form of phosphorylated SPHK1 and immune cell related proteins, such as CD4, CD8, CD68 and CD163 in the surgical specimens with high or low S1P concentration.
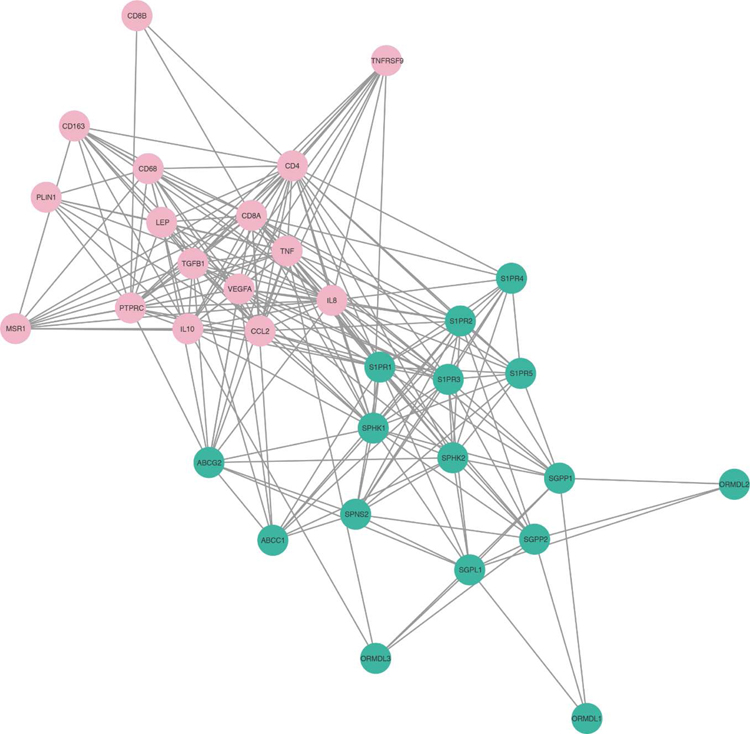
Network analysis of the S1P-related and immune-related proteins revealed that SPHK1 and many of the other S1P-related proteins are linked to immune-related proteins (Fig. 8). This analysis showed the central role of SPHK1 in the interaction of S1P signaling and expression of immune-related proteins.
Fig. 8 – Network of S1P-related and immune-related proteins.
S1P-related proteins are shown green and immune cell-related proteins are shown pink. The relationship between each protein is mapped by the association data from the STRING database, and is visualized by Cytoscape with an edge confidence score cut-off of 0.15.
Discussion
The pleiotropic bioactive lipid mediator S1P is an important new player in tumor biology. As previously published, S1P significantly affects the TME. For instance, our group and others have previously revealed that S1P is a key factor in both tumor-associated angiogenesis and lymphangiogenesis.24,33,34 We also found that S1P attracts immune cells to the wound.23,35 However, the exact mechanism as it relates to immune-cancer reactions has not been thoroughly evaluated in human breast cancer. Here, we demonstrated that S1P is mainly produced by tumor clinical samples, not peri-tumoral or normal breast clinical samples. Moreover, by utilizing the TCGA database, we revealed that tumoral samples which highly expressed SPHK1 correlated with increased expression of most of the immune-related genes in HER2-negative breast cancer. Therefore, these results support the hypothesis that SPHK1-derived S1P is significant in immune reactions mediated by cancer cells, which promotes cancer progression in breast cancer patients (Fig. 9).
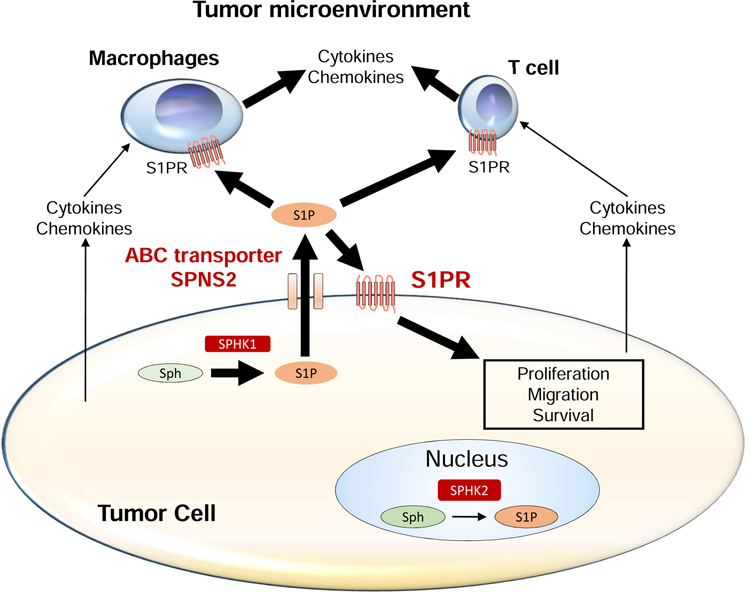
Fig. 9 – The role of S1P produced by SPHK1 in tumor microenvironment.
S1P is generated by SPHK1 from sphingosine in the cytosol of tumor cells. S1P is exported out of the cell via S1P transporters, such as ABC transporters and SPNS2. Extracellular S1P stimulates S1PRs on the surface of the tumor cell itself, and immune cells, including macrophages and lymphocytes. S1P signaling may promote the production of cytokines and chemokines in these cells within the tumor microenvironment.
We published a mass spectrometry method that sensitively and quantitatively measures S1P concentrations in vitro and in vivo.13,24,25 We have previously reported that S1P concentrations in tumoral samples are significantly more elevated than in normal breast samples utilizing mass spectrometry.26 We also demonstrated that high levels of S1P are not only seen in tumor tissue, but also in tumor interstitial fluid, indicating that S1P is part of the TME in some tumors.36 It has been suggested that cancer cells as well as normal stroma, such as vascular endothelium and immune cells in the TME, can be sources of S1P.37 However, the role of normal tissue surrounding the tumor (peri-tumor tissue) in S1P production has never been investigated. A review of the literature demonstrates that this report is novel for actually quantifying concentration of S1P in peri-tumoral sample, which is considered to be a part of the TME. Our data demonstrate that the main source of S1P is the tumor itself, and not the peri-tumor tissue, despite the fact that angiogenesis and lymphangiogenesis are occurring more in the peri-tumoral area, which implies that S1P may have more activity inside the tumor.
To explore further the function of S1P in human breast cancer, we next evaluated how tumoral versus normal breast samples differentially expressed S1P-related genes utilizing the TCGA database. Since the higher S1P levels were observed in tumoral samples versus peri-tumoral or normal breast samples, we expected the samples to express S1P-related genes in a similar pattern. Indeed, tumoral tissue expressed both SPHK1 and SPHK2 significantly more than normal breast samples. S1PR3, a known promoter of cancer progression,16 also showed higher levels in tumoral samples as expected. In contrast, S1PR2 showed lower levels in tumoral samples. This is in agreement with the notion that S1PR2 functions negatively for cancer progression by suppressing the S1PR1 and S1PR3.16 These results indicated the more complexity of S1P signaling in human cancer than expected based on the results of in vivo experiments alone.
We have previously demonstrated significantly higher S1P levels in HER2-negative versus HER2-positive disease.21 This finding indicates that the S1P signaling plays an important role in HER2-negative breast cancer. Thus, we evaluated the correlation between the expression of SPHK1 and other S1P-related genes in HER2-negative breast cancer patients. As expected, the expression of some of the S1P-related genes; S1PR2, S1PR4, S1PR5 and SPNS2, were significantly increased in cases with elevated SPHK1 versus decreased SPHK1. On the other hand, expression of some of the S1P-related genes; ABCG2, ORMDL1 and ORMDL3 were significantly decreased in cases with elevated SPHK1 versus decreased SPHK1. It has been suggested that ORMDL1 and ORMDL3 may affect metabolism of sphingolipids, which suppress S1P production (Fig. 4 and 5).31,32 Therefore, the lower levels of ORMDL1 and ORMDL3 in high-SPHK1 are reasonable, while lower levels of ABCG2 in high-SPHK1 cannot be explained clearly. Taken together, our findings provide further data supporting the impact of S1P signaling in HER2-negative breast cancer, and further research will be needed to understand the complicated function of S1P in human breast cancer.
Since one of the principal roles of S1P is recruitment of immune cells,16,23 we further examined the relationship between S1P signaling and tumor-associated immune reactions. To this end, we evaluated how HER2-negative breast cancer clinical samples differentially expressed SPHK1 and the immune-related genes. Interestingly, most of the genes we examined showed higher levels in patients with SPHK1 high. Of note, some of the genes works as pro-inflammatory, and others works as anti-inflammatory mediators, all of which will be orchestrated to modify TME. For instance, S1P is expected to promote the production of cytokines and chemokines such as TNF, TGFB1, and CCL2 in the TME.38 On the other hand, promoting IL-10 production by S1P will suppress immune function. Some reports indicated that IL-10 may suppress anti-tumor immune functions, which will promote tumor progression. Consistent with the previous report,23,35 our immunohistochemical examination suggest the association between the S1P concentration and infiltration of immune cells.
The principal limitations of the current investigation included it being a study utilizing only dry data obtained from mass spectrometry analysis and the TCGA database. Therefore, we will investigate experiments with intervention to test our hypothesis. This study expands our knowledge regarding the function of S1P in human breast cancer. A literature review demonstrated this being first report of actually quantifying concentration of S1P in peri-tumoral sample. We revealed that S1P is mainly produced by tumor tissue, rather than peri-tumoral tissue, in breast cancer patients. Another implication is the involvement of S1P signaling in the complicated immune-cancer interactions in breast cancer patients. Taken together, these findings suggest that S1P, produced by SPHK1 in breast cancer cells, actively impacts immune reactions mediated by cancer cells, which possibly promoting cancer progression in breast cancer patients. Additional investigations are clearly required to understand the involvement of S1P signaling in these complicated mechanisms.
Conclusion
We demonstrated that S1P is mainly produced by tumor tissue, rather than peri-tumoral tissue, in breast cancer patients. Our results demonstrate there is more complexity to S1P signaling in human cancer than expected otherwise by non-human studies to date. Our results suggest the involvement of S1P signaling in the complicated immune-cancer interactions in HER2-negative breast cancer cases, and additional research is required to understand the underlying functional pathways.
Supplementary Material
Acknowledgments
Disclosure
We gratefully acknowledge the VCU Lipidomics Core, which is supported in part by funding from the NIH-NCI Cancer Center Support Grant P30CA016059. This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research Grant Numbers 19K18049 for J.T., 19H03714 and 18K19576 for M. Nagahashi, 19K22651 for T.W., 17K10692 for M.Nakajima. J.T. was also supported by Tsukada Medical Foundation. M. Nagahashi was also supported by Uehara Memorial Foundation. K.T. was supported by NIH/NCI grant R01CA160688 and Susan G. Komen Investigator Initiated Research Grant IIR12222224.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Fang H, Declerck YA. Targeting the tumor microenvironment: from understanding pathways to effective clinical trials. Cancer Res. 2013;73:4965–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. [DOI] [PubMed] [Google Scholar]
- 4.Nagahashi M, Takabe K, Terracina KP, et al. Sphingosine-1-phosphate transporters as targets for cancer therapy. BioMed Research International. 2014;2014:651727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain RK. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell. 2014;26:605–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolaney SM, Boucher Y, Duda DG, Martin JD. Role of vascular density and normalization in response to neoadjuvant bevacizumab and chemotherapy in breast cancer patients. Proc Natl Acad Sci U S A. 2015;112:14325–14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. [DOI] [PubMed] [Google Scholar]
- 8.Anelli V, Gault CR, Snider AJ, Obeid LM. Role of sphingosine kinase-1 in paracrine/transcellular angiogenesis and lymphangiogenesis in vitro. FASEB J. 2010;24:2727–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagahashi M, Matsuda Y, Moro K, et al. DNA damage response and sphingolipid signaling in liver diseases. Surg Today. 2016;46:995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol. 2011;11:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takabe K, Spiegel S. Export of sphingosine-1-phosphate and cancer progression. J Lipid Res. 2014;55:1839–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitson SM, Xia P, Leclercq TM, et al. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signalling. J Exp Med. 2005;201:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takabe K, Kim RH, Allegood JC, et al. Estradiol induces export of sphingosine 1-phosphate from breast cancer cells via ABCC1 and ABCG2. J Biol Chem. 2010;285:10477–10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada A, Ishikawa T, Ota I, et al. High expression of ATP-binding cassette transporter ABCC11 in breast tumors is associated with aggressive subtypes and low disease-free survival. Breast Cancer Res Treat. 2013;137:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hisano Y, Kobayashi N, Kawahara A, et al. The sphingosine 1-phosphate transporter, SPNS2, functions as a transporter of the phosphorylated form of the immunomodulating agent FTY720. J Biol Chem. 2011;286:1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takabe K, Paugh SW, Milstien S, Spiegel S. “Inside-out” signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev. 2008;60:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchida J, Nagahashi M, Takabe K, Wakai T. Clinical Impact of Sphingosine-1-Phosphate in Breast Cancer. Mediators Inflamm. 2017;2017:2076239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruckhaberle E, Rody A, Engels K, et al. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res Treat. 2008;112:41–52. [DOI] [PubMed] [Google Scholar]
- 19.Shida D, Takabe K, Kapitonov D, et al. Targeting SphK1 as a new strategy against cancer. Curr Drug Targets. 2008;9:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim RH, Takabe K, Milstien S, Spiegel S. Export and functions of sphingosine-1-phosphate. Biochim Biophys Acta. 2009;1791:692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuchida J, Nagahashi M, Nakajima M, et al. Breast cancer sphingosine-1-phosphate is associated with phospho-sphingosine kinase 1 and lymphatic metastasis. J Surg Res. 2016;205:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoki M, Aoki H, Ramanathan R, et al. Sphingosine-1-Phosphate Signaling in Immune Cells and Inflammation: Roles and Therapeutic Potential. Mediators Inflamm. 2016;2016:8606878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagahashi M, Ramachandran S, Kim EY, et al. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012;72:726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hait NC, Allegood J, Maceyka M, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagahashi M, Tsuchida J, Moro K, et al. High levels of sphingolipids in human breast cancer. J Surg Res. 2016;204:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breslow DK, Collins SR, Bodenmiller B, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai L, Oyeniran C, Biswas DD, et al. ORMDL proteins regulate ceramide levels during sterile inflammation. J Lipid Res. 2016;57:1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon CM, Hong BS, Moon HG, et al. Sphingosine-1-phosphate promotes lymphangiogenesis by stimulating S1P1/Gi/PLC/Ca2+ signaling pathways. Blood. 2008;112:1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milstien S, Spiegel S. Targeting sphingosine-1-phosphate: a novel avenue for cancer therapeutics. Cancer Cell. 2006;9:148–150. [DOI] [PubMed] [Google Scholar]
- 35.Aoki M, Aoki H, Mukhopadhyay P, et al. Sphingosine-1-Phosphate Facilitates Skin Wound Healing by Increasing Angiogenesis and Inflammatory Cell Recruitment with Less Scar Formation. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagahashi M, Yamada A, Miyazaki H, et al. Interstitial Fluid Sphingosine-1-Phosphate in Murine Mammary Gland and Cancer and Human Breast Tissue and Cancer Determined by Novel Methods. J Mammary Gland Biol Neoplasia. 2016;21:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu Rev Immunol. 2012;30:69–94. [DOI] [PubMed] [Google Scholar]
- 38.Liang J, Nagahashi M, Kim EY, et al. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell. 2013;23:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.