Abstract
To survive within complex environmental niches, including the human host, bacteria have evolved intricate interspecies communities driven by competition for limited nutrients, cooperation via complementary metabolic proficiencies, and establishment of homeostatic relationships with the host immune system. The study of such complex, interdependent relationships is often hampered by the challenges of culturing many bacterial strains in research settings and the limited set of tools available for studying the dynamic behavior of multiple bacterial species at the microscale. Here, we utilize a microfluidic‐based co‐culture system and time‐lapse imaging to characterize dynamic interactions between Streptococcus species, Staphylococcus aureus, and Actinomyces species. Co‐culture of Streptococcus cristatus or S. salivarius in nanoliter compartments with Actinomyces graevenitzii revealed localized exclusion of Streptococcus and Staphylococcus from media immediately surrounding A. graevenitzii microcolonies. This community structure did not occur with S. mitis or S. oralis strains or in co‐cultures containing other Actinomycetaceae species such as S. odontolyticus or A. naeslundii. Moreover, fewer neutrophils were attracted to compartments containing both A. graevenitzii and Staphylococcus aureus than to an equal number of either species alone, suggesting a possible survival benefit together during immune responses.
Keywords: Actinomyces graevenitzii, cell‐microbe interactions, microbial interactions and pathogenesis, signalling, Staphylococcus
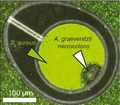
“Exclusion zone” formation around Actinomyces graevenitzii microcolonies in microfluidic nanoliter volume co‐culture chambers. This species‐specific phenomenon appears to relate to A. graevenitzii colony stress and can alter inflammatory responses.
1. INTRODUCTION
The complexity of bacterial communities that make up our microbiome mirrors the complexity of niches within the human body. Of these niches, the oral cavity is perhaps one of the most diverse, presenting extremes of tissue stiffness, surface topography, transient temperature shifts, and nutrient flux (Paster et al., 2000). Although the accessibility of the oral cavity has made it a focus of research into microbial community structure and diversity (Wilbert et al., 2020), our understanding of interspecies relationships and their role in health and disease remains limited.
A wide range of co‐culture strategies has been developed to facilitate the characterization of interspecies relationships, largely focusing on metabolic compatibility and coaggregation of species that form oral biofilms and plaque (Levin‐Sparenberg et al., 2016; Ochiai et al., 1993; Postollec et al., 2006). Coaggregation in suspension and co‐adhesion on surfaces between bacterial species appear to share common pathways (Kolenbrander & Andersen, 2000). Strong coaggregation between species can be easily identified by visual or radioactive assays (Kolenbrander, 1995), while co‐adhesion to a surface can be studied under static conditions or using flow chambers (Kolenbrander, 2000).
Two bacterial genera commonly associated with oral biofilm formation are Actinomyces (Dige et al., 2009) and Streptococcus (Dige et al., 2007). Intrageneric coaggregation of Streptococci appears more common than Actinomyces (Kolenbrander et al., 1990), while intergeneric coaggregation between Actinomyces and Streptococci species appears commonplace both in vitro (Cisar et al., 1979) and in vivo (Palmer et al., 2003).
The genus Actinomyces has recently been subdivided with the creation of the genus Schaalia (Nouioui et al., 2018) with both Actinomyces and Schaalia being members of the family Actinomycetaceae. The bacteria Schaalia odontolyticus and Streptococcal spp. are considered early colonizers, adhering directly to the salivary pellicle coating the tooth surface. This facilitates the secondary adherence of intermediate colonizers, such as Actinomyces spp., followed by late colonizers such as Fusobacterium nucleatum and Porphyromonas gingivalis during the formation of dental plaque (Kolenbrander et al., 2010). Sequential adherence of different bacterial species depends on their co‐adhesion compatibility, which is often species‐specific (Bos et al., 1996), while coaggregation of bacterial species in suspension has been shown to directly influence gene expression to induce metabolic outputs to the benefit of both species (Jakubovics et al., 2008). Some bacterial species are incompatible for co‐culture, leading to domination by one species at the expense of the other, often in a nutrient‐dependent manner (Arzmi et al., 2016; He et al., 2016).
Efforts to culture previously “unculturable” species have focused on identifying co‐culture partners that provide complementary metabolic functions to compensate for the lack of specific metabolic pathways (Stewart, 2012). Physical distances and culture volume play key roles in metabolic symbiosis, interspecies communication, and cell–cell adherence (Kolenbrander et al., 2010). Thus, multiple recent studies have used microfluidic approaches to achieve small‐volume co‐culture and to engineer co‐culture devices with defined physical constraints (Boedicker et al., 2009; Hesselman et al., 2012; Park et al., 2011).
Staphylococcus aureus is a common commensal present on the skin and upper respiratory tract of up to 50% of healthy individuals (Tong et al., 2015; Wertheim et al., 2005). It is also considered an important and dangerous opportunistic pathogen, due to high infection rates and the emergence of many antibiotic‐resistant strains (Chambers & Deleo, 2009).
Innate immune cells, especially neutrophils, are the first cellular line of defense against S. aureus infection once physical barriers are breached (Rigby & DeLeo, 2012). As such, S. aureus infections induce a robust inflammatory response, which can lead to conditions such as cellulitis in the skin (Krishna & Miller, 2012), more severe arthritic conditions following infections of the bones and joints (Siam & Hammoudeh, 1995), and sepsis. Studies have identified S. aureus in between 17% and 48% of healthy oral samples, with even higher rates of up to 64% present in young children (Smith et al., 2001). Despite this, S. aureus is not considered a significant oral pathogen, and infections in the oral cavity are usually limited to inflammatory conditions such as angular cheilitis (Ohman et al., 1986), along with rarer cases of jaw cysts (Iatrou et al., 1988) and oral mucosal lesions (Dahlen et al., 1982). Systemic dissemination of S. aureus originating from the oral cavity remains a relatively unexplored topic.
Here, we utilized a previously developed microfluidic device (Ellett et al., 2019) to perform a low‐volume co‐culture of multiple actinomyces and streptococcal species. We characterized the interactions between pairs of species in nanoliter confinement and observed the formation of exclusion zones between colonies, which were not observable in traditional co‐cultures. Exclusion zones also formed in co‐cultures of A. graevenitzii and a GFP‐expressing strain of S. aureus, and co‐culture with A. graevenitzii significantly dampened innate immune responses compared to S. aureus monocultures.
2. MATERIALS AND METHODS
2.1. Bacterial cell culture
Actinomyces graevenitzii was cultured on Chacolate II agar (GC II Agar with hemoglobin [10 g/L] and IsoVitalex™ [1% v/v]) (BD) at 37°C in an anaerobic incubator. Single colonies from agar plates were picked and separately suspended in 10 ml of brain heart infusion (BHI) broth medium. Streptococcus cristatus was incubated at 37°C with shaking overnight. Bacterial suspension concentrations were determined using a hemocytometer and the final concentration of bacteria was adjusted to 1 × 107 cells/ml by dilution in with Iscove's Modified Dulbecco's Medium IMDM supplemented with 20% fetal bovine serum (FBS).
Staphylococcus aureus strain SH1000‐GFP, which constitutively expresses green fluorescent protein (GFP), was received as a generous gift from the laboratory of Mary Mullins at the University of Sheffield (Sheffield, UK). Bacterial cultures were routinely cultivated in BHI Agar Plates with 5 µg/ml tetracycline (Teknova). Single colonies from agar plates were picked and suspended in 10 ml of BHI broth medium (Remel) with 5 µg/ml tetracycline and then incubated at 37°C in an aerobic incubator with shaking overnight. After overnight incubation, bacterial suspensions were sub‐cultured by adding 1 ml of the overnight culture into 49 ml of BHI broth with tetracycline for 4 h. Bacterial concentrations were determined using a hemocytometer. The final concentration of bacteria was adjusted to 5 × 106 cells/ml and diluted with IMDM supplemented with 20% FBS.
2.2. Fabrication of microfluidic devices
Devices were fabricated using standard soft‐lithography techniques on four‐inch wafers. Photoresist (SU‐8; Microchem) was spin‐coated onto a silicon wafer and exposed to ultraviolet (UV) light, through a photolithography mask. Briefly, two layers of negative photoresist, the first 10 μm thin, the second 50 μm thick were patterned on a silicon wafer by sequentially employing two photolithography masks and processing cycles according to instructions from the manufacturer. The silicon master wafer with photo‐patterned structures was employed to mold microchambers that were 200 μm in diameter, 50 μm in depth (Figure 1a,b). To test the effect of different depths, another silicon master wafer with photo‐patterned structures was used to mold microchambers that were 200 μm in diameter, but 10, 30, 50, and 100 μm in depth. The entrance for each microchamber was 125 µm in length, 10 µm in width, and 10 µm in depth. Polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning) was mixed with a cross‐linking agent in a ratio of 10:1 and poured onto wafers. The PDMS was cured overnight at 65°C, after which the PDMS layer was peeled off the wafer and the arrays of wells were cut from the block using a scalpel and the inlets and outlets punched using a 0.75 µm punch (Harris Uni‐Core). The microfluidic devices were bonded to glass‐bottom 6‐well plates after treating the bonding surface of PDMS and plate with oxygen plasma. The plates were heated to 70°C for 15 min to complete the PDMS‐to‐glass bonding. Each device consists of 99 chambers, uniformly distributed inside groups of three channels.
FIGURE 1.

A microfluidic device for bacterial nanoliter culture (a) Schematic shows the dimensions of the microchamber. Each egg‐shaped device consists of a central 200 µm diameter, 50 µm high cylindrical microchamber connected to the outer chamber by a 125 µm long, 10 µm wide, 10 µm high connecting channel. (b) Depending on how the device is loaded, it can be configured for mono‐culture, co‐culture, or for studying innate immune responses
2.3. Bacterial co‐culture in microfluidic devices
To perform nanoliter co‐cultures, we utilized microfluidic devices that consist of an array of 1.57 nl‐volume cylindrical chambers (200 µm diameter × 50 µm height) connected to a single 50 µm high outer channel by a 125 µm long channel with a 10 × 10 µm cross section (Figure 1a, adapted from Ellett et al. (2019). Each PDMS device contained three sample chambers containing 33 assays per chamber. When bonded to a 6‐well glass‐bottom plate, this allowed 18 conditions to be tested in parallel. We cultured a GFP‐expressing strain of Staphylococcus aureus inside the 1.57 nl chambers and observed that it achieved confluence after approximately 6 h at 37°C when loaded at a concentration of 1 × 106 cells/ml (Ellett et al., 2019).
To prepare nanoliter co‐cultures, the devices were first placed into a vacuum chamber for 20 min before loading. Once removed from the vacuum, the co‐culture chambers were primed by flowing the bacterial suspension through the outer channel of each device. The devices were then allowed to equilibrate, drawing the bacterial suspension into the co‐culture chambers. The successful loading of the inner chambers was confirmed by microscopy. If air bubbles were present, the plate was placed under vacuum for a further 5 min. During loading steps, care was taken to avoid the mixing of different samples loaded into parallel chambers on the same PDMS device. Once the chambers were loaded, the outer channel was washed with fresh media (IMDM with 20% FBS). For co‐culture experiments, the inner chamber was loaded with a pre‐mixed suspension of starter cultures from different species (Figure 1b).
To test whether the volume of the inner chamber affected bacterial growth, we fabricated a series of devices with altered chamber heights. The height of the outer chamber was adjusted to 200 µm to improved loading and washing steps, and the height of the inner chamber was tested at 10, 30, 50, and 100 µm, corresponding to 0.16, 0.94, 1.57, and 3.14 nl, respectively. In these devices, S. aureus exhibited increasingly restricted, clustered growth patterns as the volume was reduced, suggesting rapid consumption of available nutrients or accelerated sensing of quorum signaling molecules in reduced volumes (Figure A1a–e). The device with 50 µm high chambers and outer channels was utilized for all subsequent experiments unless otherwise stated.
2.4. Neutrophil isolation and neutrophil‐microbe interactions
Neutrophils were isolated from healthy donor blood (Research Blood Components, LLC) using a negative selection kit (StemCell Technologies, Inc.) according to the instructions from the manufacturer. Cells were stained with Hoechst 33342 (Thermo Fisher), washed, and resuspended at 40 × 106 cells per ml for loading into the device. Devices were loaded with mono‐ or co‐cultures of A. graevenitzii and S. aureus as described above. Following thorough washing of the main channel, neutrophils were then loaded and imaging commenced.
2.5. Image acquisition, processing, and analysis
PDMS devices are optically transparent and are bonded directly to glass coverslips, allowing detailed imaging of interactions between species on the glass surface using an inverted microscope. During the experiments, a glass‐bottom 6‐well plate (Micro Device Instruments) with six microfluidic devices was placed on a fully automated Nikon TiE microscope. The microscope was fitted with an incubator humidified and heated at 37°C. Images were acquired through 10× or 20× or 40× objectives in phase contrast. Multiple locations were imaged simultaneously. Growth of bacteria and bacteria movement were recorded using time‐lapse imaging. Individual frames were recorded at an interval of 10 min at 10×, 20×, or 40× objectives for 24 h. For detailed observations, images were also acquired every 10 or 30 s, using an oil‐immersion 100× objective, for a minimum of 90 min. The experiments for this study were repeated up to ten times, including all control experiments. Time‐lapse image sequences were analyzed by FIJI (Fiji Is Just ImageJ, NIH).
2.6. Statistics
We tested for correlations within our data by calculating the Pearson correlation coefficient. We report the correlation coefficients (r), the 95% confidence interval for r, and the p‐value testing the null hypothesis that the data were sampled from a population where there is no correlation between the two variables. When r is 0 we assumed no relationship between the variables. We compared the bacterial confluence in chambers of differing height using a one‐way ANOVA with Tukey's multiple comparison test. All statistical analysis was performed using Prism v8.4.3 (GraphPad Software). Results were plotted using Prism and Sigma Plot version 12 (Systat Software). Error bars represent mean ± SEM.
3. RESULTS
3.1. Exclusion zones form around Actinomyces graevenitzii microcolonies in co‐culture with Streptococcus
Species from the family Actinomycetaceae and genus Streptococcus are among the most common isolated from oral biofilms, particularly dental plaque (Nyvad & Kilian, 1987, 1990). Both Actinomycetaceae and Streptococcal species grew well as monocultures within our microfluidic devices. To study the co‐culture characteristics of Actinomycetaceae and Streptococcal species in our microfluidic chambers (see Methods), we co‐loaded 3 species of Actinomycetaceae (3 strains of S. odontolyticus, 3 strains of A. naeslundii, and 2 strains of A. graevenitzii) in combination with 4 species of Streptococcus (1 strain of S. salivarius, 3 strains of S. mitis, 2 strains of S. oralis, and 1 strain of S. cristatus), 56 combinations in total (Table 1, Figure 2, and Figure A2). Detailed microscopy of the microfluidic chambers was performed at 8 h.
TABLE 1.
Bacterial strains used in the study
| Bacterial strain | Source | Identifier |
|---|---|---|
| Actinomyces naeslundii F0664 | Forsyth Institute | F0664 |
| Actinomyces naeslundii FBB19 | Forsyth Institute | FBB19 |
| Actinomyces naeslundii FCC36 | Forsyth Institute | FCC36 |
| Actinomyces graeventizii F0530 | Forsyth Institute | F0530 |
| Actinomyces graeventizii F0582 | Forsyth Institute | F0582 |
| Schaalia odontolyticus H304PA2 | Forsyth Institute | H304PA2 |
| Schaalia odontolyticus H122PB30 | Forsyth Institute | H122PB30 |
| Schaalia odontolyticus ATCC 17929 | ATCC | ATCC 17929 |
| Streptococcus oralis FFB47 | Forsyth Institute | FFB47 |
| Streptococcus oralis FCB39 | Forsyth Institute | FCB39 |
| Streptococcus salivarius A64PA33 | Forsyth Institute | A64PA33 |
| Streptococcus mitis ATCC 903 | ATCC | ATCC 903 |
| Streptococcus mitis ATCC 2978 | ATCC | ATCC 2978 |
| Streptococcus mitis ATCC 49456 | ATCC | ATCC 49456 |
| Staphylococcus aureus | University of Sheffield | SH−1000 GFP |
FIGURE 2.

Species‐specific formation of “exclusion zones” in co‐cultures of Actinomyces with Streptococcal species. (a) Color map shows the results of co‐culture experiments with combinations of five strains of actinomyces and three strains of Schaalia with seven strains of streptococcus. (b) Representative image at 8 h showing co‐culture of A. naeslundii FCC36 (full yellow arrowheads) and S. salivarius A64PA33 showing no exclusion zone formation. (c) Representative image at 8 h showing co‐culture of A. graevenitzii FO582 (Empty yellow arrowheads) and S. salivarius A64PA33 showing the formation of large exclusion zones around the A. graevenitzii colonies (blue dashed circles)
Observations in nanoliter chambers with co‐cultures of either S. cristatus or S. salivarius A64PA33 with A. graevenitzii (either FO530 or FO582 strains) revealed a striking absence of physical association between the species. Otherwise confluent streptococcal cells appeared to be unable to grow in proximity to A. graevenitzii microcolonies, resulting in the formation of an "exclusion zone” bordering the A. graevenitzii (Figure 2c). Exclusion zones did not form around A. graevenitzii microcolonies in co‐culture with S. mitis (strains ATCC 903 or ATCC 49456) or S. oralis (strains FFB47 or FCB39), or around any of the other Actinomyces or Schaalia species tested (Figure 2b). These observations rule out any physical exclusion of streptococcus cells from space inhabited by actinomyces cells. Instead, the exclusion appears more likely due to the local production of a toxic metabolite or inhibitory compound with considerable species specificity. Importantly, we did not observe any separation between macroscopic co‐cultures performed using traditional co‐culture protocols (Figure A3).
3.2. Exclusion zones form in co‐cultures of A. graevenitzii and S. aureus
The formation of exclusion zones appeared to exhibit species specificity within the Streptococcus genus. To test whether this phenomenon might also occur for different major Firmicutes genera, we co‐cultured A. graevenitzii with the GFP‐expressing strain of Staphylococcus aureus (SH1000‐GFP) that had previously been shown to grow in our device (Ellett et al., 2019). We observed the formation of exclusion zones around A. graevenitzii colonies in co‐culture with S. aureus (Figure 3a,b), demonstrating that this effect is not specific to streptococcal species. Exclusion zones did not form in co‐cultures of S. aureus and A. naeslundii, suggesting that this phenomenon is not common to all actinomyces (Figure 3c). Importantly, the ability to use a GFP‐labeled strain in our studies provided considerable benefits to automated image analysis.
FIGURE 3.

Exclusion zones are formed in co‐cultures of Actinomyces graevenitzii and Staphylococcus aureus. (a) Time‐lapse images show the growth of GFP‐expressing S. aureus in the presence of A. graevenitzii FO582 microcolonies. Over 8 h, S. aureus proliferate to fill the chamber (bright green fluorescence), except for the regions bordering A. graevenitzii microcolonies (GFP‐negative regions). (b) Magnified view of the co‐culture chamber (left) showing a growth of exclusion of S. aureus from a region containing a cluster of A. graevenitzii microcolonies. The cartoon on the right depicts the area of GFP fluorescence measured (yellow) as a percentage of the chamber. (c) Average coverage of microchamber area over time by S. aureus growth in the presence of different species of Actinomyces. Suppression of S. aureus growth, corresponding to the formation of exclusion zones, was only observed in co‐culture with A. graevenitzii. Error bars: mean ± SEM. N ≥ 5 chambers measured per condition. (d) Graph shows the negative correlation between S. aureus growth and the number of A. graevenitzii microcolonies in each co‐culture (Pearson r = −0.68, ****p < 0.0001). The line shows linear regression, dashed lines are 95% confidence intervals (−0.83 to −0.45). N = 152 chambers scored. (e) Graph shows no significant correlation between S. aureus coverage and the initial ratio of S. aureus: A. graevenitzii loaded into each microchamber. N = 178 chambers scored
3.3. Exclusion zones are formed by stressed A. graevenitzii microcolonies in nutrient competition with S. aureus
Exclusion zones could be easily visualized in co‐cultures of A. graevenitzii and GFP‐expressing S. aureus (Figure 3b), allowing us to measure multiple aspects of the co‐culture that might influence exclusion zone formation.
Given that exclusion zones formed around each A. graevenitzii microcolony, we hypothesized that co‐cultures containing increasing numbers of A. graevenitzii microcolonies would have decreasing amounts of S. aureus growth. Using fluorescence microscopy, we measured the percent confluence of S. aureus growth based on the area of GFP fluorescence within each co‐culture chamber as a fraction of the total area of the chamber. S. aureus confluence ranged from 19% to 96%, depending on the number of A. graevenitzii colonies in the co‐culture. A significant negative correlation (Pearson r = −0.68 [95% CI: −0.83 to −0.45], ****p < 0.0001) was observed between the final number of A. graevenitzii microcolonies and observed suppression of S. aureus growth (Figure 3d). We did not find a significant correlation (Pearson r = 0.0067 [95% CI: −0.14 to 0.15], p = 0.93) between the initial species to species ratio of bacteria loaded and later S. aureus growth (Figure 3e).
In mono‐culture, A. graevenitzii exhibited extensive filamentous growth, formed new microcolonies, and grew to effectively fill the chamber. In contrast, the co‐culture of A. graevenitzii with S. aureus resulted in the formation of smaller microcolonies with an optically dense “core” region bordered by a radial array of relatively short filaments extending outwards into the environment (Figure 4a,b). These stunted colonies rarely produced secondary colonies (data not shown). We compared the size of exclusion zones formed around microcolonies to the size of the colony “core” and the total colony diameter, which largely reflected the length of the radial filaments extending outwards. These measurements revealed that significantly larger exclusion zones were generated around microcolonies with larger “core” regions (Pearson r = 0.75 [95% CI: 0.63 to 0.83, ****p < 0.0001) (Figure 4c), while colonies with more extensive radial filamentous growth exhibited significantly smaller exclusion zones (Pearson r = −0.46 [95% CI: −0.63 to −0.26], ****p < 0.0001) (Figure 4d).
FIGURE 4.

Exclusion zones form around stressed Actinomyces graevenitzii colonies. (a) Magnified micrograph showing details of A. graevenitzii FO582 microcolony structure and exclusion zone formation. (b) Cartoon depicting colony structure. (c) Graph showing the positive correlation between the size of the A. graevenitzii microcolony core and the size of the exclusion zone (Pearson r = 0.75, ****p < 0.0001). The line shows linear regression, and dashed lines show 95% confidence intervals (0.63–0.83). N = 75 chambers scored. (d) Graph shows a negative correlation between the length of A. graevenitzii radial filaments and exclusion zone size (Pearson r = −0.46, ****p < 0.0001). The line shows linear regression, and dashed lines show 95% confidence intervals (−0.63 to −0.26). N = 70 chambers scored
To test whether making more nutrients available affected exclusion zone formation, we compared co‐cultures performed in 50 versus 100 µm tall chambers. Relative to S. aureus monocultures in each condition, we observed less restriction of S. aureus growth in co‐culture with A. graevenitzii in 100 µm tall (9.8%) chambers compared to 50 µm tall (21.6%) chambers (Figure A1f). This observation supports our hypothesis that exclusion zone formation occurs in response to competition for nutrients.
3.4. A. graevenitzii preferentially grow as clusters in co‐culture with S. aureus
Spatial clustering of species during co‐culture on solid surfaces can provide competitive advantages and protect against environmental stresses (Nadell et al., 2016). In co‐culture with S. aureus, we observed higher numbers of clustered A. graevenitzii colonies compared to A. graevenitzii monocultures or even co‐cultures with S. cristatus (Figure 5a). However, there was no correlation between the size of the colony and the size of the surrounding exclusion zones for either “single” (Pearson r = 0.0053 [95% CI: −0.15 to 0.16], p = 0.95) or “clustered” (Pearson r = −0.012 [95% CI: −0.22 to 0.20], p = 0.91) A. graevenitzii microcolonies. The largest A. graevenitzii clusters (>160 µm diameter) did not produce significantly larger exclusion zones compared to single microcolonies (~20 µm diameter, Figure 5b,c).
FIGURE 5.

The clustering of Actinomyces graevenitzii microcolonies does not increase the exclusion zone in co‐culture with Staphylococcus aureus. (a) Graph shows an increase in the number of clustered A. graevenitzii FO582 microcolonies compared to monocultures or co‐culture with Streptococcus cristatus. Mean ± SEM. data pooled from at least 2 experiments. (b) Cartoon depicts individual versus clustered A. graevenitzii microcolonies in co‐culture with S. aureus measured in (c). Exclusion edge radius measurement is shown in red. (c) Scatterplot shows no increase in the exclusion edge radius with the increased A. graevenitzii colony size. N = 161 individual colonies scored. N = 89 clustered colonies scored
3.5. Co‐culture of S. aureus and A. graevenitzii modulates innate immune responses
Given that co‐culture with oral flora like A. graevenitzii appeared to suppress S. aureus growth, we hypothesized that this interaction might also modulate innate immune responses to S. aureus. To test this, we measured neutrophil recruitment to co‐culture of S. aureus and A. graevenitzii compared to monocultures of S. aureus or A. graevenitzii alone. As previously reported (Ellett et al., 2019), S. aureus monocultures induced robust recruitment of neutrophils into the culture chamber (Figure 6b). This response was significantly blunted for co‐cultures containing A. graevenitzii (Figure 6b). Interestingly, the recruitment of neutrophils to co‐cultures was also lower compared to A. graevenitzii monocultures (Figure 6b).
FIGURE 6.

Modulation of human neutrophil responses to co‐cultures of Actinomyces graevenitzii and Staphylococcus aureus. (a) Representative micrograph showing the experimental setup for testing neutrophil recruitment to co‐cultures of A. graevenitzii FO582 and S. aureus. (b) Bubble plots showing human neutrophil recruitment toward S. aureus and A. graevenitzii, alone and in co‐culture for different loading ratios. The diameter of each bubble represents the number of neutrophils inside each microchamber at the end of the experiment. N = 18 chambers scored per condition from 2 independent experiments
4. DISCUSSION
We utilized PDMS microfluidic devices to observe microbial mono‐ and co‐cultures of oral isolates in nanoliter volumes. The gas permeability of PDMS facilitates the loading of the dead‐end chamber through a single channel by applying vacuum, while the coverslip provides optical clarity for imaging approaches and a physical surface on which the microbes can grow. One limitation of the microfluidic chamber design used is the inability to rapidly fix and stain cells to gain a more detailed understanding of their structure, due to the excessive time taken for fixatives and labeling compounds to diffuse through the connecting channel. Nonetheless, this phenomenon provides clear visualization of an antagonistic relationship between competing bacterial species and may provide some insight regarding community structures in the oral cavity.
Detailed microscopy revealed the formation of defined “exclusion zones” surrounding A. graevenitzii microcolonies when co‐cultured with S. cristatus or S. salivarius but not S. oralis or S. mitis. Additionally, exclusion zones were not observed with S. odontolytica or A. naeslundii in co‐culture with any Streptococcal species tested, suggesting that the phenomenon exhibits a degree of species specificity.
Dissecting the complexity of microbial community structure and its role in the health and disease in the oral cavity remains a focus of ongoing research. Physical and metabolic characteristics facilitate successful colonization of various oral surfaces and ongoing survival in this complex and dynamic environment (Kolenbrander et al., 2010). Many oral microbes have proven challenging to culture, often because they require the presence of one or more species in consortia to process specific metabolites (Vartoukian et al., 2016). In addition to such relationships, the microbial community structure is also dictated by antagonism driven by competition for space and nutrients (Nadell et al., 2016).
It remains unclear how the formation of exclusion zones relates to readouts from alternative co‐culture methods; however, it is worth noting that they did not form between A. naeslundii and S. salivarius, which do not co‐aggregate in suspension (Kitada & Oho, 2012). Spatial organization of Actinomyces and Streptococci in vivo suggest that these genera often coexist in proximity (Mark Welch et al., 2016); however, these observations provide limited information at species resolution. Sequencing approaches suggest that A. graevenitzii and S. salivarius both reside on the tongue dorsum (Mark Welch et al., 2019); however, this approach provides limited spatial resolution.
Formation of exclusion zones around A. graevenitzii microcolonies was also observed in co‐culture with a GFP‐expressing strain of Staphylococcus aureus, allowing us to perform a detailed analysis of exclusion zone formation using automated imaging approaches. The key factor influencing the outcome appears to be the number of A. graevenitzii microcolonies present in the co‐culture, with an increased number of microcolonies resulting in increased suppression of S. aureus growth. The ratio of bacteria inoculated into the chamber did not significantly affect exclusion zone formation, likely because the rapid doubling time of S. aureus overcame any initial difference.
The compact A. graevenitzii microcolony morphology observed in co‐culture may reflect a state of stress for the bacterium, which might also be related to the formation of exclusion zones. Microcolonies under less stress might exhibit more extensive radial filamentous growth (as observed in mono‐culture), while colonies under more stress might exhibit restricted filamentous growth, resulting in the formation of a larger dense “core” region.
While it appears that the exclusion zone represents a physical space containing no living bacteria, it is unclear whether this is due to physical exclusion for matrix deposition, suppression of proliferation in this area by quorum signaling, or active killing of invading cells by a toxic metabolite. While the formation of exclusion zones only occurred between Actinomyces graevenitzii with S. cristatus, S. salivarius, and S. aureus, it is unlikely that the formation of a simple physical barrier explains the interactions. Additionally, the kinetics of S. aureus coverage in co‐culture with A. graevenitzii, which appear to show an initial increase followed by a decrease, suggest that the formation of the exclusion zone involves the death of existing S. aureus cells in that area. Importantly, macroscopic co‐culture on solid media did not result in any visible cross‐inhibition between A. graevenitizii and S. aureus colonies, highlighting the importance of small‐volume culture and high‐resolution analysis for identification of such interactions.
These interactions are particularly interesting in the context of well‐known opportunistic pathogens such as S. aureus. Modulation of inflammatory responses in these experiments may be a direct response to compounds released by the bacteria during co‐culture or may simply follow from suppression of S. aureus proliferation, which we previously demonstrated to be important for effective neutrophil recruitment (Ellett et al., 2019). It is also possible that the two species become less active / more quiescent, and thus, they stimulate neutrophils less than exponentially growing monocultures.
In cases where S. aureus is identified in oral lesions, it is often isolated in the company of other opportunistic pathogens such as Candida albicans (Ohman et al., 1986), infections with which are generally associated with loss of microbiome stability. Thus, the exclusion of S. aureus by A. graevenitzii builds on the concept that established and stable commensal communities are important to prevent the colonization of a niche by pathogens.
Our results support a model in which the formation of an exclusion zone is triggered by the interaction of specific species in proximity. Low‐volume techniques such as microchambers and droplet‐based microfluidics may enhance quorum sensing and competition for nutrients (Boedicker et al., 2009), better mimicking in vivo conditions. Microfluidic approaches overcome some limitations of traditional bulk suspension co‐culture approaches, which provide limited spatiotemporal information regarding interspecies interactions and often simply result in domination by the faster‐growing species.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTION
Fatemeh Jalali: Data curation (lead); Investigation (lead); Methodology (equal); Writing‐original draft (supporting). Felix Ellett: Conceptualization (equal); Formal analysis (lead); Investigation (supporting); Methodology (equal); Supervision (supporting); Visualization (lead); Writing‐original draft (lead); Writing‐review & editing (equal). Pooja Balani: Methodology (equal); Resources (equal). Margaret J. Duncan: Conceptualization (equal); Funding acquisition (equal); Resources (equal); Writing‐review & editing (equal). Floyd E. Dewhirst: Conceptualization (equal); Funding acquisition (equal); Resources (equal); Writing‐review & editing (equal). Gary G. Borisy: Conceptualization (equal); Funding acquisition (equal); Resources (equal); Supervision (equal); Writing‐review & editing (equal). Daniel Irimia: Conceptualization (equal); Funding acquisition (equal); Project administration (equal); Supervision (equal); Writing‐review & editing (equal).
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
This project was supported by grants from the National Institutes of Health, National Institute of Dental and Craniofacial Research (DE024468 and DE022586), National Institute of General Medical Sciences (GM092804), and Shriners Hospital for Children. Microfluidic devices were fabricated at the Massachusetts General Hospital BioMEMS Resource Center, supported by a grant from the National Institute of Biomedical Imaging and Bioengineering (EB002503).
APPENDIX A.
FIGURE A1.

Culture of Staphylococcus aureus in chambers with different volumes in the presence and absence of Actinomyces graevenitzii. (a–d) Chamber designs with a 200 µm outer chamber and inner chambers of 10 µm (a), 30 µm (b), 50 µm (c), and 100 µm (d). (e) Growth of S. aureus in chambers of different height, in the presence (upper panels) and absence (lower panels) of A. graevenitzii FO582 compared to growth observed in the 50 µm design (Figure 1). The growth of S. aureus does not cover the entire area in chambers with heights lower than 50 µm. (f) Graph shows the measurements of S. aureus coverage in the presence and absence of A. graevenitzii in different height chambers. No difference is observed in shallow chambers where S. aureus growth is already restricted by the small volume. A comparison of 50 and 100 µm high chambers demonstrated that smaller exclusion zones are generated in larger volume co‐cultures. Error bars: mean ± SD, n ≥ 5 chambers scored per condition. One‐way ANOVA with Tukey's multiple comparison test
FIGURE A2.
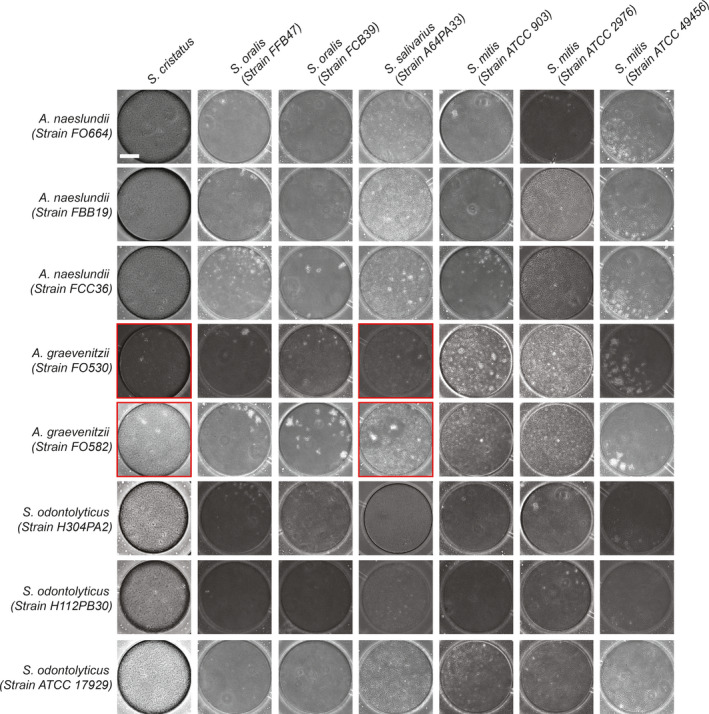
Co‐culture of Actinomyces and Schaalia with streptococcal species in microchambers. Representative images from microfluidic co‐culture experiments, supplemental to Figure 2a. Scale bar: 50 µm
FIGURE A3.
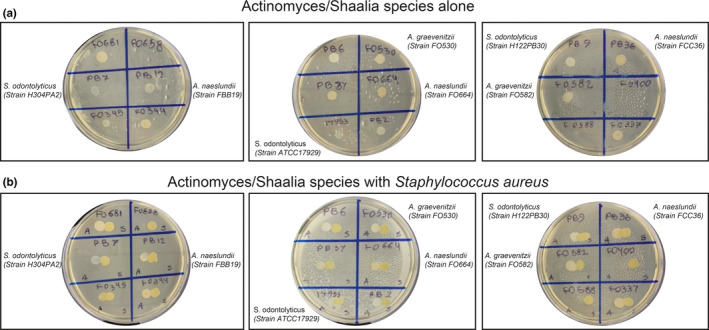
Macroscopic co‐culture of Actinomyces and Schaalia species with Staphylococcus aureus. (a) Spot monocultures of Actinomyces and Schaalia species on BHI agar plates. 2/18 strains failed to grow. (b) Co‐culture of actinomyces or Schaalia species with Staphylococcus aureus (Strain SH1000‐GFP) did not show macroscopic cross‐species inhibition
Fatemeh Jalali and Felix Ellett Contributed equally.
Contributor Information
Felix Ellett, Email: fellett@mgh.harvard.edu.
Daniel Irimia, Email: dirimia@mgh.harvard.edu.
DATA AVAILABILITY STATEMENT
All data are included in this published article.
REFERENCES
- Arzmi, M. H. , Alnuaimi, A. D. , Dashper, S. , Cirillo, N. , Reynolds, E. C. , & McCullough, M. (2016). Polymicrobial biofilm formation by Candida albicans, Actinomyces naeslundii, and Streptococcus mutans is Candida albicans strain and medium dependent. Medical Mycology, 54(8), 856–864. [DOI] [PubMed] [Google Scholar]
- Boedicker, J. Q. , Vincent, M. E. , & Ismagilov, R. F. (2009). Microfluidic confinement of single cells of bacteria in small volumes initiates high‐density behavior of quorum sensing and growth and reveals its variability. Angewandte Chemie (International Ed. in English), 48(32), 5908–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, R. , van der Mei, H. C. , & Busscher, H. J. (1996). Co‐adhesion of oral microbial pairs under flow in the presence of saliva and lactose. Journal of Dental Research, 75(2), 809–815. [DOI] [PubMed] [Google Scholar]
- Chambers, H. F. , & Deleo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature Reviews Microbiology, 7(9), 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar, J. O. , Kolenbrander, P. E. , & McIntire, F. C. (1979). Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii . Infection and Immunity, 24(3), 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen, G. , Linde, A. , Moller, A. J. , & Ohman, A. (1982). A retrospective study of microbiologic samples from oral mucosal lesions. Oral Surgery, Oral Medicine, Oral Pathology, 53(3), 250–255. [DOI] [PubMed] [Google Scholar]
- Dige, I. , Nilsson, H. , Kilian, M. , & Nyvad, B. (2007). In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. European Journal of Oral Sciences, 115(6), 459–467. [DOI] [PubMed] [Google Scholar]
- Dige, I. , Raarup, M. K. , Nyengaard, J. R. , Kilian, M. , & Nyvad, B. (2009). Actinomyces naeslundii in initial dental biofilm formation. Microbiology, 155(Pt 7), 2116–2126. [DOI] [PubMed] [Google Scholar]
- Ellett, F. , Jalali, F. , Marand, A. L. , Jorgensen, J. , Mutlu, B. R. , Lee, J. et al (2019). Microfluidic arenas for war games between neutrophils and microbes. Lab on a Chip, 19(7), 1205–1216. 10.1039/C8LC01263F [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J. , Hwang, G. , Liu, Y. , Gao, L. , Kilpatrick‐Liverman, L. , Santarpia, P. et al (2016). l‐Arginine modifies the exopolysaccharide matrix and thwarts Streptococcus mutans outgrowth within mixed‐species oral biofilms. Journal of Bacteriology, 198(19), 2651–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselman, M. C. , Odoni, D. I. , Ryback, B. M. , de Groot, S. , van Heck, R. G. , Keijsers, J. , Kolkman, Pim , Nieuwenhuijse, David , van Nuland, Y. M. , Sebus, E. , Spee, R. , de Vries, H. , Wapenaar, M. T. , Ingham, C. J. , Schroën, K. , Vítor, A. P. , dos Santos, M. , Spaans, S. K. , Hugenholtz, Floor , & van Passel, M. W. J. (2012). A multi‐platform flow device for microbial (co‐) cultivation and microscopic analysis. PLoS One, 7(5), e36982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatrou, I. A. , Legakis, N. , Ioannidou, E. , & Patrikiou, A. (1988). Anaerobic bacteria in jaw cysts. British Journal of Oral and Maxillofacial Surgery, 26(1), 62–69. [DOI] [PubMed] [Google Scholar]
- Jakubovics, N. S. , Gill, S. R. , Iobst, S. E. , Vickerman, M. M. , & Kolenbrander, P. E. (2008). Regulation of gene expression in a mixed‐genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii . Journal of Bacteriology, 190(10), 3646–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada, K. , & Oho, T. (2012). Effect of saliva viscosity on the co‐aggregation between oral streptococci and Actinomyces naeslundii . Gerodontology, 29(2), e981–e987. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. (1995). Coaggregations among oral bacteria. Methods in Enzymology, 253, 385–397. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. (2000). Oral microbial communities: Biofilms, interactions, and genetic systems. Annual Review of Microbiology, 54, 413–437. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , & Andersen, R. N. (2000). Coaggregation and coadhesion in community structure and co‐operation in biofilms. Water Research, 59, 65. [Google Scholar]
- Kolenbrander, P. E. , Andersen, R. N. , & Moore, L. V. (1990). Intrageneric coaggregation among strains of human oral bacteria: Potential role in primary colonization of the tooth surface. Applied and Environment Microbiology, 56(12), 3890–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , Palmer, R. J. Jr , Periasamy, S. , & Jakubovics, N. S. (2010). Oral multispecies biofilm development and the key role of cell‐cell distance. Nature Reviews Microbiology, 8(7), 471–480. [DOI] [PubMed] [Google Scholar]
- Krishna, S. , & Miller, L. S. (2012). Host‐pathogen interactions between the skin and Staphylococcus aureus . Current Opinion in Microbiology, 15(1), 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin‐Sparenberg, E. , Shin, J. M. , Hastings, E. M. , Freeland, M. , Segaloff, H. , Rickard, A. H. , & Foxman, B. (2016). High‐throughput quantitative method for assessing coaggregation among oral bacterial species. Letters in Applied Microbiology, 63(4), 274–281. [DOI] [PubMed] [Google Scholar]
- Mark Welch, J. L. , Dewhirst, F. E. , & Borisy, G. G. (2019). Biogeography of the oral microbiome: The site‐specialist hypothesis. Annual Review of Microbiology, 73, 335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch, J. L. , Rossetti, B. J. , Rieken, C. W. , Dewhirst, F. E. , & Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences of the United States of America, 113(6), E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell, C. D. , Drescher, K. , & Foster, K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nature Reviews Microbiology, 14(9), 589–600. [DOI] [PubMed] [Google Scholar]
- Nouioui, I. , Carro, L. , García‐López, M. , Meier‐Kolthoff, J. P. , Woyke, T. , Kyrpides, N. C. , Pukall, R. , Klenk, H.‐P. , Goodfellow, M. , & Göker, M. (2018). Genome‐based taxonomic classification of the phylum Actinobacteria. Frontiers in Microbiology, 9, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad, B. , & Kilian, M. (1987). Microbiology of the early colonization of human enamel and root surfaces in vivo. Scandinavian Journal of Dental Research, 95(5), 369–380. [DOI] [PubMed] [Google Scholar]
- Nyvad, B. , & Kilian, M. (1990). Comparison of the initial streptococcal microflora on dental enamel in caries‐active and in caries‐inactive individuals. Caries Research, 24(4), 267–272. [DOI] [PubMed] [Google Scholar]
- Ochiai, K. , Kurita‐Ochiai, T. , Kamino, Y. , & Ikeda, T. (1993). Effect of co‐aggregation on the pathogenicity of oral bacteria. Journal of Medical Microbiology, 39(3), 183–190. [DOI] [PubMed] [Google Scholar]
- Ohman, S. C. , Dahlen, G. , Moller, A. , & Ohman, A. (1986). Angular cheilitis: A clinical and microbial study. Journal of Oral Pathology, 15(4), 213–217. [DOI] [PubMed] [Google Scholar]
- Palmer, R. J. Jr , Gordon, S. M. , Cisar, J. O. , & Kolenbrander, P. E. (2003). Coaggregation‐mediated interactions of streptococci and actinomyces detected in initial human dental plaque. Journal of Bacteriology, 185(11), 3400–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kerner, A. , Burns, M. A. , & Lin, X. N. (2011). Microdroplet‐enabled highly parallel co‐cultivation of microbial communities. PLoS One, 6(2), e17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster, B. J. , Olsen, I. , Aas, J. A. , & Dewhirst, F. E. (2000). The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000, 42(1), 80–87. [DOI] [PubMed] [Google Scholar]
- Postollec, F. , Norde, W. , de Vries, J. , Busscher, H. J. , & van der Mei, H. C. (2006). Interactive forces between co‐aggregating and non‐co‐aggregating oral bacterial pairs. Journal of Dental Research, 85(3), 231–234. [DOI] [PubMed] [Google Scholar]
- Rigby, K. M. , & DeLeo, F. R. (2012). Neutrophils in innate host defense against Staphylococcus aureus infections. Seminars in Immunopathology, 34(2), 237–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam, A. R. , & Hammoudeh, M. (1995). Staphylococcus aureus triggered reactive arthritis. Annals of the Rheumatic Diseases, 54(2), 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. J. , Jackson, M. S. , & Bagg, J. (2001). The ecology of Staphylococcus species in the oral cavity. Journal of Medical Microbiology, 50(11), 940–946. [DOI] [PubMed] [Google Scholar]
- Stewart, E. J. (2012). Growing unculturable bacteria. Journal of Bacteriology, 194(16), 4151–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, S. Y. , Davis, J. S. , Eichenberger, E. , Holland, T. L. , & Fowler, V. G. Jr (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clinical Microbiology Reviews, 28(3), 603–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian, S. R. , Adamowska, A. , Lawlor, M. , Moazzez, R. , Dewhirst, F. E. , & Wade, W. G. (2016). In vitro cultivation of ‘unculturable’ oral bacteria, facilitated by community culture and media supplementation with siderophores. PLoS One, 11(1), e0146926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim, H. F. , Melles, D. C. , Vos, M. C. , van Leeuwen, W. , van Belkum, A. , Verbrugh, H. A. , & Nouwen, J. L. (2005). The role of nasal carriage in Staphylococcus aureus infections. Lancet Infectious Diseases, 5(12), 751–762. [DOI] [PubMed] [Google Scholar]
- Wilbert, S. A. , Mark Welch, J. L. , & Borisy, G. G. (2020). Spatial ecology of the human tongue dorsum microbiome. Cell Reports, 30(12), 4003–4015.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in this published article.


