Abstract
The capacity for macrophages to polarize into distinct functional activation states (e.g., M1, M2) is critical to tune an inflammatory response to the relevant infection or injury. Alternative or M2 polarization of macrophages is most often achieved in vitro in response to IL-4/IL-13 and results in the transcriptional up-regulation of a constellation of characteristic M2 marker genes. In vivo, additional signals from the inflammatory milieu can further increase or decrease M2 marker expression. Particularly, activation of cAMP-generating G protein-coupled receptors is reported to increase M2 markers, but whether this is strictly dependent upon cAMP production is unclear. We report herein that increased cAMP alone can increase IL-4-dependent M2 marker expression through a PKA/C/EBPβ/CREB dependent pathway in murine macrophages.
Keywords: Adenylate cyclase, alternative activation, cAMP, IL-4, M2 macrophages
Introduction
For many years, a paradigm of macrophage activation has evolved that proposed that macrophages can be differentially activated in a stimulus-dependent fashion. The notion of functional “plasticity” of macrophages as a mechanism by which these cells adapt to environmental changes was proposed by Stout and Suttles1 and has since been extensively studied. The two major activation states, known as “Classically Activated” (or M1), and “Alternatively Activated” (or M2), have been extensively characterized.2 Classically activated macrophages are typically characterized by the ability to produce an array of pro-inflammatory cytokines (e.g., TNF-α, IL-6) and are typically differentiated in vitro by stimulating naive macrophages with microbial constituents, such as LPS, and the Th1 cytokine IFN-γ. In contrast, alternatively activated (M2) macrophages, originally described by Martinez et al.,3 are commonly generated following stimulation of cells with the Th2 cytokines IL-4 or IL-13 and do not generate inflammatory cytokines. Rather, M2 macrophages up-regulate certain markers including CD206 (mannose receptor), arginase-1 (Arg1), and others.2 As an additional distinction, M1 macrophages obtain energy through glycolysis, whereas M2 macrophages use oxidative metabolism to fuel their functions.4 In vivo, M1 macrophages predominate in conditions of bacterial, protozoan, and virus infection where they are necessary to clear the pathogen, but may result in tissue damage.5 In contrast, M2 macrophages are abundant during helminth infections or in allergic airway diseases such as asthma,6 and are important for tissue repair in response to certain infectious agents.7 However, this simplistic differentiation model suffers from the fact that functionally distinct subsets of M1 and M2 macrophages have been identified. These subsets differ further with respect to the source of macrophages being studied, including their ontogeny, the stimuli used, the transcription factors activated, the cytokines and chemokines up-regulated, and other physiological and metabolic changes that are not always conserved between mice and humans,2,8,9 and multiple reviews of the activated macrophage transcriptomes and the associated transcription factors have been published.10,11 Of significance for this study, there is evidence that macrophage activation phenotypes may change upon exposure to multiple, as compared with single, stimuli.12
In vitro, M2 marker expression occurs rapidly following stimulation of macrophages with exogenous IL-4 or IL-13 and requires activation of the transcription factor STAT6 which is activated downstream of the IL-4/IL-13 receptor.13 However, unlike in vitro experimental systems, macrophages in vivo are often exposed to simultaneous stimulation through a number of receptors in addition to IL-4/IL-13R, and how the combinatorial effects of these multiple signals modulate M2 marker expression and polarization is much less studied. In particular, multiple G protein-coupled receptors (GPCRs) that share the ability to activate cellular cAMP production, including prostaglandin receptors, adenosine receptors, and atypical chemokine receptors, have all been shown to enhance M2 marker expression in different contexts.14,15 However, the specific role of cAMP production, if any, in this effect has not been previously defined. In this study, we specifically address the role of cAMP in regulating M2 marker expression in primary murine macrophages. Our data show that increased cAMP augments IL-4-mediated M2 marker expression, and we identify specific transcription factors that contribute to this synergistic effect. Our data support the hypothesis that simultaneous exposure of macrophages to IL-4 and agents that augment intracellular cAMP may be a relevant mechanism to tune macrophage polarization in vivo.
Materials and methods
Reagents
Protein-free Escherichia coli K235 LPS (< 0.008% protein) was prepared by modification of the hot phenol–water extraction method described previously.16 Anti-β-actin, anti-IRF-4, anti-PPARγ, anti-pCREB, and anti-pSTAT6 Abs were from Cell Signaling (Beverly, MA, USA), and total anti-arginase-1 Ab from Invitrogen (Carlsbad, CA, USA). Anti-Stat6, were obtained from EMD Millipore (MA). H-89 and 8-bromo cAMP were obtained from Sigma Aldrich (St Louis, MO). Adenylate Cyclase Toxin was a gift from Dr Erik Hewlett and was purified as described previously.17
Cell culture
Primary murine peritoneal macrophages were obtained by peritoneal lavage from wild type (WT) 6 to 8-wk-old C57BL/6J or BALB/cByJ mice (The Jackson Laboratory, Bar Harbor, ME), IRF-4−/−, PPARγ−/−, and Stat6−/− mice 4 d after i.p. injection with sterile thioglycollate as described previously.18 Stat6−/− mice were kindly provided by Dr Achsah Keegan, University of Maryland, Baltimore (UMB) (on a BALB/c background), IRF-4−/− macrophages were kindly provided by Dr Tak Mak (University of Toronto), were on a C57BL/6 background. PPARγ−/− mice were provided by Dr. Mary Jane Thomassen (East Carolina University) and were on a C57BL/6 background. Macrophages were cultured in RPMI supplemented with 2% FCS, 2 mM glutamine, penicillin, and streptomycin as described previously.19 The mouse macrophage-like cell line, RAW 264.7, was purchased from the American Type Culture Collection (Manassas, VA). Rh CREB1 RAW 264.7 cells20 were obtained from Dr Ian Fraser (NIH), and C/EBPβ+/+ and C/EBPβ–/– immortalized macrophages were the kind gift of Dr Dhan Kalvakolanu, (UMB) and were generated as described.21 RAW 264.7 macrophages and CREB1 stable knock-down cells were cultured in DMEM (BioWhittaker), supplemented with 10% (vol/vol) FBS (HyClone Laboratories), glutamine (2 mM), penicillin (10,000 U/ml), and streptomycin (10,000 µg/ml) at 37°C in 5% CO2 in air.
Cell stimulation
Primary murine macrophages, immortalized bone marrow-derived macrophages, and RAW 264.7 cells were cultured (2 × 106 cells/well) in 12-well plates. After overnight incubation, medium was replaced by fresh medium and cells were stimulated with medium only or IL-4 (40 ng/ml) in the absence or presence of cyclic AMP-activating reagents for the times indicated.
Western analysis
Macrophages were washed with PBS and then lysed in buffer (1% Triton X-100, 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 10 mM TRIS with protease inhibitor cocktail (Roche) and 1 mM sodium vanadate), and boiled for 5 min with Laemmli lysis buffer for SDS-PAGE and Western analysis. Twenty µg of total protein in Laemmli buffer was boiled for 5 min, resolved by 10% SDS-PAGE in Tris/glycine/SDS buffer (25 mM Tris, 250 mM glycine, 0.1% SDS) from Bio-Rad (Hercules, CA), and then electro-transferred onto Immobilon-P transfer membranes (Millipore, Bedford, MA) at 100 V for 1.5 h (4°C). After blocking for 1 h in TBS-T (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat milk, membranes were washed three times in TBS-T and probed for 20 h at 4°C with the respective Abs, according to the manufacturer’s instructions. Following washing in TBS-T, membranes were incubated with secondary HRP-conjugated, anti-rabbit IgG or anti-mouse IgG from Cell Signaling (1: 2,000 dilution) for 1 h at room temperature, washed three times in TBS-T, and bands were detected using ECL Plus reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Measurement of steady-state mRNA by quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent from Invitrogen (Carlsbad, CA, USA) as specified by the manufacturer’s instructions and quantified by spectrophotometric analysis. The cDNA was prepared from 1 μg2 of total RNA using iScript reverse transcriptase (Bio-Rad, Hercules, CA, USA) and both poly-oligo dT and random primer mix, as recommended by the manufacturer’s instructions. The resulting cDNA was quantified by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and ABI Prism 7900HT cycler as described previously (reference). Primers for detection of arginase 1,19 mannose receptor (MR), Mgl2, and GAPDH (housekeeping gene) mRNAs were designed using the Primer Express 2.0 program (Applied Biosystems). Asterisks represent a significant difference between the gene expression levels of the samples by t-test (P < 0.05). Primer sequences used in this study: arginase-1 sense (5_-CAGAAGAATGGAAGAGTCAG-3′) and antisense (5-CAGATATGCAGGCAGGGAGTCACC-3′); Man receptor, sense (5-GATATGAAGCCATGTACTCCTTACTGG-3′) and antisense (5-GGCAGAGGTGCAGTCTGCAT-3′); Mgl2 sense (5-AGGCACCCTAAGAGCCATTT-3′) and antisense (5-CCCTCTTCTCCAGTGTGCTC-3′); CREB sense (5-CCAGTCTCCACAAGTCCAAACAG-3′) and antisense (5-GGCACTGTTACAGTGGTGATGG-3′); IL-10 sense (5-ATTTGAATTCCCTGGGTGAGAAG-3′) and antisense (5-CACAGGGGAGAAATCGATGACA-3′); IL-12 p40 sense (5-TCTTTGTTCGAATCCAGCGC-3′) and antisense (5-GGAACGCACCTTTCTGGTTACA-3′); and GAPDH sense (5-AGCCTC GTCCCGTAGACAAAAT-3′) and antisense (5-TGGCAACAATCTCCACTTTGC-3′).
Results
Effect of cAMP-regulated pathways on the induction of alternatively activated (M2) macrophage markers
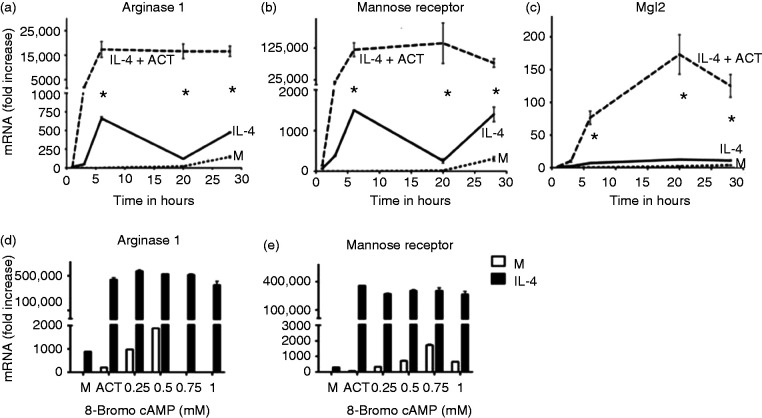
To define the role of cAMP-responsive pathways in macrophage polarization, thioglycollate-elicited peritoneal macrophages were stimulated with rIL-4 to elicit transcription of alternative activation genes in the absence or presence of the cAMP-generating adenylate cyclase toxin (ACT) purified from Bordetella pertussis. Use of ACT is an efficient way to rapidly elevate cytosolic cAMP levels without activating host cell enzymes.17 Figure 1a shows that IL-4-induced expression of mRNA for genes encoding the M2 markers arginase 1 (Arg1), MR, and Mgl2 were greatly increased in the presence of ACT, suggesting that cAMP-responsive pathways may positively regulate key genes of the M2 transcriptional program. To confirm a role for cAMP in the regulation of these M2-associated genes, macrophages were stimulated with IL-4 in the absence or presence of increasing concentrations of the cell-permeable cAMP analog 8-bromo-cAMP (Figure 1b). 8-Bromo-cAMP enhanced IL-4-dependent transcription of Arg1 and MR mRNA to a degree comparable to that of ACT treatment. Thus, increased intracellular cAMP augments cytokine-inducible mRNA expression of these M2 markers.
Figure 1.
cAMP regulates induction of alternatively activated (M2) macrophage markers. (a-c) Peritoneal macrophages were stimulated with media alone (M), or rIL-4 (40 ng/ml) in the absence or presence of adenylate cyclase toxin (ACT) (10 nM) for the indicated time intervals and mRNA was measured by qRT-PCR. for Arginase 1, MR, or Mgl2. (d-e) Peritoneal macrophages were stimulated with rIL-4 (40 ng/ml) in the absence or presence of ACT or increasing concentrations of 8-bromo-cAMP (0.25-1mM) and mRNA was measured by qRT-PCR. for Arg 1 and MR. Results are represented as mean ± SEM from three experiments.
Role of cAMP-PKA pathway in regulation of alternative activation markers in macrophages
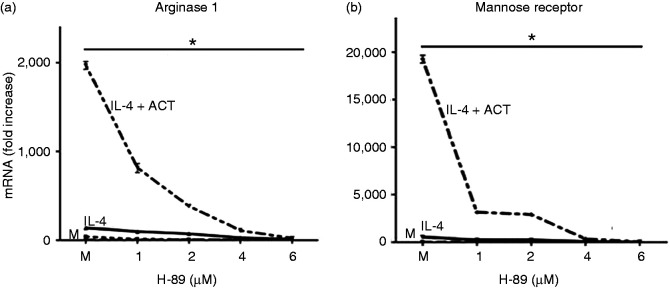
In macrophages, intracellular cAMP potently activates the serine/threonine kinase protein kinase A (PKA) by directly interacting with the PKA regulatory subunits.22 Activated PKA has been shown to affect transcription in a number of cellular contexts by direct phosphorylation of transcription factors.23 We hypothesized that if PKA activity were contributing to the observed effects of cAMP on M2 gene transcription, the PKA inhibitor, H-89, may reverse the synergistic effects on M2 markers induced by cAMP-activating agents in IL-4-stimulated macrophages. The PKA catalytic antagonist, H-89, suppressed the elevated transcription of Arg1 and MR mRNA induced by the combination of IL-4 + ACT in a concentration-dependent manner (Figure 2a and 2b). These data suggest a role for the PKA pathway in the cAMP-increased expression of these two alternative activation markers.
Figure 2.
Role of cAMP-PKA pathway in regulation of alternatively activated (M2) markers in macrophages. (a,b) Peritoneal macrophages were stimulated for 6 h with media alone (M) or rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM), without or with the PKA inhibitor, H-89, at the indicated concentration. mRNA was measured by qRT-PCR for Arg1 (a) and MR (b). Results are represented as mean ± SEM from three experiments.
Role of transcription factors IRF-4 and STAT6 in the synergistic regulation of M2 markers by IL-4 and cAMP
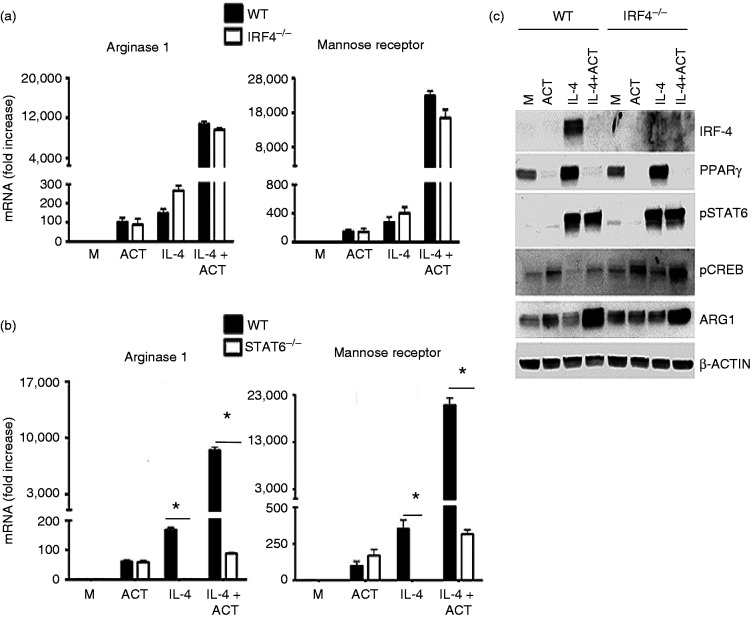
Helminth infection of mice has been shown to elicit M2 markers, the expression of which was reported to be regulated by the transcription factor IRF-4,24 and cAMP has been shown to positively regulate IRF-4 in adipocytes.25 To test the possible role of IRF-4 in the transcriptional effects of cAMP on M2 marker induction, macrophages from WT C57BL/6J and IRF-4−/− mice were compared for induction of M2 markers after treatment with IL-4 in the absence or presence of ACT. IL-4 induced Arg1 and MR mRNA comparably in WT and IRF-4−/− macrophages (Figure 3a). ACT enhanced IL-4-induced Arg1 and MR mRNA levels to the same extent in WT and IRF-4−/− macrophages, arguing against a role for IRF-4 in the action of cAMP (Figure 3a). As expected, M2 markers Arg1 and MR were not induced by IL-4 treatment in STAT6−/− macrophages (Figure 3b). To confirm that elevated cAMP did not confer STAT6-independent M2 marker induction, WT and STAT6−/− macrophages were stimulated with IL-4 +/− ACT treatment. Synergistic induction of Arg 1 and MR mRNA by IL-4 and ACT required expression of STAT6 (Figure 3b).
Figure 3.
cAMP regulation of alternatively activated (M2) markers in macrophages is independent of IRF-4, but STAT6-dependent. (a) WT (solid bars) and IRF-4−/− (empty bars) peritoneal macrophages were stimulated for 6 h with media alone (M) or rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM) and Arg1 and MR mRNA was measured by qRT-PCR. (b) WT (solid bars) and STAT6−/− (empty bars) peritoneal macrophages were stimulated for 6 h with media alone (M) rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM) and Arg1 and MR mRNA was measured by qRT-PCR. P < 0.01. (c) Whole-cell lysates were prepared from macrophages after stimulation with rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM) for 6 h. These protein lysates were subjected to Western analysis using anti-IRF-4, anti-PPARγ, anti-phospho-STAT-6, anti-phospho-CREB, anti-Arg1, and anti-β-actin Abs. The β-actin was used as a loading control. Data shown are representative of two separate experiments. Results are represented as mean ± SEM from three experiments.
Total cell lysates extracted from macrophages treated with IL-4 in the absence or presence of ACT were subjected to Western analysis (Figure 3c). IL-4-induced IRF-4 was significantly increased in WT macrophages; however, IRF-4 accumulation was markedly reduced in the presence of ACT. Similarly, levels of PPARγ, a transcriptional regulator known to be induced by IL-4 and required for M2 macrophage differentiation, were similar in IL-4-treated WT and IRF-4−/− macrophages, whereas ACT treatment strongly inhibited IL-4-induced PPARγ in both WT and IRF-4−/− macrophages. Activation of the master M2 transcription factor STAT6, which is phosphorylated downstream of the IL-4 and IL-13 receptors and is required for induction of M2 markers,13 was not affected by ACT treatment. Activation of the transcription factor CREB, known to be activated by cAMP pathways through PKA,23 was increased by ACT in WT macrophages and to a greater extent in IRF-4−/− macrophages (Figure 3c). The synergistic induction of Arg 1 protein induced by IL-4 in the presence of ACT was comparable in WT and IRF-4−/− macrophages, supporting the mRNA data in Figure 3a. The β-actin protein, used as a loading control, was comparable in all samples.
Partial dependence on CREB expression for M2 marker regulation by cAMP
cAMP is a potent activator of PKA, and we have shown a requirement for PKA in the synergistic regulation of M2 markers by IL-4 and 8-bromo-cAMP (Figure 2). One way by which PKA regulates transcription is through direct phosphorylation and activation of the transcription factor CREB.26 Activated CREB binds to a CREB response element (CRE) and is then bound by CREB binding protein that serves as a co-activator, leading to increased or decreased transcription.27,28 CREB has been shown to regulate many genes during macrophage differentiation.15
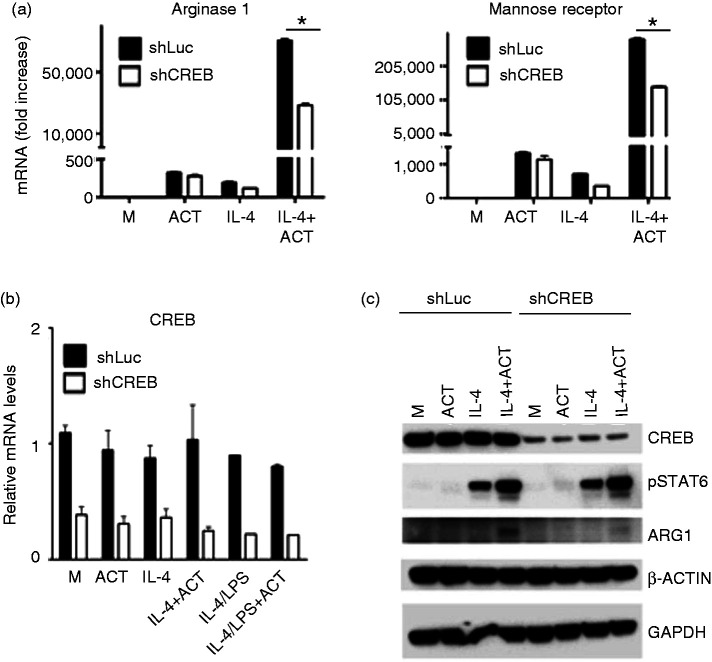
To evaluate the potential role of CREB expression in the synergistic regulation of select M2 markers, we obtained RAW 264.7 macrophages in which CREB expression was knocked-down through stable transfection with control miR-shRNA (luc) or miR-shCreb1a specific for CREB,29 and examined these cells for induction of M2 markers after treating with IL-4 in the absence or presence of ACT. IL-4 treatment of stably transfected control shRNA (Luc) cells resulted in augmented Arg 1 and MR mRNA expression in the presence of ACT. We observed a diminished induction of these same genes in the CREB knock-down cell line, indicating a partial CREB dependence in the synergistic actions of IL-4 and ACT (Figure 4a). CREB mRNA was not affected by treatment in the stably transfected control miR-shRNA (Luc) RAW 264.7 cells; however, CREB mRNA levels were reduced by ∼80% in the stably expressing miR-shRNA Creb1a RAW 264.7 cells in all the treatments (Figure 4b). Although the knock-down in total CREB in our cell lines is significant, it is not complete, and therefore it is possible these experiments under-represent the contribution of CREB. Total lysates were prepared from these cells and subjected to Western analysis. As shown in Figure 4c, CREB protein was detected comparably in the cells transfected with control miR-shRNA (Luc), regardless of treatment, and CREB protein was reduced by ∼80% in the stable cell line expressing miR-shRNA Creb1a. IL-4-induced phospho-STAT6 was comparable in stably expressing control miR-shRNA (luc) or miR-shRNA Creb1a and it was slightly increased in the presence of ACT in both cell lines. No phosphorylation of STAT6 proteins was detected in the lysates with stimulated with medium or ACT, but pSTAT6 was increased by IL-4 treatment, and further increased by IL-4 + ACT. Arg-1 protein was detected in lysates from cells stably transfected with miR-RNA (Luc) treated with IL-4+ACT; however, this protein was detected at much lower amounts in lysates obtained from cells expressing miR-shRNA specific for Creb1a. β-Actin and GAPDH proteins were expressed comparably and used as loading controls.
Figure 4.
M2 marker regulation by cAMP displays partial dependence on CREB expression. (a, b) RAW 264.7 cell lines stably transfected with a control miR-shRNA (Luc) or miR-shRNA specific for Creb1a (shCreb) were stimulated for 6 h with media alone (M) or rIL-4 (40 ng/ml) in the absence or presence ACT (10 nM) and mRNA was measured by qRT-PCR: (a) Arg1 and MR (b) CREB mRNA. (c) Western analysis: whole-cell lysates were prepared from RAW 264.7 cell lines after stimulation with rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM) and were analyzed by Western analysis using with anti-total CREB, anti-phospho-STAT-6, anti-Arg1, and β-actin. β-Actin was used as loading control. Representative of two separate experiments.
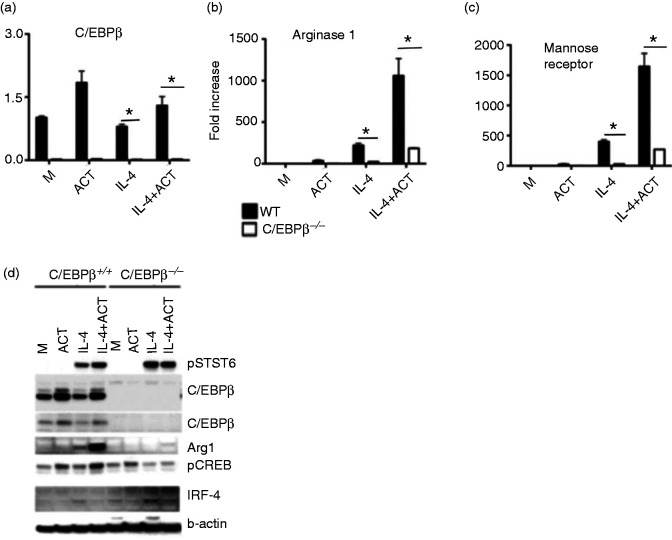
Strong dependence on C/EBPβ expression in the regulation of M2 markers by cAMP and IL-4
There are several reports that the transcription factor C/EBPβ is involved in alternative activation of macrophages. It has been reported that the transcription factor C/EBPβ is proportionately increased along with alternative activation markers in macrophages.30 It has also been shown that activated CREB directly binds to two CRE-binding sites in the promoter of LAP/C/EBPβ and can regulate its transcription.31 We therefore hypothesized that C/EBPβ may have a specific role in IL-4-induced alternative activation in the presence of cAMP. To test the possible role of C/EBPβ in our studies, we compared immortalized bone marrow-derived macrophages from WT and C/EBPβ−/− mice and examined induction of alternatively active markers after treating with IL-4 in the absence or presence of ACT. C/EBPβ mRNA was increased in WT macrophages in the presence of ACT and, as expected, there was no detectable C/EBPβ mRNA in C/EBPβ-deficient macrophages (Figure 5a). This figure also shows that IL-4 stimulation did not enhance C/EBPβ mRNA levels. Figure 5b shows that Arg 1 and MR mRNA were induced by IL-4 in bone marrow-derived immortalized WT macrophages and that expression of both genes was almost completely ablated in C/EBPβ−/− macrophages. IL-4-induced Arg-1 and MR mRNA were synergistically increased in the presence of ACT, but only in WT macrophages (Figure 5b), suggesting that C/EBPβ mediates ACT-induced M2 gene expression. Total lysates from WT and C/EBPβ−/− macrophages were prepared after treatment and subjected to Western analysis. As shown in the Figure 5c, no phosphorylation of STAT6 proteins was detected in the lysates from macrophages stimulated with medium or ACT only, while IL-4-induced p-STAT6 was comparable in WT and C/EBPβ−/− macrophages and was not further enhanced by the presence of ACT. Two forms of C/EBPβ protein were detected in the WT macrophages and were slightly increased by ACT (without or with IL-4); as expected, there was no detectable C/EBPβ protein in C/EBPβ−/− macrophages. IL-4-induced Arg-1 protein was detected in IL-4-treated WT macrophage lysates and was strongly increased in the presence of ACT. However, Arg 1 protein was weakly present in C/EBPβ−/− macrophages. Phospho-CREB was observed in WT macrophages and it was synergistically increased in the presence of IL-4 plus ACT. However, in C/EBPβ−/− macrophages, phospho-CREB was not up-regulated by IL-4 or IL-4+ACT. As we observed earlier, IRF-4 protein was induced in both WT and C/EBPβ−/− macrophages by IL-4 treatment; however, IRF-4 protein was inhibited in the presence of ACT. The β-actin was comparable among lysates and used as loading control.
Figure 5.
Role of C/EBPβ in the regulation of M2 markers by cAMP and IL-4. (a-c) Immortalized bone marrow-derived macrophages derived from WT mice or mice deficient in C/EBPβ were stimulated were stimulated for 6 h with media alone (M) or rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM) and mRNA was measured by qRT-PCR. (a) C/EBPβ, (b) Arg1 and (c) MR (d) Western blot analysis. Whole-cell lysates were prepared from immortalized bone marrow-derived macrophages (C/EBPβ+/+ and C/EBPβ−/−) after stimulation with rIL-4 (40 ng/ml) in the absence or presence of ACT (10 nM) for 6 h and protein lysates subjected to Western analysis using anti-phospho-STAT-6, anti-C/EBPβ, anti-Arg1, anti-phospho-CREB, anti-IRF-4, and anti-β-actin Abs. β-Actin was used as a loading control. Data shown are representative of two separate experiments.
Discussion
Macrophage polarization into distinct functional states (M1 vs. M2) is initiated by cues from the extracellular milieu and enables macrophages to respond appropriately to different classes of pathogens or to sterile injury. Alternative activation of macrophages (M2) can be achieved in vitro by exposure to exogenous cytokine (IL-4/IL-13), but the extent of M2 marker expression can also be “tuned” by the presence of additional secondary signals including those transmitted through cAMP-generating GPCRs. A recent report found that the cAMP analog dibutyryl-cAMP could increase expression of select M2 markers in bone marrow-derived macrophages through unknown transcription factors.32 We have found that elevating levels of intracellular cAMP either by treatment of macrophages with the ACT or by addition of cAMP analog can enhance transcription of a canonical M2 markers in the presence of IL-4/IL-13 without a strict requirement for additional GPCR-regulated signaling pathways. It should be noted that the levels of exogenous cAMP analog used in this study are in line with previous reports.33,34 ACT did not exhibit a clear pattern of effect on known M1 genes during stimulation by LPS + IFN-γ, with some being moderately suppressed (e.g., IL-12 p40) (Supplemental Figure 1A), while others were not affected. We did observe increased IL-4/ACT-dependent transcription of the anti-inflammatory gene, IL-10, in line with reports of IL-10 production from M2 cells.35 Mechanistically, cAMP does not appear to act primarily by enhancing the activity of the key M2 transcription factor STAT6, but rather, likely acts, in part, by driving increased activity of the C/EBPβ/CREB axis utilizing the PKA kinase. Although we observe a significant effect of the H89 PKA inhibitor in our study, it should be noted that we have only utilized this single antagonist that specifically inhibits the catalytic kinase functions of PKA, and a complete knockout in PKA could conceivably produce a distinct phenotype. There may, of course, be additional cAMP-responsive pathways that amplify M2 marker expression such as post-transcriptional stability or epigenetic modifications.36 Our molecular observations suggest that the combinatorial action of surface receptors coupled to adenylate cyclase activation, in conjunction with cytokine receptors (e.g. IL-4/13R) may be a generalizable mechanism for increasing the extent of macrophage M2 polarization.
The potential in vivo relevance of our findings is apparent when considering that several of the GPCRs that couple to GαS complexes and have the capacity to generate cAMP also have ligands with well-described roles in inflammation, including contexts where M2 macrophage polarization is important. Among these GPCRs are the receptors for prostaglandins E2, D2, and I2, adenosine receptors A2a and A2b, and the histamine H2 receptor. For example, both prostaglandin and histamine receptors, in particular, are important mediators of vascular and airway contraction and cytokine responses in the lung during conditions such as allergic asthma in which the extent of M2 macrophage polarization is also critical.37 The abundant production of prostaglandins and histamine in inflamed lungs may be shaping the macrophage polarization profile by regulating intracellular cAMP levels.
Supplemental Material
Supplemental material, sj-pdf-1-ini-10.1177_1753425920975082 for cAMP levels regulate macrophage alternative activation marker expression by Swamy Polumuri, Darren J Perkins and Stefanie N Vogel in Innate Immunity
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH Grants AI123371, AI125215 and AI143845 (SNV).
ORCID iD: Darren J Perkins https://orcid.org/0000-0003-1368-6366
Supplemental material: Supplemental material for this article is available online.
References
- 1.Stout RD, Suttles J. Functional plasticity of macrophages: Reversible adaptation to changing microenvironments. J Leukocyte Biol 2004; 76: 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014; 41: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol 2009; 27: 451–483. [DOI] [PubMed] [Google Scholar]
- 4.Galvan-Pena S, O'Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol 2014; 5: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karp CL, Murray PJ. Non-canonical alternatives: What a macrophage is 4. J Exp Med 2012; 209: 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keegan AD, Shirey KA, Bagdure D, et al. Enhanced allergic responsiveness after early childhood infection with respiratory viruses: Are long-lived alternatively activated macrophages the missing link? Pathog Dis 2016; 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirey KA, Pletneva LM, Puche AC, et al. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Mucosal Immunol 2010; 3: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez FO, Helming L, Milde R, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: Similarities and differences. Blood 2013; 121: e57–e69. [DOI] [PubMed] [Google Scholar]
- 9.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8: 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novoselov VV, Sazonova MA, Ivanova EA, et al. Study of the activated macrophage transcriptome. Exp Mol Pathol 2015; 99: 575–580. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat Rev Immunol 2011; 11: 750–761. [DOI] [PubMed] [Google Scholar]
- 12.Tarique AA, Logan J, Thomas E, et al. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am J Respir Cell Mol Biol 2015; 53: 676–688. [DOI] [PubMed] [Google Scholar]
- 13.Heller NM, Qi X, Junttila IS, et al. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci Signal 2008; 1: ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbera-Cremades M, Baroja-Mazo A, Pelegrin P. Purinergic signaling during macrophage differentiation results in M2 alternative activated macrophages. J Leukocyte Biol 2016; 99: 289–299. [DOI] [PubMed] [Google Scholar]
- 15.Luan B, Yoon YS, Le Lay J, et al. CREB pathway links PGE2 signaling with macrophage polarization. Proc Natl Acad Sci USA 2015; 112: 15642–15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntire FC, Sievert HW, Barlow GH, et al. Chemical, physical, biological properties of a lipopolysaccharide from Escherichia coli K-235. Biochemistry 1967; 6: 2363–2372. [DOI] [PubMed] [Google Scholar]
- 17.Perkins DJ, Gray MC, Hewlett EL, et al. Bordetella pertussis adenylate cyclase toxin (ACT) induces cyclooxygenase-2 (COX-2) in murine macrophages and is facilitated by ACT interaction with CD11b/CD18 (Mac-1). Mol Microbiol 2007; 66: 1003–1015. [DOI] [PubMed] [Google Scholar]
- 18.Dobrovolskaia MA, Medvedev AE, Thomas KE, et al. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: Effects of TLR “homotolerance” versus “heterotolerance” on NF-kappa B signaling pathway components. J Immunol 2003; 170: 508–519. [DOI] [PubMed] [Google Scholar]
- 19.Perkins DJ, Richard K, Hansen AM, et al. Autocrine-paracrine prostaglandin E2 signaling restricts TLR4 internalization and TRIF signaling. Nat Immunol 2018; 19: 1309--1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Li N, Oh KS, et al. Comprehensive RNAi-based screening of human and mouse TLR pathways identifies species-specific preferences in signaling protein use. Sci Signal 2016; 9: ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gade P, Roy SK, Li H, et al. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol Cell Biol 2008; 28: 2528–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghigo A, Mika D. cAMP/PKA signaling compartmentalization in cardiomyocytes: Lessons from FRET-based biosensors. J Mol Cell Cardiol 2019; 131: 112–121. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg D, Groussin L, Jullian E, et al. Role of the PKA-regulated transcription factor CREB in development and tumorigenesis of endocrine tissues. Ann N Y Acad Sci 2002; 968: 65–74. [DOI] [PubMed] [Google Scholar]
- 24.Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010; 11: 936–944. [DOI] [PubMed] [Google Scholar]
- 25.Kong X, Banks A, Liu T, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell 2014; 158: 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delghandi MP, Johannessen M, Moens U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal 2005; 17: 1343–1351. [DOI] [PubMed] [Google Scholar]
- 27.Herzig S, Long F, Jhala US, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001; 413: 179–183. [DOI] [PubMed] [Google Scholar]
- 28.Long F, Schipani E, Asahara H, et al. The CREB family of activators is required for endochondral bone development. Development 2001; 128: 541–550. [DOI] [PubMed] [Google Scholar]
- 29.Wall EA, Zavzavadjian JR, Chang MS, et al. Suppression of LPS-induced TNF-alpha production in macrophages by cAMP is mediated by PKA-AKAP95-p105. Sci Signal 2009; 2: ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamkin DM, Srivastava S, Bradshaw KP, et al. C/EBPbeta regulates the M2 transcriptome in beta-adrenergic-stimulated macrophages. Brain Behav Immun 2019; 80: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niehof M, Manns MP, Trautwein C. CREB controls LAP/C/EBP beta transcription. Mol Cell Biol 1997; 17: 3600–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negreiros-Lima GL, Lima KM, Moreira IZ, et al. Cyclic AMP regulates key features of macrophages via PKA: Recruitment, reprogramming and efferocytosis. Cells 2020; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katakami Y, Nakao Y, Koizumi T, et al. Regulation of tumour necrosis factor production by mouse peritoneal macrophages: The role of cellular cyclic AMP. Immunology 1988; 64: 719–724. [PMC free article] [PubMed] [Google Scholar]
- 34.Matsui T, Nakao Y, Koizumi T, et al. Effect of prostaglandin E2 on gamma-interferon and 1,25(OH)2D3 vitamin D3-induced c-myc reduction during HL-60 cell differentiation. Leuk Res 1988; 12: 597–605. [DOI] [PubMed] [Google Scholar]
- 35.Shirey KA, Cole LE, Keegan AD, et al. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J Immunol 2008; 181: 4159–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perkins DJ, Patel MC, Blanco JC, et al. Epigenetic mechanisms governing innate inflammatory responses. J Interferon Cytokine Res 2016; 36: 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girodet PO, Nguyen D, Mancini JD, et al. Alternative macrophage activation is increased in asthma. Am J Respir Cell Mol Biol 2016; 55: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ini-10.1177_1753425920975082 for cAMP levels regulate macrophage alternative activation marker expression by Swamy Polumuri, Darren J Perkins and Stefanie N Vogel in Innate Immunity