Abstract
Bovine coronavirus (BCoV) is associated with severe diarrhea, dehydration, and depression, which result in significant economic damages in the dairy and beef cattle industries worldwide. However, differences in the gut microbiota structure and their correlations with differing physiological parameters between BCoV-infected calves with diarrhea and recovered calves are not well understood. In this study, fecal specimens were collected from 10 post-weaned calves, before and after 2 months of fluid therapy, and the samples were used for microbiota analysis. Following recovery, the alpha-diversity profiles (observed operational taxonomic units [OTUs], and Chao1, Shannon, and Simpson indices) changed significantly when compared with those of calves with diarrhea. Beta-diversity analysis exhibited significant differences in gut microbiota compositions between calves with diarrhea and those in the recovered state. The abundances of eight phyla and thirteen genera in feces changed markedly after restoration of BCoV diarrhea. In addition, our correlation study clearly revealed that increased abundances of the genera Caproiciproducens, Pseudoflavonifractor, and Oscillibacter negatively correlated with serum glucose, and phosphorus levels, but positively correlated with serum chloride in calves with diarrhea, whereas increased abundances of the genera Peptostreptococcaceae;Clostridium (Clostridium cluster XI), Intestinibacter, Cellulosilyticum, Ruminococcus, Romboutsia, Paeniclostridium, Clostridiaceae;Clostridium and Turicibacter in recovered calves showed the opposite pattern. These results suggest that structural changes of the gut microbiota after recovery from BCoV infection correlate with changes in physiological parameters. In conclusion, our data provide evidence of gut microbiota-composition changes and their correlations with the physical profiles of post-weaned calves, before and after fluid therapy for BCoV-related diarrhea.
Keywords: Bovine coronavirus, Gut microbiota, Physiological parameters, Post-weaned calf
INTRODUCTION
Coronaviruses can cause respiratory and enteric infections in humans and other domestic animals including pigs, chickens, and cattle. Bovine coronavirus (BCoV) is an RNA virus belonging to the Coronaviridae family, and BCoV infection results in the development of long-lasting or severe disease [1]. BCoV infection of cattle is highly prevalent globally, and the virus is recognized as the causative agent of neonatal calf diarrhea, winter dysentery in adult cattle, and respiratory infections in calves and feedlot cattle [2]. BCoV infection raises serious health problems that impede weight gain or milk production in cattle of various ages, leading to large economic damages in the dairy and beef cattle industries worldwide [3]. BCoV infection occur via the respiratory or fecal–oral routes, and BCoV isolates following both routes of infection have shown similar genotypes [4]. After enteric infection, BCoV replicates in the epithelial cells of the small and large intestines, destroys the villi, and causes degeneration and necrosis of the crypt epithelium and petechiae, which collectively interfere with the absorption of electrolytes and water [5]. This process leads to dehydration, electrolyte imbalance (usually decreased sodium level and increased or decreased potassium level), strong ion (metabolic) acidosis, increased production of D-lactate, and a negative energy balance (nutrient malabsorption) [6]. These symptoms induce severe and often bloody diarrhea in calves and can be life-threatening. Treatment for diarrhea mainly focuses on correcting dehydration, electrolyte imbalance, and acidosis through fluid therapy [7]. Prompt and appropriate treatment via fluid therapy is imperative for successful recovery from diarrhea associated with BCoV infection.
The commensal gut microbiota plays important roles in maintaining host homeostasis and health [8]. This host-gut microbiota system also impacts the control of infectious diseases. The microbiota can directly protect against infection by competing with pathogens at mucosal entry sites or by releasing soluble molecules that inhibit pathogenic colonization, or can provide indirect protection by stimulating host immune responses [9]. Thus, alteration of the gut microbiota has been associated with a decreased effectiveness of the protective barrier against infection, which can promote host infection [10]. Although previous studies have focused on individual pathogens that induce calf diarrhea, recent data have suggested that the gut microbiota plays an important role in controlling and eliminating infectious diseases. Moreover, recent studies have demonstrated dynamic changes in the gut microbiota composition in cattle under various growth stages and conditions [11,12].
However, despite the importance of the gut microbiota in modulating host health, little is known regarding changes in the gut microbiota and their correlations with physiological parameters in post-weaned calves between the diarrheic state and the recovered state. Therefore, we compared the compositional changes in the fecal microbiota before and after treatment in post-weaned calves infected with BCoV and investigated possible correlations between certain microbiota and physiological parameters before and after recovery.
MATERIALS AND METHODS
Animals and sampling
The study was performed in a Holstein farm located in Jiri Mountain, Korea. The experimental samples were obtained from ten female Holstein calves aged between 117 and 138 days with diarrhea for ≥ 3 days. In this study, fecal samples from selected animals were previously tested for the presence of common diarrhea-causing pathogens, such as Escherichia coli K99, Salmonella spp., Cryptosporidium spp., BCoV, bovine rotavirus, and bovine viral diarrhea virus, and as a result, only BCoV was positively detected [13]. Sampling was performed twice from the same animals before and after 2 months of treatment, once in October 2019 and once in December 2019. During the 2 months, intravenous fluid therapy was conducted to avoid electrolyte imbalance and dehydration related diarrhea. All post-weaned calves that previously had BCoV-associated diarrhea were not detected BCoV and were diagnosed with health by a veterinarian after 2 months of treatment. Fecal and serum samples were frozen at −80°C or −20°C until they were processed and analyzed. All animal procedures were performed according to ethical guidelines and approved by the Institutional Animal Care and Use Committee of Jeonbuk National University (JBNU 2016-00026).
Evaluation of physiological parameters
Serum biochemistry tests were performed to evaluate eight physiological parameters (total serum protein, albumin, blood urea nitrogen, glucose, sodium, potassium, chloride, and phosphorus) using a Catalyst One™ chemistry analyzer (IDEXX Laboratories, Westbrook, ME, USA).
Fecal DNA extraction and 16s rRNA gene sequencing library construction
For bacterial gene sequencing, total DNA extraction was performed from frozen feces using a FastDNA Spin Kit (MP Biomedicals, Irvine, CA, USA) following their instructions. NanoDrop 2000 UV-vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used for measuring of final DNA concentration. For amplification of the 16S rRNA gene, the 341F/805R primer pair targeting the V3–V4 variable region were used for the process of polymerase chain reaction (PCR). PCR was conducted as described conditions: denaturation at 94°C for 3 min; annealing at 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min; and a final elongation step at 72°C for 5 min, and then PCR products purification of each samples was conducted with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). After pooling of mixed amplicons, sequencing was carried out by using the Illumina MiSeq Sequencing system (Illumina, San Diego, CA, USA).
Analysis of 16s rRNA sequencing data
The 16S rRNA gene amplicon sequencing was processed by the open-source software pipeline, and raw reads were subjected to quality-control analysis, where low-quality (< Q25) reads were filtered using Trimmomatic (version 0.32). The filtered paired-end reads were joined into single sequences using PANDAseq. Primer sequences were then trimmed at similarity cut-off of 0.8 using an in-house program of ChunLab. Denoising of non-redundant reads were conducted using DUDE-Seq. The EzBioCloud database was used to obtain taxonomic assignments and to identify chimeric reads with best-hit similarity rate below 97%. The sequences were clustered into observed operational taxonomic units (OTUs) at a 97% similarity using CD-HIT and UCLUST. Microbial diversity was analyzed using CLcommunity software (version 3.46, ChunLab, Seoul, Korea). Linear discriminant analysis (LDA) effect size (LEfSe) analysis was conducted using the Galaxy framework (https://huttenhower.sph.harvard.edu/galaxy/) to identify significantly enriched bacterial taxa in the feces of diarrheic calves between before and after fluidic treatment, based on a p-value of < 0.05 and an LDA score of > 2.0.
Statistical analysis
Spearman’s correlation coefficient analysis was conducted based on the relative abundance of genus taxa with respect to blood-parameters. Correlation with a p-value < 0.05 were selected to construct the network map. All statistical analyses and graphical procedures were carried out using GraphPad Prism 8 software (GraphPad, La Jolla, CA, USA).
RESULTS
Changes in blood-parameter values in post-weaned calves between the diarrheic and recovered states
BCoV is a respiratory and enteric virus that is frequently isolated from fecal samples of diarrheic calves [4]. In the present study, fecal samples collected from ten post-weaned calves with diarrhea before fluid therapy (referred to as the pre-treatment group) were positive for BCoV. However, after 2 months of treatment, BCoV was not detected in the fecal samples of the post-treatment group, and the symptoms of diarrhea were also improved (Table 1).
Table 1. Metadata of the samples.
| Group | Sample name | Age (day) | Sex | Breed | BCoV detection | Appearance of feces |
|---|---|---|---|---|---|---|
| Pre-treatment (n = 10) | Pre-1 | 122 | Female | Holstein Friesian cattle | + | 2 |
| Pre-2 | 139 | + | 3 | |||
| Pre-3 | 128 | + | 2 | |||
| Pre-4 | 136 | + | 2 | |||
| Pre-5 | 148 | + | 2 | |||
| Pre-6 | 117 | + | 3 | |||
| Pre-7 | 135 | + | 2 | |||
| Pre-8 | 117 | + | 2 | |||
| Pre-9 | 124 | + | 2 | |||
| Pre-10 | 147 | + | 2 | |||
| Post-treatment (n = 10) | Post-1 | 178 | Female | Holstein Friesian cattle | − | 0 |
| Post-2 | 195 | − | 0 | |||
| Post-3 | 184 | − | 0 | |||
| Post-4 | 192 | − | 0 | |||
| Post-5 | 204 | − | 0 | |||
| Post-6 | 173 | − | 0 | |||
| Post-7 | 191 | − | 0 | |||
| Post-8 | 173 | − | 0 | |||
| Post-9 | 180 | − | 0 | |||
| Post-10 | 203 | − | 0 |
Data from Chae et al. with CC-BY [13].
Fecal appearance - normal (0); semi-solid (1); loose (2); watery (3).
BCoV, bovine coronavirus.
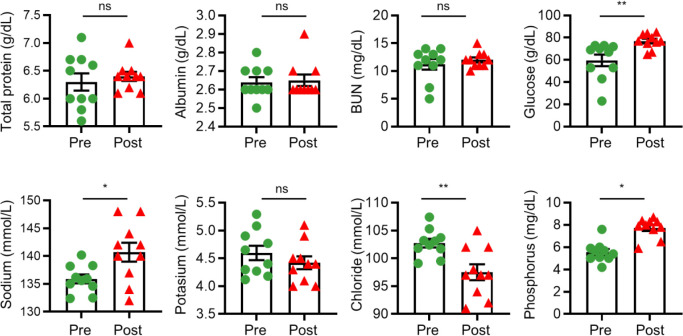
Since electrolyte disturbances and metabolic changes are the most significant consequences of diarrhea in calves [14], blood-serum parameters were analyzed to compare the physiological conditions of calves in the diarrheic and recovered states. The levels of blood-serum parameters, such as glucose, sodium, and phosphorous, which showed a lower concentration in post-weaned calves with diarrhea than in those that had recovered, were significantly enhanced, whereas the chloride concentration in the serum was significantly reduced after 2 months of treatment. However, total serum protein, albumin, and BUN concentrations remained unaffected (Fig. 1). These data indicate that fluid therapy effectively altered the physiological parameters, and that these changes were related the recovery of diarrhea caused by BCoV infection.
Fig. 1. Principal hematological and biochemical parameters in post-weaned calves with BCoV-related diarrhea, before and after treatment.
Serum concentrations of total protein, albumin, BUN, glucose, and electrolytes (sodium, potassium, chloride and phosphorus) were measured in post-weaned calves with diarrhea, before and after treatment. BCoV, bovine coronavirus; BUN, blood urea nitrogen.
Comparing the 16S rRNA gene-sequencing data in post-weaned calves between the diarrheic and recovered states
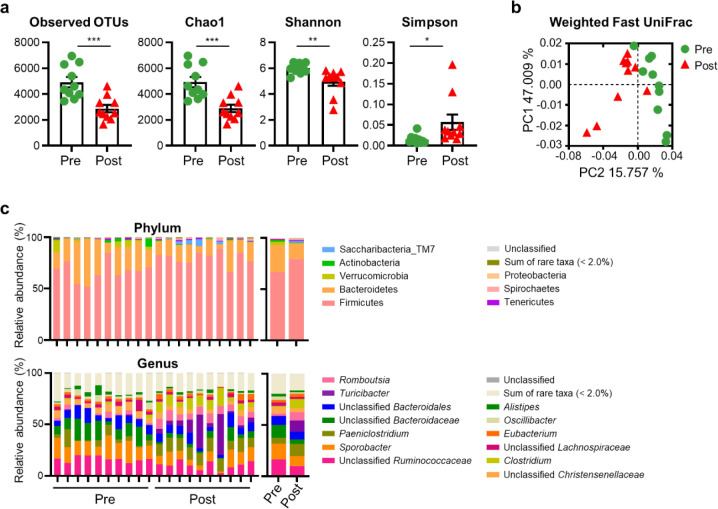
To investigate changes in the gut microbiota structure of post-weaned calves in the diarrheic and recovered states, we analyzed their gut microbiota using 16S rRNA gene amplicon sequencing. A total of 2,052,580 reads were generated, with an average of 102,629 reads per calf. The Good’s coverage of fecal samples was > 99.45%, indicating that our sample coverage was sufficiently high to analyze microbial diversity. Alpha diversity, as determined using the observed OTUs, and Chao1 and Shannon indices, were significantly lower in the post-treatment group than in the pre-treatment group. However, an increased Simpson index was observed after 2 months of fluid treatment (Fig. 2a). We next measured beta diversity using principal coordinates analysis based on the weighted fast UniFrac distance matrix. The results showed that the gut microbiota in calves with BCoV-mediated diarrhea clustered separately from those in recovered calves (Fig. 2b).
Fig. 2. Comparison of gut microbiota profiles in post-weaned calves with BCoV-related diarrhea, before and after treatment.
(a) Alpha-diversity (observed OTUs, and Chao1, Shannon, and Simpson indices) in each microbiota, as determined by comparing diarrheic and recovered calves. (b) Discriminant analysis of beta diversity among different samples. Microbial alterations between diarrheic (green) and recovered calves (red) are illustrated by the line or circle, and individual data are also depicted. *p < 0.05. (c) Bar chart representing the microbiota compositions at the phylum and genus levels in post-weaned calves with diarrhea, before and after treatment. Mean relative abundances of the main bacterial phyla and genera (> 2.0% of all sequences) that were distinctive between the diarrheic and recovered calves. BCoV, bovine coronavirus; OTUs, operational taxonomic units.
The OTUs mapped to 28 phyla, 63 classes, 131 orders, 272 families, and 798 genera. The top eight phyla (Firmicutes, Bacteroidetes, Verrucomicrobia, Actinobacteria, Saccharibacteria_TM7, Tenericutes, Spirochaetes, and Proteobacteria) and the top 13 genera Ruminococcaceae_uc (unclassified genus in the Ruminococcaceae family), Sporobacter, Paeniclostridium, Bacteroidaceae_uc, Bacteroidales_uc_g, Turicibacter, Romboutsia, Christensenellaceae_uc, Clostridium, Lachnospiraceae_uc, Eubacterium, Oscillibacter, and Alistipes are displayed in Fig. 2c. These genera accounted for 80.78% and 83.59% of the reads in the pre- and post-treatment groups, respectively.
At the phylum level, the abundances of Bacteroidetes, Verrucomicrobia, Actinobacteria, and Proteobacteria were significantly reduced, while those of Firmicutes, Saccharibacteria_TM7, and Spirochaetes were elevated in the post-treatment group compared to those in the pre-treatment group (p < 0.05). In addition, several significant differences were also observed at the genus level. Compared with the pre-treatment group, the abundances of Ruminococcaceae_uc, Oscillibacter, Bacteroidaceae_uc, Sporobacter, and Christensenellaceae_uc were significantly lower in the post-treatment group, while those of Romboutsia, Clostridium, Paeniclostridium, and Turicibacter showed the opposite pattern (p < 0.05) (Fig. 2c).
Alterations of the gut microbiome community in post-weaned calves after recovery from diarrhea
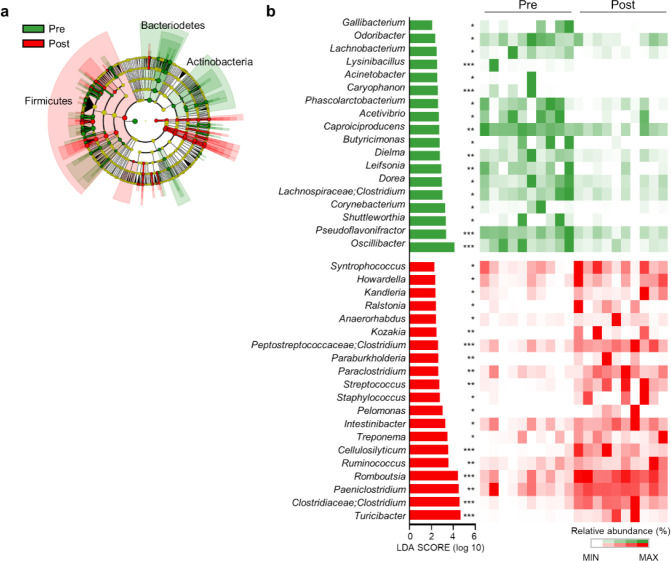
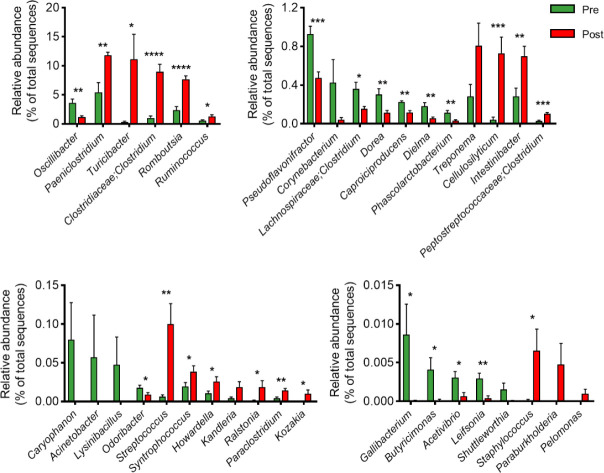
Next, we conducted LEfSe analysis to identify the top discriminatory microbiota at the species level between the diarrheic and recovery states. A cladogram displaying the taxonomic hierarchical structure of the fecal microbiota (from the phylum to genus levels) indicated significant differences in phylogenetic distributions between the microbiota of the pre- and post-treatment groups (Fig. 3a). Using a logarithmic LDA score cut-off of 2.0, we identified 18 and 20 genera that were abundant in the microbiota of the pre- and post-treatment groups, respectively (Fig. 3b). We also identified 28 genera showing significant differences in relative abundance between the pre- and post-treatment group. The genera Oscillibacter, Pseudoflavonifractor, Lachnospiraceae, Clostridium, Dorea, Caproiciproducens, Dielma, Phascolarctobacterium, Odoribacter, Gallibacterium, Butyricimonas, Acetivibrio, and Leifsonia were significantly abundant in the pre-treatment group. However, the genera Paeniclostridium, Turicibacter, Clostridiaceae;Clostridium, Romboutsia, Ruminococcus, Cellulosilyticum, Intestinibacter, Peptostreptococcaceae;Clostridium, Streptococcus, Syntrophococcus, Howardella, Ralstonia, Paraclostridium, Kozakia, Staphylococcus, and Turicibacter were significantly abundant in the post-treatment group (Fig. 4). These data suggested that the composition of fecal microbiota was significantly altered in response to 2 months of treatment in post-weaned calves with BCoV-mediated diarrhea.
Fig. 3. Microbiota composition in the feces of diarrheic and recovered calves.
(a) Phylogenetic cladogram generated by performing LEfSe analysis, depicting taxonomic associations between the microbiome communities of the pre- and post-treatment groups. Each node represents a specific taxonomic type. (b) The rankings of significantly different genera (based on the LEfSe method) were reflected in the log LDA scores of both groups. LEfSe analysis was based on the nonparametric factorial Kruskal–Wallis rank-sum test, followed by the Wilcoxon signed-rank test. Featured LDA scores > 2.0 are plotted (*p < 0.05, **p < 0.01, ***p < 0.001). LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis.
Fig. 4. Relative abundances of bacterial genera in the feces of diarrheic and recovered calves.
The relative abundances of bacterial genera that averaged > 1%, between 1% and 0.1%, between 0.1% and 0.01%, and < 0.01% of the relative abundance observed in post-weaned calves with diarrhea, before and after treatment. The error bars represent standard errors. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Correlations between gut microbiota compositions and clinical characteristics
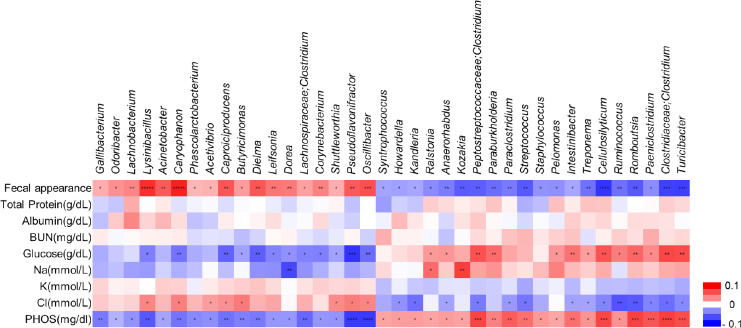
Based on our longitudinal gut microbiome results, we attempted to analyze the functional correlations between differential gut microbiota compositions and blood parameters. Spearman’s correlation coefficient (r) was computed for nine different parameters and 38 bacterial taxa (Fig. 5). The results showed 126 statistically significant interactions between eight blood parameters and 38 bacterial taxa (p < 0.05). The genera Caproiciproducens, Pseudoflavonifractor, and Oscillibacter, which were more abundant in the post-weaned calves with diarrhea, showed a negative correlation with blood parameters, such as the glucose, and phosphorus levels, but it positively correlated with chloride levels. However, the genera Peptostreptococcaceae;Clostridium, Intestinibacter, Cellulosilyticum, Ruminococcus, Romboutsia, Paeniclostridium, Clostridiaceae, Clostridium, and Turicibacter, which were more abundant in post-weaned calves after treatment of BCoV-mediated diarrhea, showed positive correlation with serum glucose and phosphorus levels but negative correlation with serum chloride levels. The increased abundance of Dorea showed negative correlations with glucose, sodium, and phosphorus level in pre-treatment group, while a positive correlation was observed between those parameters and increasing abundance of Ralstonia genus in the post-treatment group. These results suggest that alterations of gut microbiota compositions between the pre- and post-treatment groups are correlated with changes in their physiological parameters.
Fig. 5. Correlations between the gut microbiome and physiological parameters.
Spearman’s correlation analysis was used to investigate the relationships between fecal bacterial populations and blood parameters, considering longitudinal results obtained with all groups. The red and blue cells indicate positive and negative correlations, respectively. Correction for multiple comparisons used the false discovery rate (FDR; threshold of 0.05). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. BUN, blood urea nitrogen; PHOS, phosphorus.
DISCUSSION
Coronaviruses cause severe respiratory and enteric infectious diseases that pose health risks to humans and domestic animals. Although previous studies have shown changes in the gut microbiota caused by intestinal infectious diseases in cattle [15,16], no reports have described an altered gut microbiota in calves with diarrhea caused by BCoV. Our results showed that the gut microbiota was different before and after diarrhea recovery and indicated a clear correlation between the abundances of taxa (at the genus level) and the blood parameters for the first time.
We collected blood and fecal samples from the same ten post-weaned calves with diarrhea, before and after fluid therapy. After two months of treatment, BCoV was no longer detected in the feces, and the symptoms of diarrhea had improved. In addition, since diarrhea leads to symptoms, such as dehydration, abnormalities in terms of electrolytes and the acid–base balance, and nutrient malabsorption, it is important to measure the concentrations of total protein, BUN, glucose, electrolytes in the serum as a diagnostic index of diarrhea [17,18]. In this regard, our results indicated that blood parameters associated with diarrheic symptoms, such as lower levels of serum glucose, sodium, and phosphorus, were enhanced in the post-weaned calves after treatment. Furthermore, although higher concentrations of total serum protein, serum albumin, and BUN are generally observed in calves with diarrhea [19,20], our results showed that these blood parameters were maintained at low concentrations even during the diarrheic state, and no differences were found before and after treatment. These data indicate that values of diagnostic serum physiological parameters of calves with diarrhea vary depending on their causative agents, suggesting that analysis of blood parameters only is inadequate as a method for diagnosing BCoV-mediated diarrhea. Our previous study suggested that monitoring the monocyte count and haptoglobin concentration along with clinical examination and PCR can be used to assess the prognosis of post-weaned calves with diarrhea caused by BCoV infection [13].
Previous studies have shown that diarrhea often leads to reduced microbial diversity in humans and animals [21–23]. In our research, alpha diversity indices were found to be significantly lower in the post-treatment group than that in the pre-treatment group, which implied that the diversity of the intestinal microbiota might not be properly recovered in post-weaned calves after treatment. An explanation for these findings would be that infectious diarrhea causes serious damage to the intestines, accompanied by inflammation [24]. Thus, two months of fluid treatment might not be enough to restore the gut microbial diversity, suggesting that further study on changes in the gut microbiota after long-term treatment is necessary.
In case of rotavirus-mediated diarrhea, upon comparing the main differences in bacterial phyla between infected and uninfected calves, the abundances of Firmicutes and Bacteroidetes were found to be reduced and that of Proteobacteria was found to be elevated in the rotavirus diarrheic calves [16]. However, in our study, only Firmicutes increased in abundance after recovery in calves with BCoV-related diarrhea, indicating that the alteration of the gut microbiota may differ depending on the type of virus that induces diarrhea.
We identified differences in core gut microbiota between calves in the diarrheic and recovered states through LEfSe analysis. Then, we further investigated the correlation between the abundances of taxa (at the genus level) and the blood parameters. Our data showed that 14 genera were significantly correlated with three different blood parameters simultaneously. In particular, the abundances of the genera Caproiciproducens, Pseudoflavonifractor, and Oscillibacter, which were more abundant in the pre-treatment group, were found to be negatively correlated with the serum glucose and phosphorus levels but positively correlated with the serum chloride level. Notably, only Dorea showed negative correlations with serum glucose, sodium, and phosphorus levels. Previous results revealed that the genus Caproiciproducens is currently found in hens with gut dysbiosis induced by Salmonella Typhimurium infection [25], and the abundance of the genus Pseudoflavonifractor is increased in chickens infected with influenza A virus subtype H9N2 [26]. Besides, though Dorea is often found in healthy individuals, its elevated abundance is also observed in patients with multiple sclerosis and colon cancer [27,28], indicating that this genus may play a deteriorative role in immune regulation. In diarrheic calves, the abundances of opportunistic pathogens were observed to be correlated with diarrhea symptoms, suggesting that changes in the gut microbiota caused by BCoV-mediated diarrhea may lead to chronic diseases.
On the contrary, the abundances of the genera Peptostreptococcaceae;Clostridium, Intestinibacter, Cellulosilyticum, Ruminococcus, Romboutsia, Paeniclostridium, Clostridiaceae;Clostridium, and Turicibacter increased in recovered calves, and they showed positive correlations with the serum glucose and phosphorus levels, but negative correlations with serum chloride level. Among these genera, Ruminococcus has been shown to easily digest plant cellulose and produce short-chain fatty acids, such as acetate, formate, and succinate, which are associated with anti-inflammatory effects [29]. The genus Cellulosilyticum has been reported to be able to break down both fiber and protein. Peptostreptococcaceae;Clostridium was found to be predominant in the perinatal intestinal microbiota of cattle [30]. Moreover, the genus Ralstonia was detected at higher levels in the milk microbiomes of healthy dairy cows than those of mastitic dairy cows [31]. Taken together, our results demonstrate that fluidic treatment tended to increase some beneficial bacteria associated with fiber digestion in healthy calves and reduce the opportunistic pathogens correlated with the diarrheic symptoms in post-weaned calves with BCoV-mediated diarrhea, suggesting that the improvement of the intestinal dysfunction by fluid therapy might influence on the microbial changes.
The data generated in this study indicated that the gut microbiota of diarrheic and recovered calves were different. Significant differences in the microbiota structure and composition between these two groups suggest that the microbial environment could be changed by two months of fluid therapy, which is commonly used for calves with diarrhea. Furthermore, these results provide new information regarding the relationship between the gut microbiota and the physiological parameters of calves with BCoV-mediated diarrhea.
Acknowledgements
Not applicable.
Competing interests
No potential conflict of interest relevant to this article was reported.
Funding sources
This research was supported by a grant from the World Institute of Kimchi (KE2001-1), which is funded by the Ministry of Science and ICT, and partially supported by Technology Development Program (Project No. 1116043-1) for Bio-industry, Ministry for Agriculture, Food and Rural Affairs, Korea.
Availability of data and material
Upon reasonable request, the datasets of this study can be available from the corresponding author.
Authors’ contributions
Conceptualization: Kwon MS, Jo HE, Choi HJ.
Data curation: Kwon MS, Jo HE.
Formal analysis: Kwon MS, Jo HE.
Methodology: Kwon MS, Jo HE, Lee J.
Software: Jo HE.
Validation: Park J, Choi HJ.
Investigation: Kwon MS, Jo HE, Lee J.
Writing - original draft: Kwon MS, Jo HE, Choi KS, Yu D, Oh Y.
Writing - review & editing: Kwon MS, Park J, Choi HJ.
Ethics approval and consent to participate
All procedures were performed according to ethical guidelines for the use of animal samples, as approved by the Jeonbuk National University (Institutional Animal Care and Use Committee decision number JBNU 2016-00026).
REFERENCES
- 1.Hodnik JJ, Ježek J, Starič J. Coronaviruses in cattle. Trop Anim Health Prod. 2020 doi: 10.1007/s11250-020-02354-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin North Am Food Anim Pract. 2010;26:123–46. doi: 10.1016/j.cvfa.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JR. Effect of bovine viral diarrhea virus in the feedlot. Vet Clin North Am Food Anim Pract. 2004;20:39–50. doi: 10.1016/j.cvfa.2003.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho KO, Hoet AE, Loerch SC, Wittum TE, Saif LJ. Evaluation of concurrent shedding of bovine coronavirus via the respiratory tract and enteric route in feedlot cattle. Am J Vet Res. 2001;62:1436–41. doi: 10.2460/ajvr.2001.62.1436. [DOI] [PubMed] [Google Scholar]
- 5.Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tello L, Perez-Freytes R. Fluid and electrolyte therapy uring vomiting and diarrhea. Vet Clin North Am Small Anim Pract. 2017;47:505–19. doi: 10.1016/j.cvsm.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berchtold J. Treatment of calf diarrhea: intravenous fluid therapy. Vet Clin North Am Food Anim Pract. 2009;25:73–99. doi: 10.1016/j.cvfa.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abt MC, Artis D. The intestinal microbiota in health and disease: the influence of microbial products on immune cell homeostasis. Curr Opin Gastroenterol. 2009;25:496–502. doi: 10.1097/MOG.0b013e328331b6b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, et al. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan L, Hensley C, Mahsoub HM, Ramesh AK, Zhou P. Chapter two: microbiota in viral infection and disease in humans and farm animals. Prog Mol Biol Transl Sci. 2020;171:15–60. doi: 10.1016/bs.pmbts.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oikonomou G, Teixeira AGV, Foditsch C, Bicalho ML, Machado VS, Bicalho RC. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLOS ONE. 2013;8:e63157. doi: 10.1371/journal.pone.0063157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh P, Teal TK, Marsh TL, Tiedje JM, Mosci R, Jernigan K, et al. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome. 2015;3:45. doi: 10.1186/s40168-015-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae JB, Park J, Jung SH, Kang JH, Chae JS, Choi KS. Acute phase response in bovine coronavirus positive post-weaned calves with diarrhea. Acta Vet Scand. 2019;61:36. doi: 10.1186/s13028-019-0471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SH, Choi EW, Kim D. Relationship between the values of blood parameters and physical status in Korean native calves with diarrhea. J Vet Sci. 2020;21:e17. doi: 10.4142/jvs.2020.21.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez DE, Arroyo LG, Costa MC, Viel L, Weese JS. Characterization of the fecal bacterial microbiota of healthy and diarrheic dairy calves. J Vet Intern Med. 2017;31:928–39. doi: 10.1111/jvim.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang JY, Kim S, Kwon MS, Lee J, Yu DH, Song RH, et al. Rotavirus-mediated alteration of gut microbiota and its correlation with physiological characteristics in neonatal calves. J Microbiol. 2019;57:113–21. doi: 10.1007/s12275-019-8549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayers RG, Kennedy A, Krump L, Sayers GP, Kennedy E. An observational study using blood gas analysis to assess neonatal calf diarrhea and subsequent recovery with a European Commission-compliant oral electrolyte solution. J Dairy Sci. 2016;99:4647–55. doi: 10.3168/jds.2015-10600. [DOI] [PubMed] [Google Scholar]
- 18.Smith GW. Treatment of calf diarrhea: oral fluid therapy. Vet Clin North Am Food Anim Pract. 2009;25:55–72. doi: 10.1016/j.cvfa.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Walker PG, Constable PD, Morin DE, Drackley JK, Foreman JH, Thurmon JC. A reliable, practical, and economical protocol for inducing diarrhea and severe dehydration in the neonatal calf. Can J Vet Res. 1998;62:205–13. [PMC free article] [PubMed] [Google Scholar]
- 20.Gånheim C, Alenius S, Waller KP. Acute phase proteins as indicators of calf herd health. Vet J. 2007;173:645–51. doi: 10.1016/j.tvjl.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guard BC, Barr JW, Reddivari L, Klemashevich C, Jayaraman A, Steiner JM, et al. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLOS One. 2015;10:e0127259. doi: 10.1371/journal.pone.0127259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorkiewicz G, Thallinger GG, Trajanoski S, Lackner S, Stocker G, Hinterleitner T, et al. Alterations in the colonic microbiota in response to osmotic diarrhea. PLOS One. 2013;8:e55817. doi: 10.1371/journal.pone.0055817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, Zeng D, Wang Q, Wang N, Zeng B, Niu L, et al. Diarrhea-associated intestinal microbiota in captive Sichuan golden snub-nosed monkeys (Rhinopithecus roxellana) Microbes Environ. 2018;33:249–56. doi: 10.1264/jsme2.ME17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chigerwe M, Heller MC. Diagnosis and treatment of infectious enteritis in adult ruminants. Vet Clin North Am Food Anim Pract. 2018;34:119–31. doi: 10.1016/j.cvfa.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan S, Chousalkar KK. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J Anim Sci Biotechnol. 2020;11:29. doi: 10.1186/s40104-020-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yitbarek A, Weese JS, Alkie TN, Parkinson J, Sharif S. Influenza A virus subtype H9N2 infection disrupts the composition of intestinal microbiota of chickens. FEMS Microbiol Ecol. 2018;94 doi: 10.1093/femsec/fix165. [DOI] [PubMed] [Google Scholar]
- 27.Shahi SK, Freedman SN, Mangalam AK. Gut microbiome in multiple sclerosis: the players involved and the roles they play. Gut Microbes. 2017;8:607–15. doi: 10.1080/19490976.2017.1349041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen XJ, Rawls JF, Randall TA, Burcal L, Mpande C, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–47. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10:e02566–18. doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weese JS, Jelinski M. Assessment of the fecal microbiota in beef calves. J Vet Intern Med. 2017;31:176–85. doi: 10.1111/jvim.14611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addis MF, Tanca A, Uzzau S, Oikonomou G, Bicalho RC, Moroni P. The bovine milk microbiota: insights and perspectives from -omics studies. Mol Biosyst. 2016;12:2359–72. doi: 10.1039/C6MB00217J. [DOI] [PubMed] [Google Scholar]