Abstract
Interleukin-10 (IL10), a pleiotropic cytokine secreted by type-2 helper (Th2) T cells, contributes to the oncogenic activation or inactivation of tumor-suppressor genes. The present study investigated whether hypomethylation of IL10 CpG island (CGI) was associated with the risk of developing gastric cancer (GC) and the prognosis of patients with GC. A fragment (hg18, chr1: 206945638-206945774) at the CGI of IL10 was selected for the present methylation assay. Quantitative methylation-specific PCR was used to evaluate the methylation of IL10 CGI in 117 tumor samples from patients with GC. The results demonstrated that IL10 CGI methylation was significantly lower in the tumor tissues compared with that in the paired adjacent non-tumor tissues (median percentage of methylated reference, 29.16 vs. 42.82%, respectively; P=4×10−8). Furthermore, results from receiver operating characteristic curve analysis identified a significant area under the curve of 0.706, with a sensitivity and a specificity of 77.8 and 58.1%, respectively, between cancer tissues and paired adjacent non-tumor tissues. Furthermore, the methylation of IL10 CGI was significantly associated with patients' age at diagnosis (r=−0.201; P=0.03). Subgroup analyses demonstrated that the association between IL10 CGI hypomethylation and the risk of GC was specific for patients with low differentiation (P=1×10−7) and Borrmann types III+IV (P=1×10−7). In addition, IL10 CGI hypomethylation was significantly associated with the risk of GC for patients without smoking history (P=3×10−7) or a family history of cancer (P=2×10−7). The results from Kaplan-Meier survival analysis demonstrated that IL10 CGI hypomethylation was associated with a significantly shorter overall survival of patients with GC (P=0.041). Similar results were identified for patients with GC who did not have smoking history (P=0.037) or a family history of cancer (P=0.049). The results from this study demonstrated that IL10 CGI hypomethylation may be considered as a potential biomarker for the diagnosis and prognosis of patients with GC in the Chinese population.
Keywords: IL10, CpG island, gastric cancer, DNA methylation, quantitative methylation-specific PCR
Introduction
Although the incidence of gastric cancer (GC) is declining, GC caused ~1 million new cases and 781,000 deaths in 2018 and remains the third leading cause of death in the world (1). Numerous types of cancer, including GC have a background of chronic inflammatory processes caused by infection or exposure to environmental factors (2). Understanding the molecular mechanisms of inflammation during GC is therefore crucial for the development of novel therapeutic strategies against GC (1). H. pylori infects >50% of the world population and may affect gastric physiology, via triggering gastritis and canceration processes (3). Treatment of H. pylori infection may therefore help prevent GC in the general population (4,5); however, this treatment appears to be unrelated to the prognosis of patients with early GC (5,6). Gastric biopsy of patients with gastrointestinal-related symptoms demonstrated that in addition to the common H. pylori-related gastritis, several other modes of mucosal damage are also increasing (7).
Interleukin-10 (IL10) is one of the most important anti-inflammatory cytokines (8). IL10 is associated with oncogenic activation or inactivation of tumor-suppressor genes (9). High levels of circulating IL10 were identified in patients with digestive cancers (10–12). In addition, the persistence of circulating IL10 in patients with colorectal cancer following surgery could predict a high tendency to relapse in these patients (13). High levels of circulating IL10 were also reported to be associated with an unfavorable prognosis for patients with GC (14,15). In addition, a meta-analysis demonstrated that a high level of circulating IL10 resulted in poor survival in patients with various types of cancer (16).
IL10 tends to be hypomethylated in several cancer types (17), including pancreatic (18), breast (19) and cervical cancer (20), as well as chronic lymphocytic leukemia (12). TET-2 catalyzes the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (21), which is an important intermediate in the process of active DNA demethylation in primary human monocytes (22). The expression of IL10 and TET-2 was found to increase in Vicenin-2 treatment (23), which indicated that IL10 expression is increased during active DNA demethylation. Demethylation with 5′-AZA-deoxycytidine (5-AZA) results in an upregulation of IL10 expression in CD4+ T cells, and CpG-DNA methylation of the DNA methyltransferase 3α gene was demonstrated to silence IL10 transcription in macrophages, indicating the crucial role of CpG methylation in the regulation of IL10 expression (24–26).
According to these previous findings, the present study determined the association between IL10 CpG island (CGI) methylation and the risk of GC. This study aimed to evaluate the diagnostic and prognostic value of IL10 CGI methylation for Chinese patients with GC.
Materials and methods
Subjects
A total of 117 patients with GC (mean age, 56.40 years; age range, 21–83 years) treated at the Taihu Hospital of Wuxi, China, between January 2008 and February 2015 were included in the present study. Gastric tumor and adjacent non-tumor tissues (5 cm from the tumor) were collected. Patients were diagnosed by pathologists' histological diagnosis, and none had received radiation or chemotherapy prior to sample collection. The time between primary surgery and the death of the patient or the last follow-up was defined as the overall survival (OS) (27). This study was approved by the Ethics Committee of Taihu Hospital of Wuxi (approval number: 20160026) (Wuxi, China). All patients provided written informed consent.
Sanger sequencing, capillary electrophoresis and DNA methylation assay
Genomic DNA extraction (QIAamp DNA mini kit; Qiagen GmbH) and subsequent bisulfite conversion (EpiTech Bisulfite kits; Qiagen Benelux B.V.) were performed as previously described (28,29). The program of qMSP was as follows: Pre-denaturation at 95°C for 10 min; 45 cycles of denaturation at 95°C for 20 sec, annealing at 58°C for 20 sec and extension at 72°C for 30 sec. Sanger sequencing (BGI Biotech Co., Ltd) was used to determine randomly selected sodium bisulfite-modified DNA sequences, as previously described (30). A fully automated high-resolution capillary electrophoresis apparatus (Qsep100; BIOptic Inc.) was used to conduct the fragment size of the quantitative methylation-specific PCR (qMSP) product. This fragment size was compared with the theoretical fragment length to verify the uniqueness of IL10. The methylation level of IL10 CGI was assessed by qMSP as previously described (12,31). The qMSP primer sequences of IL10 were as follows: Forward 5′-AAGGTATTTCGGAGATTTC-3′ and reverse 5′-AACTCAACACTACTCTATTAC-3′. The qMSP primer sequences of actin β (ACTB) were as follows: Forward 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′. The percentage of methylated reference (PMR) was applied to quantitatively represent the methylation level (28). The PMR in each sample was calculated by the 2−ΔΔCq quantification method to represent the methylation level of IL10 CGI, and the ΔΔCq equation was as follows: ΔΔCq=sample DNA (CqIL10 -CqACTB control)-fully methylated DNA (CqIL10-CqACTB control) (32).
Data mining study
To evaluate the association between mRNA expression and IL10 methylation, the gene expression and methylation data from 371 stomach adenocarcinoma samples were collected from the stomach adenocarcinoma dataset (TCGA, PanCancer Altas) (33) through cBioPortal (http://www.cbioportal.org/). Furthermore, the data of the relative mRNA expression of IL10 in 4 oral squamous cell carcinoma cell lines (OC3, SAS, SCC and HSC3) before and after treatment with 10 µm 5-AZA for 4 days was retrieved from the Gene Expression Omnibus (GEO) database (GSE38823; http://www.ncbi.nlm.nih.gov/geo).
Statistical analysis
Wilcoxon rank sum test was used to compare the methylation level of IL10 CGI obtained from Chinese cohort and TCGA cohort in tumor tissues with adjacent non-tumor tissues. A Spearman correlation test was used to evaluate the correlation between IL10 methylation and patients' age, and between IL10 methylation and IL10 expression from TCGA dataset. A χ2 test was used to investigate whether IL10 CGI methylation of cancer tissues was associated with other clinical parameters. Independent t-test was used to compare mRNA expression of IL10 in 4 carcinoma cell lines before and after treatment with 10 µm 5-AZA for 4 days. Receiver operating characteristic (ROC) analysis was used to determine the diagnostic value of IL10 CGI methylation in GC. With the mean methylation value (PMR=32.0%) as the cutoff value, the OS of TCGA patients with gastric adenocarcinoma was divided into 2 groups according to the IL10 CGI methylation status. Subsequently, the Kaplan-Meier method and log-rank test was used to test whether IL10 CGI methylation status could affect patients' OS. Two-sided P<0.05 was considered to indicate a statistically significant difference.
Results
DNA methylation analysis
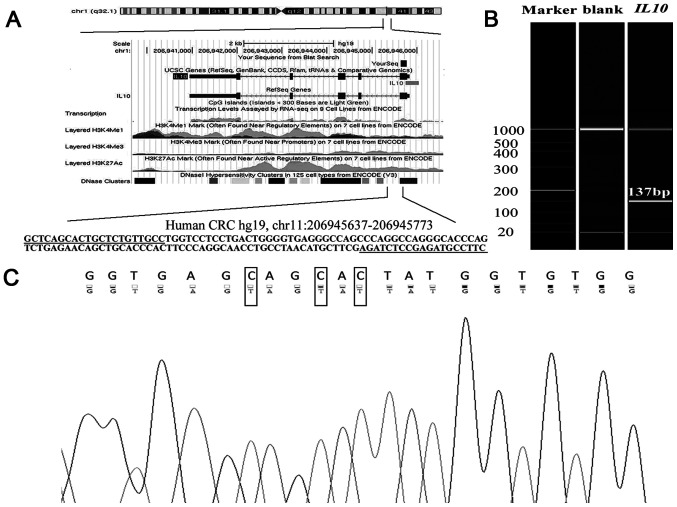
As shown in Fig. 1A, the amplified fragment is located on chr11:206945637-206945773 based on the genomic region from the University of California Santa Cruz genome browser according to Human 2013 (GRCh38/hg38) assembly (34). The methylation of CpG sites on the amplification fragment of IL10 CGI was measured (Fig. 1A). The results from capillary electrophoresis demonstrated that the qMSP product was unique and its length was 137 bp, as expected (Fig. 1B). In addition, the qMSP product was sequenced with the expected bisulphite-converted sequence according to the Sanger sequencing result (Fig. 1C).
Figure 1.
Target sequence on IL10 promoter region. (A) Genomic position and functional annotation of the amplified fragment. The primers for the quantitative methylation-specific PCR were underlined, and five CpG sites were identified (in grey). (B) Capillary electrophoresis of the amplified fragment (137 bp). (C) Sanger sequencing results. Top row of the sequence represents the original sequence of the fragment. Second row of the sequence represents the converted sequences. C nucleobase with corresponding converted T were in black boxes. A, adenine; C, cytosine; F, forward; G, guanine; IL10, interleukin 10; R, reverse; T, thymine.
Association between IL10 CGI hypomethylation and the risk of GC and patients' clinicopathological characteristics
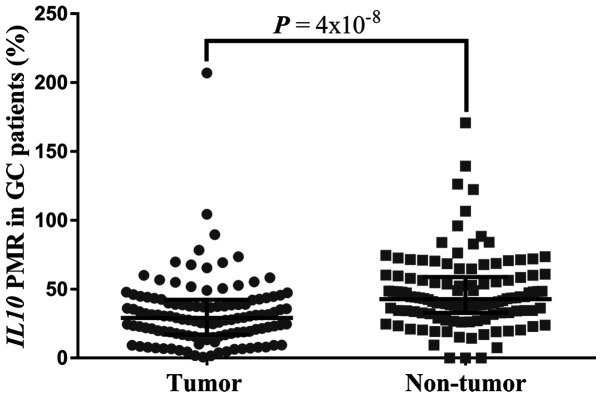
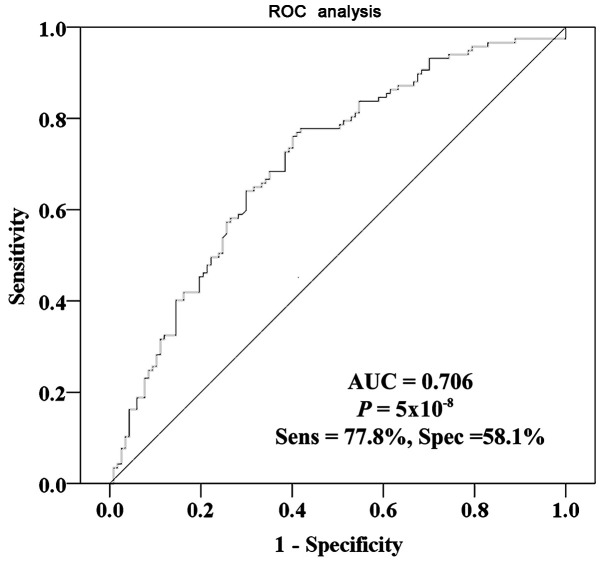
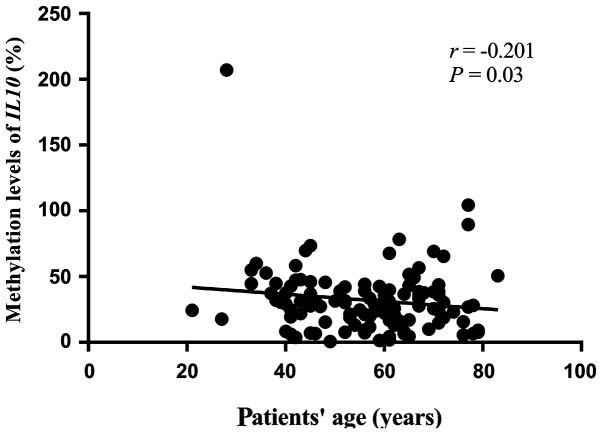
The results demonstrated that IL10 CGI methylation was significantly lower in tumor tissues compared with that in adjacent non-tumor tissues (median PMR, 29.16 vs. 42.82%; P=4×10−8; Fig. 2). Furthermore, results from ROC curve analysis confirmed that IL10 CGI hypomethylation may be a potential marker of GC diagnosis. In particular, a significant AUC of 0.706 with a sensitivity of 77.8% and a specificity of 58.1% was identified between cancer tissues and para-tumor tissues (Fig. 3). In addition, IL10 CGI methylation in tumor tissues was significantly associated with patient's age at diagnosis (r=−0.201; P=0.03; Fig. 4). Using the average IL10 CGI methylation level as the cutoff value, 117 patients were divided into either IL10 CGI hypermethylated patients (n=48) or IL10 CGI hypomethylated patients (n=69). Next, c2 test was used to investigate whether IL10 CGI methylation of cancer tissues was associated with other clinical parameters. It was found that IL10 CGI methylation was not associated with other parameters, including gender, surgical procedures, differentiation, lymph node metastasis, TNM stage, Borrmann type, disease recurrence, nerve invasion, family history of cancer, drinking history or smoking history (P>0.05; Table SI).
Figure 2.
Comparisons of IL10 CpG island methylation between tumor tissues and paired adjacent normal tissues. IL10, interleukin 10, PMR, percentage of methylated reference. GC, gastric cancer.
Figure 3.
Diagnostic value of IL10 CGI hypomethylation between gastric cancer tissues and paired para-tumor tissues. ROC curve analysis confirmed the potential diagnostic value of IL10 CGI hypomethylation, and yielded a significant AUC of 0.706 with a sensitivity of 77.8% and a specificity of 58.1% between cancer tissues and para-tumor tissues. AUC, area under the curve; CGI, CpG island; IL10, interleukin 10; ROC, receiver operating characteristic; Sens, sensitivity; Spe, specificity.
Figure 4.
Association of IL10 CGI methylation with age of patients with GC. An inverse correlation was found between IL10 methylation in tumor and age of patients with GC. The P-value was calculated by Spearman correlation test. GC, gastric cancer.
Stratified analyses by clinical phenotypes
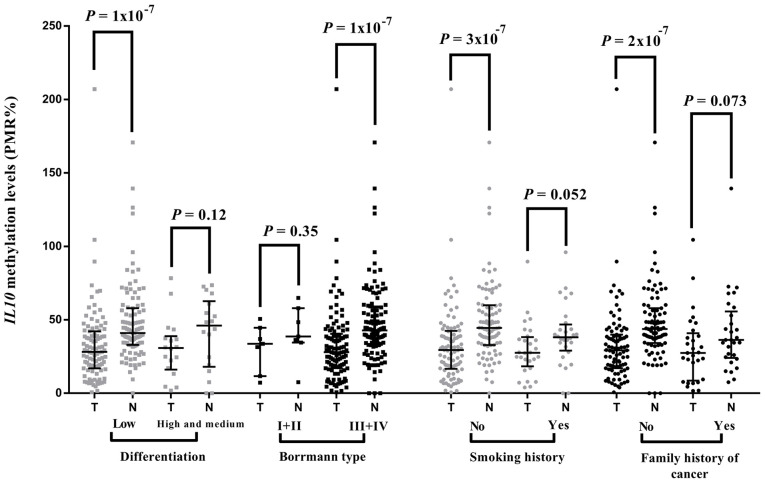
Stratified analyses demonstrated that the association between IL10 CGI hypomethylation and the risk of GC was specific for patients with low differentiation (P=1×10−7; Fig. 5) and III+IV Borrmann types (P=1×10−7; Fig. 5), but not in high and medium differentiations (P=0.12) or in I+II Borrmann types (P=0.35). In addition, IL10 CGI hypomethylation was significantly associated with the risk of GC for patients without smoking history (P=3×10−7; Fig. 5) or a family history of cancer (P=2×10−7; Fig. 5).
Figure 5.
Stratified comparisons of IL10 CpG island methylation between tumor tissues and paired para-tumor tissues. P-values were calculated using Wilcoxon rank sum test. Stratified tests were performed according to tumor differentiation, Borrmann type, smoking history and family history of cancer. Plots are presented as median with interquartile range. A significant P-value was identified in the following subgroups: Low differentiation, III+IV Borrmann types, non-smoker patients and patients with no family history of cancer. IL10, interleukin 10; N, normal adjacent tissue; PMR, percentage of methylated reference; T, tumor tissue.
IL10 CGI hypomethylation is correlated with poor prognosis in patients with GC
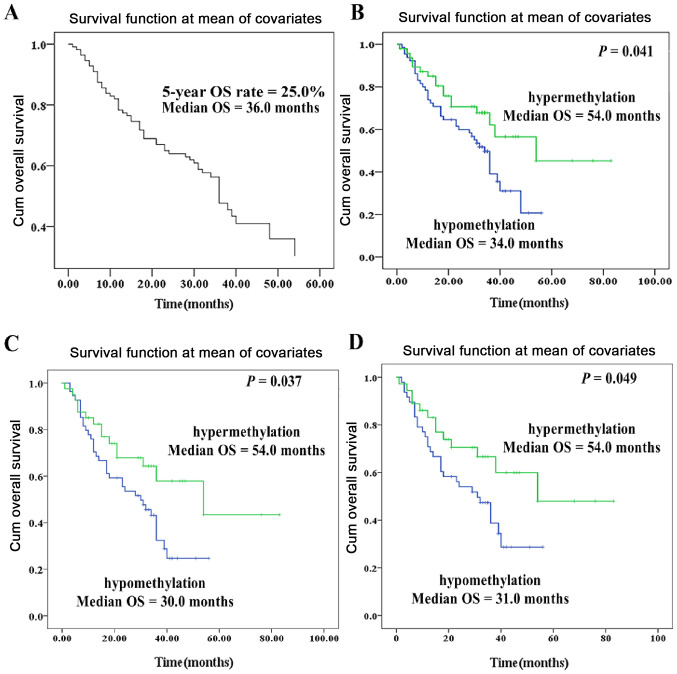
OS analysis was performed among the 117 patients. As presented in Fig. 6A, the 5-year OS rate and the median survival time were 25.0% and 36.0 months for the these patients, respectively (Fig. 6A). By using the average IL10 CGI methylation level as a cut-off value (PMR=32.0%), the 117 patients were divided into patients with IL10 CGI hypermethylation (n=48) or patients with IL10 CGI hypomethylation (n=69). The results from Kaplan-Meier survival analysis demonstrated that IL10 CGI hypomethylation was associated with a significantly shorter OS of patients with GC (P=0.041; Fig. 6B). Furthermore, Kaplan-Meier survival analysis of patients with GC and no and low differentiation demonstrated that IL10 CGI hypomethylation was associated with a significantly worse prognosis (P=0.037; Fig. 6C). Kaplan-Meier survival analysis of non-smoker patients with GC demonstrated that IL10 CGI hypomethylation was associated with a significantly worse prognosis (P=0.049; Fig. 6D).
Figure 6.
Association between aberrant IL10 CGI methylation and the prognosis of patients with GC. (A) Cox regression model of OS analysis demonstrated that the median OS time was 36.0 months. (B) Kaplan-Meier and log-rank analysis of the OS of patients with GC. (C) Kaplan-Meier and log-rank analysis of the survival in patients with GC with no and low differentiation. (D) Kaplan-Meier and log-rank analysis of the survival in non-smoker patients with GC. The cut-off value of IL10 CGI hypermethylation was set at 32.0%. GC, gastric cancer; OS, overall survival; CGI, CpG island; IL10, interleukin 10.
Data mining of public databases
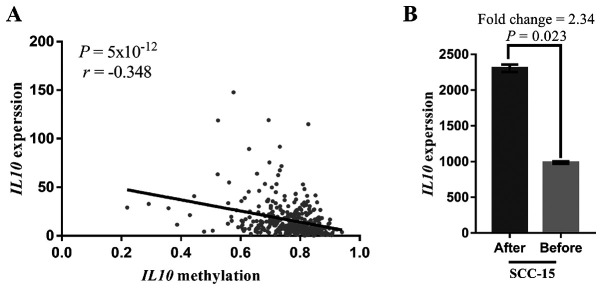
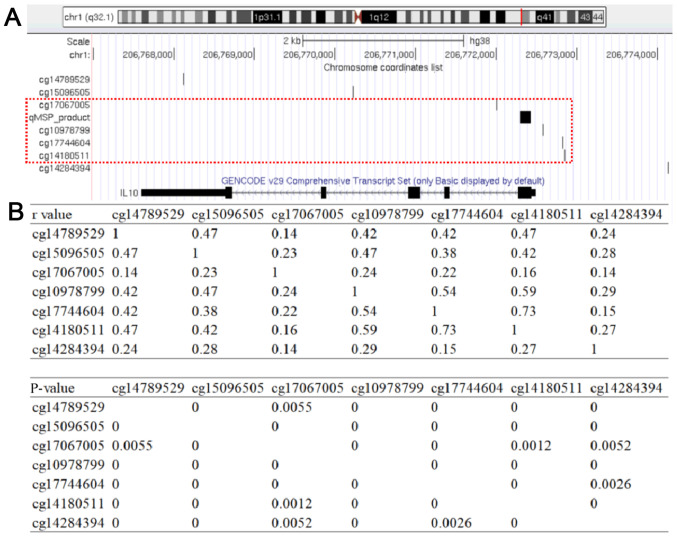
The analysis of data from 372 GC tissues from TCGA dataset demonstrated a significantly negative correlation between IL10 methylation and gene expression (r=−0.348; P=5×10−12; Fig. 7A), suggesting a pivotal role of IL10 methylation in the regulation of IL10 gene function. In addition, the results from GEO data analysis demonstrated that the relative expression level of IL10 significantly increased in cell lines treated with the demethylation agent 5-AZA (fold-change=2.34; P=0.023; Fig. 7B). As shown in Fig. 8, the analysis of the TCGA dataset demonstrated that the methylation levels of the seven CpG sites among 372 TCGA GC tissue samples were positively correlated with each other (r>0.13; P<0.006) especially for the two CpG sites cg17067005 and cg10978799 that are near the two ends of the qMSP product (r=0.33; P<0.0001).
Figure 7.
Association between IL10 methylation and IL10 expression. (A) An inverse correlation was found between IL10 methylation and mRNA expression in 372 samples of the stomach adenocarcinoma dataset (TCGA, PanCancer Altas) (r=−0.348, P=5×10−12). (B) Gene Expression Omnibus data mining results showed that the relative expression level of IL10 increased when OC3, SAS, SCC and HSC3 (oral squamous cell carcinoma cell lines) cells were treated with the demethylation agent 5′-AZA.
Figure 8.
Association of CGI methylation with other 7 CpG sites at the IL10 locus based on bioinformatics analysis. (A) The genomic positions (GRCh38/hg38 version) of 7 CpG sites at the IL10 locus. (B) The table shows the association analysis of 7 CpG loci on the IL10 locus. The top table represents the correlation coefficients between 7 CpG sites at the IL10 locus, while the bottom table represents the P-values between 7 CpG sites at the IL10 locus.
Discussion
The present study demonstrated for the first time that IL10 CGI hypomethylation was associated with the risk of GC and with worse OS in patients with GC. IL10 CGI methylation in tumor tissues was significantly associated with patient's age at diagnosis Furthermore, the results from this study demonstrated that the contribution of IL10 CGI hypomethylation to GC was specific to patients with low differentiation, patients with Borrmann types III and IV, non-smoker patients or patients with no family history of cancer.
As an essential anti-inflammatory cytokine, IL10 can be produced in response to pro-inflammatory signals in most immune cells, including macrophages, and T, B and dendritic cells (35). IL10 expression is highly dynamic and requires strict regulation (36). High level of IL10 is observed in the human monocytic cell line following stimulation by coagulation factors, leading to GC cell migration and invasion via transformation of macrophages into tumor-associated macrophage-like cells (37). It was also reported that IL10 can stimulate MCF-7 breast cancer cell proliferation by activating the IL10-signal transducer and activator of transcription 3-neutrophil gelatinase-associated lipocalin axis, which may contribute to tumor progression (38).
A previous study demonstrated that increased IL10 production in chronic lymphocytic leukemia cells was associated with decreased DNA methylation at the IL10 locus (12). Another study also reported that IL10 hypomethylation in cancerous tissues from patients with breast cancer can activate IL10 gene expression (19). It was demonstrated that DNA methylation of the IL10 promoter can inhibit IL10 expression in Th1 cells (39), CD4+ T lymphocytes, macrophages (39), blood cells and gingival tissues (25). In addition, transfection of 1 and 0.6-kb non-CpG methylated IL10 proximal promoter fragments into HeLa cells cannot increase the reporter gene expression, which can be reversed by cassette methylation of these promoter fragments (20). Furthermore, upregulation of IL10 expression in tumor tissues is associated with poor prognosis in patients with breast cancer (40) and laryngeal squamous cell carcinoma (41).
IL10 acts as a cellular molecule with broad-spectrum antiinflammatory activity (42). IL10 overexpression inhibits the phagocytosis of effector cells by macrophages, therefore promoting cancer progression (43). Due to the limited sample size, the present study did not perform correlation analysis between IL10 CGI methylation and IL10 expression level. Therefore, the data from a TCGA dataset were analyzed to determine the association between IL10 methylation and expression level. The data mining in 372 GC tissues from TCGA confirmed that IL10 methylation was negatively correlated with IL10 expression. IL10 hypomethylation may therefore serve a crucial role in the development and progression of GC by upregulating IL10 expression level. The present study demonstrated that IL10 hypomethylation was associated with poor OS in patients with GC, in particular in non-smoker patients and those with poorly differentiated tumor tissues. This study demonstrated that IL10 CGI hypomethylation was associated with the risk of developing GC and the worse prognosis of GC, suggesting its potential role as a diagnostic and prognostic biomarker.
It was previously reported that smoking was associated with increased methylation of microRNA-124a-3 in the gastric mucosa of healthy Japanese volunteers (44), and with increased methylation of transmembrane protein 106A gene (45), mutL homolog 1 gene (46,47), O-6-methylguanine-DNA methyltransferase gene (47), methylated in tumors 25 (MINT25) CGI (47), tumor-suppressor candidate 3 gene (48) and cadherin 1 gene (49) in patients with GC. The present study demonstrated that IL10 CGI hypomethylation was associated with the risk of GC and worse OS in non-smoker patients with GC. The findings from the present study suggested that IL10 CGI methylation may be considered as a biomarker independent of smoking. In this study, 87 patients had no family history of cancer. The results demonstrated that the association between IL10 CGI methylation and the risk of GC was specific to patients with no family history of cancer. These findings suggested that epigenetics may serve a crucial role in the development of cancer for individuals with no family history of cancer (50).
The present study only examined the CpG site within the qMSP product of IL10. Further investigation is therefore required to confirm whether the results from this study corresponded to the methylation of the CpG sites in other IL10 regions. There are seven CpG sites at the IL10 locus in the Infinium Human Methylation 450K BeadChip (Illumina, Inc.). The analysis of the TCGA dataset further demonstrated that the methylation levels of the seven CpG sites among 372 TCGA GC tissue samples were positively correlated with each other, especially for the two CpG sites cg17067005 and cg10978799 that are near the two ends of the qMSP product. However, further investigation is required to verify whether the IL10 CGI methylation corresponds to the promoter methylation of IL10.
In conclusion, the present study provided a potential epigenetic cause for the contribution of IL10 to the risk of developing GC and the OS of patients with GC. Future work may confirm that IL10 CGI hypomethylation is specific to patients with low differentiation, Borrmann types III and IV, a family history of cancer, and non-smokers.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This study was supported by the Zhejiang Medical and Health Science and Technology Project (grant no. 2017KY646), the Suzhou Science and Technology Project (grant no. SYSD2018057) and the K. C. Wong Magna Fund in Ningbo University.
Funding
This study was supported by the Zhejiang Medical and Health Science and Technology Project (grant no. 2017KY646), the Suzhou Science and Technology Project (grant no. SYSD2018057) and the K. C. Wong Magna Fund in Ningbo University.
Availability of data and materials
The IL10 methylation data generated and/or analyzed during the current study are not publicly available due to patient privacy restrictions on this joint project but are available from the corresponding author on reasonable request.
Authors' contributions
JW and SD conceived and designed the study. JT, RP, LX, QM, XY, JZ, HZ, LM and YX performed the experiments. JT, RP and LX analyzed the data. JT, RP, JW and SD wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Zhejiang Province Cancer Hospital. Written consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Senol K, Ozkan MB, Vural S, Tez M. The role of inflammation in gastric cancer. Adv Exp Med Biol. 2014;816:235–257. doi: 10.1007/978-3-0348-0837-8_10. [DOI] [PubMed] [Google Scholar]
- 2.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob Health. 2016;4:e609–e616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196–202. doi: 10.1016/j.canlet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Robinson K, Kaneko K, Andersen LP. Helicobacter: Inflammation, immunology and vaccines. Helicobacter. 2017;22(Suppl 1) doi: 10.1111/hel.12406. doi: 10.1111/hel.12406. [DOI] [PubMed] [Google Scholar]
- 5.Shams AZ, Haug U. Strategies for prevention of gastrointestinal cancers in developing countries: A systematic review. J Glob Health. 2017;7:020405. doi: 10.7189/jogh.07.020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varga MG, Wang T, Cai H, Xiang YB, Gao YT, Ji BT, Pawlita M, Waterboer T, Zheng W, Shu XO, Epplein M. Helicobacter pylori blood biomarkers and gastric cancer survival in China. Cancer Epidemiol Biomarkers Prev. 2018;27:342–344. doi: 10.1158/1055-9965.EPI-17-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi WT, Lauwers GY. Patterns of gastric injury: Beyond Helicobacter pylori. Surg Pathol Clin. 2017;10:801–822. doi: 10.1016/j.path.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Mingomataj EC, Bakiri AH. Regulator versus effector paradigm: Interleukin-10 as indicator of the switching response. Clin Rev Allergy Immunol. 2016;50:97–113. doi: 10.1007/s12016-015-8514-7. [DOI] [PubMed] [Google Scholar]
- 9.Rivas-Ortiz CI, Lopez-Vidal Y, Arredondo-Hernandez LJR, Castillo-Rojas G. Genetic alterations in gastric cancer associated with Helicobacter pylori infection. Front Med (Lausanne) 2017;4:47. doi: 10.3389/fmed.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu W, Pang Q, Lin T, Wang Z, Zhang J, Tai M, Zhang L, Zhang L, Gu M, Liu C, Qu K. A Causal role of genetically elevated circulating interleukin-10 in the development of digestive cancers: Evidence from mendelian randomization analysis based on 29,307 subjects. Medicine (Baltimore) 2016;95:e2799. doi: 10.1097/MD.0000000000002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kindlund B, Sjoling A, Yakkala C, Adamsson J, Janzon A, Hansson LE, Hermansson M, Janson P, Winqvist O, Lundin SB. CD4+ regulatory T cells in gastric cancer mucosa are proliferating and express high levels of IL-10 but little TGF-β. Gastric Cancer. 2017;20:116–125. doi: 10.1007/s10120-015-0591-z. [DOI] [PubMed] [Google Scholar]
- 12.Drennan S, D'Avola A, Gao Y, Weigel C, Chrysostomou E, Steele AJ, Zenz T, Plass C, Johnson PW, Williams AP, et al. IL-10 production by CLL cells is enhanced in the anergic IGHV mutated subset and associates with reduced DNA methylation of the IL10 locus. Leukemia. 2017;31:1686–1694. doi: 10.1038/leu.2016.356. [DOI] [PubMed] [Google Scholar]
- 13.Giacomelli L, Gianni W, Belfiore C, Gandini O, Repetto L, Filippini A, Frati L, Aglianò AM, Gazzaniga P. Persistence of epidermal growth factor receptor and interleukin 10 in blood of colorectal cancer patients after surgery identifies patients with high risk to relapse. Clin Cancer Res. 2003;9:2678–2682. [PubMed] [Google Scholar]
- 14.Chang WJ, Du Y, Zhao X, Ma LY, Cao GW. Inflammation-related factors predicting prognosis of gastric cancer. World J Gastroenterol. 2014;20:4586–4596. doi: 10.3748/wjg.v20.i16.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeguchi M, Hatada T, Yamamoto M, Miyake T, Matsunaga T, Fukumoto Y, Yamada Y, Fukuda K, Saito H, Tatebe S. Serum interleukin-6 and −10 levels in patients with gastric cancer. Gastric Cancer. 2009;12:95–100. doi: 10.1007/s10120-009-0509-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhao S, Wu D, Wu P, Wang Z, Huang J. Serum IL-10 predicts worse outcome in cancer patients: A meta-analysis. PLoS One. 2015;10:e0139598. doi: 10.1371/journal.pone.0139598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X, He Z, Li H, Yao C, Zhang Y, He L, Li S, Huang J, Guo Z. Distinct functional patterns of gene promoter hypomethylation and hypermethylation in cancer genomes. PLoS One. 2012;7:e44822. doi: 10.1371/journal.pone.0044822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen KS, Bamlet WR, Oberg AL, de Andrade M, Matsumoto ME, Tang H, Thibodeau SN, Petersen GM, Wang L. Leukocyte DNA methylation signature differentiates pancreatic cancer patients from healthy controls. PLoS One. 2011;6:e18223. doi: 10.1371/journal.pone.0018223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son KS, Kang HS, Kim SJ, Jung SY, Min SY, Lee SY, Kim SW, Kwon Y, Lee KS, Shin KH, Ro J. Hypomethylation of the interleukin-10 gene in breast cancer tissues. Breast. 2010;19:484–488. doi: 10.1016/j.breast.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Szalmas A, Banati F, Koroknai A, László B, Fehér E, Salamon D, Gergely L, Minárovits J, Kónya J. Lineage-specific silencing of human IL-10 gene expression by promoter methylation in cervical cancer cells. Eur J Cancer. 2008;44:1030–1038. doi: 10.1016/j.ejca.2008.02.046. [DOI] [PubMed] [Google Scholar]
- 21.Mancini M, Veljkovic N, Leo E, Aluigi M, Borsi E, Galloni C, Iacobucci I, Barbieri E, Santucci MA. Cytoplasmatic compartmentalization by Bcr-Abl promotes TET2 loss-of-function in chronic myeloid leukemia. J Cell Biochem. 2012;113:2765–2774. doi: 10.1002/jcb.24154. [DOI] [PubMed] [Google Scholar]
- 22.Klug M, Schmidhofer S, Gebhard C, Andreesen R, Rehli M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013;14:R46. doi: 10.1186/gb-2013-14-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan N, Ali A, Withycombe C, Ahluwalia M, Al-Nasseri RH, Tonks A, Morris K. TET-2 up-regulation is associated with the anti-inflammatory action of Vicenin-2. Cytokine. 2018;108:37–42. doi: 10.1016/j.cyto.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann SR, Moller J, Rauen T, Paul D, Gahr M, Rösen-Wolff Z, Brenner S, Hedrich CM. Dynamic CpG-DNA methylation of Il10 and Il19 in CD4+ T lymphocytes and macrophages: Effects on tissue-specific gene expression. Klin Padiatr. 2012;224:53–60. doi: 10.1055/s-0031-1291359. [DOI] [PubMed] [Google Scholar]
- 25.Larsson L, Thorbert-Mros S, Rymo L, Berglundh T. Influence of epigenetic modifications of the interleukin-10 promoter on IL10 gene expression. Eur J Oral Sci. 2012;120:14–20. doi: 10.1111/j.1600-0722.2011.00917.x. [DOI] [PubMed] [Google Scholar]
- 26.Verschoor CP, McEwen LM, Kohli V, Wolfson C, Bowdish DM, Raina P, Kobor MS, Balion C. The relation between DNA methylation patterns and serum cytokine levels in community-dwelling adults: A preliminary study. BMC Genet. 2017;18:57. doi: 10.1186/s12863-017-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D, Xu J, Yu G, Zhang B, Wang H, Wang C, Ru G, Sun A, Shen L, Wei Q. Expression status of fatty acid synthase (FAS) but not HER2 is correlated with the differentiation grade and prognosis of esophageal carcinoma. Hepatogastroenterology. 2013;60:99–106. doi: 10.5754/hge12415. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Chen C, Bi X, Zhou C, Huang T, Ni C, Yang P, Chen S, Ye M, Duan S. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi: 10.1016/j.gene.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Yang Y, Liu J, Li B, Xu Y, Li C, Xu Q, Liu G, Chen Y, Ying J, Duan S. NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population. Oncotarget. 2017;8:8105–8119. doi: 10.18632/oncotarget.14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C, Pan R, Li B, Huang T, Zhao J, Ying J, Duan S. GPX3 hypermethylation in gastric cancer and its prognostic value in patients aged over 60. Future Oncol. 2019;15:1279–1289. doi: 10.2217/fon-2018-0674. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Chen X, Hu H, Jiang Y, Yu H, Dai J, Mao Y, Duan S. Elevated UMOD methylation level in peripheral blood is associated with gout risk. Sci Rep. 2017;7:11196. doi: 10.1038/s41598-017-11627-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q, Han L, Liu Y, Wang C, Duan D, Lu N, Wang K, Zhang L, Gu K, Duan S, Mai Y. Elevation of PTPN1 promoter methylation is a significant risk factor of type 2 diabetes in the Chinese population. Exp Ther Med. 2017;14:2976–2982. doi: 10.3892/etm.2017.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, Gibbs DL, Weerasinghe A, Huang KL, Tokheim C, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell. 2018;173:305–320.e10. doi: 10.1016/j.cell.2018.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 36.Rutz S, Ouyang W. Regulation of interleukin-10 expression. Adv Exp Med Biol. 2016;941:89–116. doi: 10.1007/978-94-024-0921-5_5. [DOI] [PubMed] [Google Scholar]
- 37.Ma YY, He XJ, Wang HJ, Xia YJ, Wang SL, Ye ZY, Tao HQ. Interaction of coagulation factors and tumor-associated macrophages mediates migration and invasion of gastric cancer. Cancer Sci. 2011;102:336–342. doi: 10.1111/j.1349-7006.2010.01795.x. [DOI] [PubMed] [Google Scholar]
- 38.Jung M, Weigert A, Tausendschon M, Mora J, Ören B, Sola A, Hotter G, Muta T, Brüne B. Interleukin-10-induced neutrophil gelatinase-associated lipocalin production in macrophages with consequences for tumor growth. Mol Cell Biol. 2012;32:3938–3948. doi: 10.1128/MCB.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang W, Lee CG, Lee C, Verma R, Rudra D, Park ZY, Im SH. Locus-specific reversible DNA methylation regulates transient IL-10 expression in Th1 cells. J Immunol. 2018;200:1865–1875. doi: 10.4049/jimmunol.1701162. [DOI] [PubMed] [Google Scholar]
- 40.Bhattacharjee HK, Bansal VK, Nepal B, Srivastava S, Dinda AK, Misra MC. Is interleukin 10 (IL10) expression in breast cancer a marker of poor prognosis? Indian J Surg Oncol. 2016;7:320–325. doi: 10.1007/s13193-016-0512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Lian M, Ma H, Wang R, Ma Z, Wang H, Zhai J, Meng L, Feng L, Bai Y, et al. Competing endogenous RNA network analysis of CD274, IL10 and FOXP3 co-expression in laryngeal squamous cell carcinoma. Mol Med Rep. 2018;17:3859–3869. doi: 10.3892/mmr.2017.8307. [DOI] [PubMed] [Google Scholar]
- 42.Mosser DM, Zhang X. Interleukin-10: New perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015;367:103–107. doi: 10.1016/j.canlet.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Shimazu T, Asada K, Charvat H, Kusano C, Otake Y, Kakugawa Y, Watanabe H, Gotoda T, Ushijima T, Tsugane S. Association of gastric cancer risk factors with DNA methylation levels in gastric mucosa of healthy Japanese: A cross-sectional study. Carcinogenesis. 2015;36:1291–1298. doi: 10.1093/carcin/bgv125. [DOI] [PubMed] [Google Scholar]
- 45.Xu D, Qu L, Hu J, Li G, Lv P, Ma D, Guo M, Chen Y. Transmembrane protein 106A is silenced by promoter region hypermethylation and suppresses gastric cancer growth by inducing apoptosis. J Cell Mol Med. 2014;18:1655–1666. doi: 10.1111/jcmm.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wani M, Afroze D, Makhdoomi M, Hamid I, Wani B, Bhat G, Wani R, Wani K. Promoter methylation status of DNA repair gene (hMLH1) in gastric carcinoma patients of the Kashmir valley. Asian Pac J Cancer Prev. 2012;13:4177–4181. doi: 10.7314/APJCP.2012.13.8.4177. [DOI] [PubMed] [Google Scholar]
- 47.Hong SH, Kim HG, Chung WB, Kim EY, Lee JY, Yoon SM, Kwon JG, Sohn YK, Kwak EK, Kim JW. DNA hypermethylation of tumor-related genes in gastric carcinoma. J Korean Med Sci. 2005;20:236–241. doi: 10.3346/jkms.2005.20.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuasa Y, Nagasaki H, Oze I, Akiyama Y, Yoshida S, Shitara K, Ito S, Hosono S, Watanabe M, Ito H, et al. Insulin-like growth factor 2 hypomethylation of blood leukocyte DNA is associated with gastric cancer risk. Int J Cancer. 2012;131:2596–2603. doi: 10.1002/ijc.27554. [DOI] [PubMed] [Google Scholar]
- 49.Poplawski T, Tomaszewska K, Galicki M, Morawiec Z, Blasiak J. Promoter methylation of cancer-related genes in gastric carcinoma. Exp Oncol. 2008;30:112–116. [PubMed] [Google Scholar]
- 50.Verma M. The role of epigenomics in the study of cancer biomarkers and in the development of diagnostic tools. Adv Exp Med Biol. 2015;867:59–80. doi: 10.1007/978-94-017-7215-0_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The IL10 methylation data generated and/or analyzed during the current study are not publicly available due to patient privacy restrictions on this joint project but are available from the corresponding author on reasonable request.