Abstract
Breast cancer (BC) is the leading cause of death in females worldwide. Although cisplatin is a strong-effect and broad-spectrum chemotherapy drug, resistance to cisplatin remains a significant factor effecting clinical efficacy. The underlying mechanism of cancer cell resistance to cisplatin is not fully understood. MicroRNAs (miRs/miRNAs), as a regulator, are involved in regulating chemosensitivity to numerous chemotherapeutic drugs. The present study aimed to investigate the function of miR-181a-5p as a potential tumor suppressor in improving the efficiency of cisplatin in BC. The IC50 of cisplatin and miR-181a-5p expression were determined in five BC cell lines, and HS578T was selected as an appropriate cell line for subsequent experiments. The sensitivity of HS578T cells to cisplatin was assessed using cell proliferation, migration and apoptosis assays. Western blotting was performed to detect the expression of vitamin D receptor (VDR) and autophagy in HS578T cells. It was found that the increase in autophagy resulted in increased apoptosis and sensitivity to cisplatin in HS578T cells. miR-181a-5p transfection also inhibited the proliferation and migration ability of HS578T cells and induced apoptosis. Meanwhile, HS578T cells have increased sensitivity to cisplatin. VDR, as a target gene and autophagy regulator of miR-181a-5p, was negatively regulated by miR-181a-5p. Upon the decrease in VDR expression, the autophagy in HS578T cells was increased. These results indicate that the increase in autophagy enhanced the chemosensitivity of cisplatin by inducing apoptosis of HS578T cells and by inhibiting proliferation and migration. The present study showed that miR-181a-5p increased the chemical sensitivity of HS578T cells to cisplatin by inhibiting VDR to promote autophagy. The use of miR-181a-5p/autophagy/VDR-based treatment strategies may be a potential method to overcome cisplatin resistance in BC.
Keywords: microRNA-181a-5p, autophagy, cisplatin, breast cancer
Introduction
As the most common cause of death worldwide, breast cancer (BC) mortality ranks first among Chinese women with malignant tumors (1). Although comprehensive treatments with surgery and radiotherapy are used for BC, chemotherapy has been deemed safer and more essential for prolonging survival time or decreasing metastases (2). Cisplatin is an effective strong-effect and broad-spectrum chemotherapy agent for the therapy of BC (3). Cisplatin induces apoptosis in cancer cells by forming platinum-DNA adducts (4). However, cisplatin resistance remains a significant factor limiting clinical efficacy (5). Therefore, sensitivity to cisplatin in BC must be improved.
MicroRNAs (miRs/miRNAs) are a type of endogenously expressed non-coding small RNA that can regulate the expression of various genes (6). miRNAs have been determined to play key roles in various biological processes, including cell proliferation, differentiation and apoptosis (7). In recent years, a growing number of studies have shown that the imbalance of miRNAs may be responsible for resistance (8–10). miR-181a was reported to be differentially expressed in BC (11,12). Therefore, targeted regulation of miRNAs have become a potential method to decrease cisplatin resistance in BC.
Autophagy plays a major homeostatic role in controlling the quality and quantity of proteins and organelles, which can help cells resist poor growth and promote cell survival (13,14). Protective autophagy can also promote chemotherapy-induced apoptosis (15). Vitamin D receptor (VDR), as a regulator of autophagy, is a ubiquitous nuclear receptor that can regulate the expression of numerous genes involved in cell differentiation, proliferation and calcium/phosphate homeostasis (16,17). Studies indicated that VDR acts as a major transcriptional regulator and plays a crucial role in chemotherapy sensitivity (18).
The present study investigated the function and downstream genes of miR-181a-5p, a potential tumor suppressor, in order to improve the efficiency of cisplatin in BC.
Materials and methods
Cell culture and transfection
Human breast cancer cell lines HS578T, HCC70, MDA-MB-231, MDA-MB-468 and BT549 were cultured in corresponding culture medium (HyClone; Cytiva) containing 10% fetal bovine serum (Capricorn Scientific GmbH) and 1% penicillin-streptomycin mixture (MDA-MB-231-L15, Base; MDA-MB-468, L15 medium; Hst578, high sugar DMEM, HCC70, RPMI-1640; and BT549, RPMI-1640) at 37°C and 5% CO2 and sub-cultured every 4 days.
The five cell lines were treated with different concentrations of cisplatin (0.8, 1.6, 3.1, 6.2, 12.5, 25, 50, 100 and 200 µM) for 48 h in order to determine their IC50 of cisplatin. Upon reaching 70% confluence, miR-181a inhibitor (100 pmol, 5′-ACUCACCGACAGCGUUGAAUGUU-3′), miR-181a mimics (100 pmol, 5′-AACAUUCAACGCUGUCGGUGAGUUCACCGACAGCGUUGAAUGUUUU-3′) or control (100 pmol, mimics-NC: 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′; and Inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′), was transfected into HS578T cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. After 8–12 h of cell culture, the medium was replaced with serum-containing medium that was preheated at 37°C and then cultured for another 24 h. Finally, cells were treated with cisplatin according to different experimental groups (Table I). Cells were collected for functional assays after 48 h.
Table I.
Experimental grouping.
| Group | a | b | c | d | e | f | g | h | i | j |
|---|---|---|---|---|---|---|---|---|---|---|
| Cisplatin-NC | + | + | + | + | + | – | – | – | – | – |
| Cisplatin | – | – | – | – | – | + | + | + | + | + |
| Mimics-NC | – | + | – | – | – | – | + | – | – | – |
| Mimics-181a | – | – | + | – | – | – | – | + | – | – |
| Inhibitor-NC | – | – | – | + | – | – | – | – | + | – |
| Inhibitor-181a | – | – | – | – | + | – | – | – | – | + |
NC, negative control.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® (Roche Biotech) was used to isolate total RNA from all cells. cDNA was synthesized using the RevertAid First Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. qPCR was performed on the ABI 7500 Real-Time system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following temperature protocol: 95°C for 10 min; 95°C for 15 sec, 60°C for 20 sec, 72°C for 25 sec for 40 cycles; and 72°C for 5 min. U6 expression was used to normalize the relative expression of miR-181a. The following primer pairs were used for the qPCR: U6 forward, 5′-CTCACTTCGGCAGCACATA-3′ and U6 reverse, 5′-AACTCTTCACGATTTTGTCTGTC-3′; miR-181a forward, 5′-AGCCAACATTCAACGCTGTCG-3′ and miR-181a reverse, 5′-CAGTGCAGGGTCCGAGGTATTC-3′. Data was analyzed from at least three independent experiments and calculated using the 2−ΔΔCq method (19).
Cell proliferation assay
Cells were seeded on a 96-well plate at a density of 5×103 cells/well and cultured overnight in 100 ml of the corresponding medium. Following treatment with the respective cisplatin IC50 for 48 h, Cell Counting Kit-8 (Biosharp Life Sciences) was used to determine cell proliferation, according to the manufacturer's protocol. The results are reported as the ratio of cell proliferation/inhibition, and each experiment was repeated three times. The formula used was as follows: Suppression of proliferation rate (%)=(1-absorbance value of experimental group/absorbance value of control group) ×100%.
Transwell assay
Following treatment with or without cisplatin for 48 h, cells were collected in each group after trypsin digestion. The Matrigel-coated upper chamber of the Transwell insert was inoculated with 1 ml cell suspension at a density of 5×105 cells/ml, while the lower chamber was filled with 500 µl culture medium. Cells were incubated for 24 h. After washing with PBS, cells were fixed with polymethanol at 37°C for 30 min, air dried and stained with 0.1% crystal violet at 37°C for 30 min. The experiment was repeated three times. Cells were randomly observed and counted under a light microscope (magnification, ×100; Olympus BX53; Olympus Corporation).
Flow cytometry analysis of apoptosis
Following treatment with or without cisplatin for 48 h, cells were collected and stained with propidium iodide and Annexin V-FITC according to the manufacturer's protocol (Sangon Biotech Co., Ltd.). Stained cells were analyzed by flow cytometry (Guava EasyCyte; InCyte Software; EMD Millipore). Experiments were repeated three times.
Western blotting
Following treatment with or without cisplatin for 48 h, cells were lysed with RIPA lysis buffer (Sangon Biotech Co., Ltd.). A BCA protein quantitation kit (Sangon Biotech Co., Ltd.) was used to determine protein concentration. After 12% SDS-PAGE, protein samples (30 µg/well) were transferred to PVDF membranes and blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) at room temperature for 1 h. Membranes were incubated with primary antibodies against VDR (1:100; cat. no. ab3508; Abcam) and microtubule-associated proteins 1A/1B light chain 3B (LC3B; 1:2,000; cat. no. ab192890; Abcam) at 37°C for 1 h and then overnight at 4°C. After washing with PBS, membranes were incubated for 1 h at room temperature with HRP-conjugated goat anti-rabbit secondary antibody (1:5,000; cat. no. ab6789; Abcam). Protein signals were visualized using an ECL system (EMD Millipore). GAPDH (1:5,000; cat. no. ab8245; Abcam) was used as the internal control. A multimode microplate reader (Varioskan LUX; Thermo Fisher Scientific, Inc.) was used for densitometry.
Immunofluorescence
Following treatment with or without cisplatin for 48 h, cells were seeded onto coverslips and then fixed in 4% paraformaldehyde at room temperature for 15 min. Cells were washed three times with PBS, permeabilized at room temperature for 20 min with 0.5% Triton X-100 and blocked at room temperature for 1 h with 1% BSA. Cells were incubated at 4°C overnight with rabbit polyclonal anti-VDR antibody (1:100; cat. no. ab3508; Abcam). After washing with PBS, cells were incubated for 2 h at room temperature with FITC-labeled anti-rabbit secondary antibody (1:5,000; cat. no. ab6721; Abcam). Fluorescence microscopy (Olympus Corporation) (magnification, 100×) was used to acquire images.
Statistical analysis
All experiments were repeated at least three times. The data are reported as the mean ± standard deviation. Data were analyzed using SPSS 19.0 (IBM Corp.). One-way ANOVA and Tukey's HSD test were used to analyze significant differences between groups or among groups. P<0.05 was considered to indicate a statistically significant difference.
Results
HS578T as a cell model for experiments
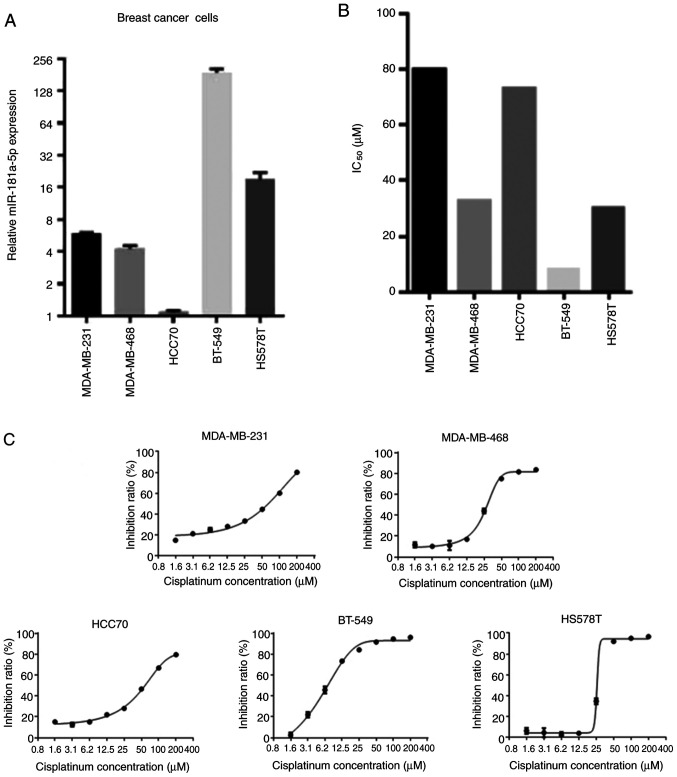
To select the most appropriate cell line suitable for subsequent experiments, the relative expression of miR-181a-5p and the IC50 of cisplatin was assessed in the five types of BC cells (Fig. 1). Since HS578T showed average miR-181a-5p expression and IC50 values compared with other cells, this cell line was chosen for subsequent experimentation.
Figure 1.
HS578T as a cell model for experiments. (A) Relative expression of miR-181a-5p in the five breast cancer cell lines. (B) The IC50 of cisplatin and (C) inhibition ratio of different concentrations of cisplatin in the five breast cancer cell lines. miR, microRNA.
miR-181a-5p enhances the chemosensitivity of cisplatin by regulating biological processes in HS578T cells
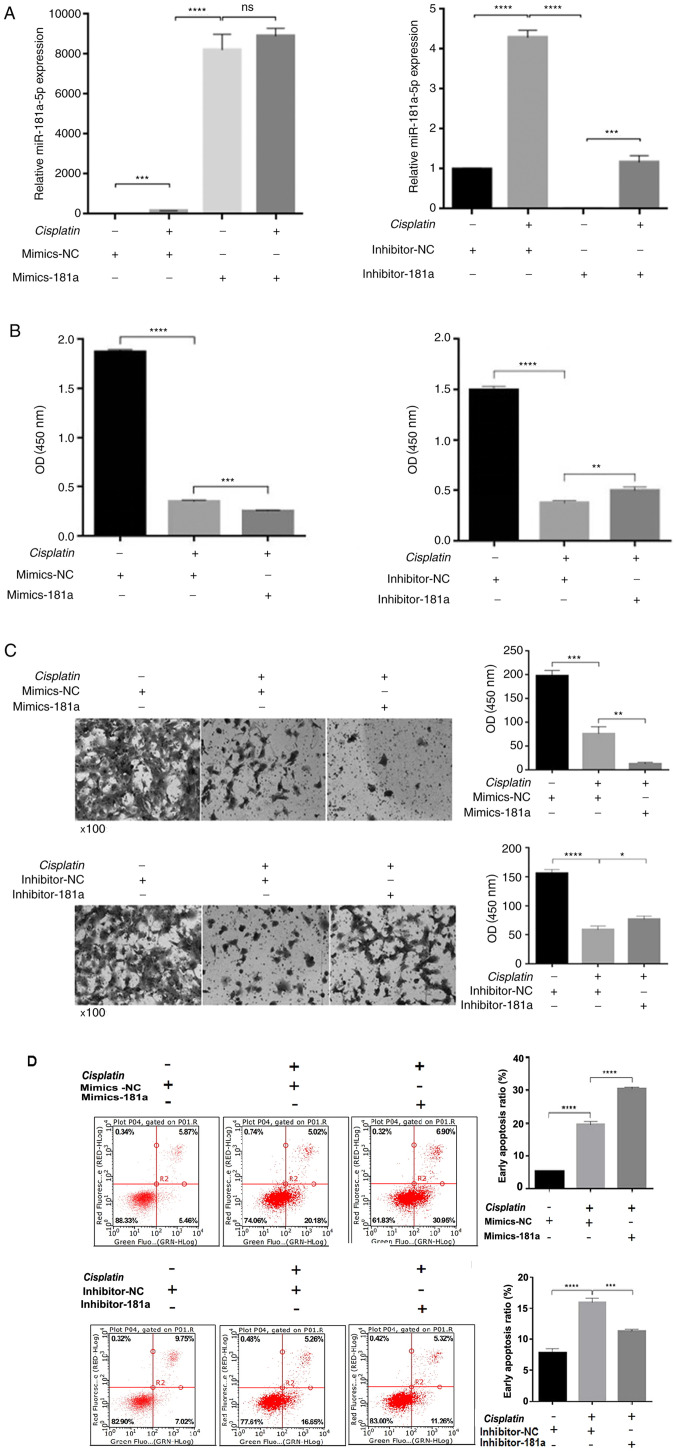
To investigate whether miR-181a-5p affected the therapeutic effects of cisplatin by regulating proliferation, migration and apoptosis, miR-181a-5p mimics, miR-181a-5p inhibitor or control was transfected into HS578T cells and then treated with or without 30.26 µM cisplatin for 48 h. As shown in Fig. 2A, miR-181a-5p expression was significantly different among the groups, indicating successful transfection. Cisplatin treatment increased miR-181a-5p expression, which may be due to the synergy of the drug. Cell viability was then detected using a cell proliferation assay. Following 48 h of treatment with cisplatin, compared with the control group, cell viability was significantly inhibited in the mimics-181a group, while the cell viability recovered to some extent in the inhibitor-181a group (Fig. 2B). When miR-181a-5p is overexpressed, the cell invasion ability was weaker compared with the control group. Cell invasion ability was recovered in the inhibitor-181a group compared with the control group (Fig. 2C). Subsequently, flow cytometry was performed to determine whether miR-181a-5p overexpression can enhance cisplatin-induced apoptosis. miR-181a-5p overexpression increased cisplatin-induced apoptosis compared with the control group. Meanwhile, in the inhibitor-181a group, apoptosis was significantly decreased compared with the control group (Fig. 2D).
Figure 2.
miR-181a-5p enhances cisplatin chemosensitivity by regulating biological processes in HS578T cells. (A) miR-181a-5p expression among the groups. (B) Cell proliferation was determined using a Cell Counting Kit-8 assay. (C) Cell invasion ability was determined using a Transwell assay. (D) Cellular apoptosis was determined by flow cytometry. Data were reported as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. control group. NC, negative control; miR, microRNA.
miR-181a-5p potentially enhances cisplatin chemosensitivity by negatively regulating VDR
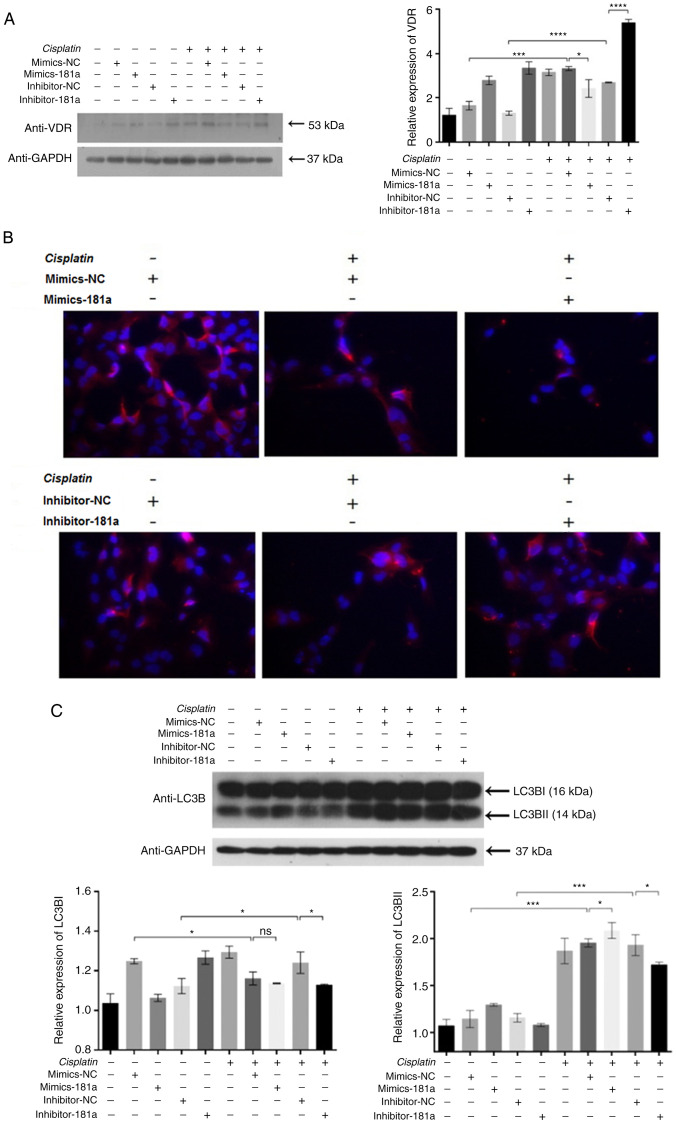
miR-181a-5p overexpression inhibited the VDR expression in HS578T cells, as shown by western blotting (Fig. 3A). However, in the absence of cisplatin, miR-181a-5p mimics enhanced VDR expression, which may be due to the limited inhibitory effect of miR-181a-5p alone. Meanwhile, miR-181a-5p and cisplatin exerts synergistic effects to effectively inhibit VDR expression. Inhibition of miR-181a-5p expression resulted in an increase in VDR expression (column 5 vs. column 10). TargetScan prediction revealed VDR as a potential target gene of miR-181a-5p. Since vitamin D signals via VDR and is involved with preventing cancer (20), the present study focused on investigating VDR. To verify the effects of miR-181a-5p on VDR expression, western blotting and immunofluorescence was performed. Western blotting results showed that miR-181a-5p overexpression inhibited the expression of VDR levels in HS578T cells (Fig. 3A). Immunofluorescence assay showed that VDR is located in the cytoplasm, and the expression levels were consistent with those observed in western blot assays (Fig. 3B). These results indicated that following the administration of cisplatin, miR-181a-5p directly suppressed the endogenous expression of VDR in HS578T cells. Based on the association between miR-181a-5p with VDR, and since VDR signaling is an essential mediator of autophagy, the present study hypothesized that miR-181a-5p might inhibit or induce autophagy through regulating VDR. Subsequently, western blotting was used for detecting the levels of autophagy. Following cisplatin treatment, autophagy levels were lower in the inhibitor-181a group compared with the control group (Fig. 3C). These data showed that VDR downregulation enhanced the level of autophagy. In conclusion, the results support that miR-181a-5p overexpression could increase the chemosensitivity of HS578T cells to cisplatin by inhibiting VDR-mediated cell autophagy.
Figure 3.
VDR potentially enhances cisplatin chemosensitivity in HS578T cells. (A) VDR expression determined by western blotting. (B) VDR expression determined by immunofluorescence assay (magnification, ×100). (C) LC3BI and LC3BII expression in cells was determined by western blotting. Data reported as the mean ± standard deviation of three independent experiments. *P<0.05, ***P<0.001 and ****P<0.0001 vs. control group. VDR, vitamin D receptor; NC, negative control; LC3BI/II, microtubule-associated proteins 1A/1B light chain 3B.
Discussion
There are 280,000 new cases of BC and 66,000 deaths each year in China, accounting for 12.2% of all newly diagnosed cases and 9.6% of all deaths from BC worldwide (1). With the advancement in medical technology, more treatment options are available for improving the quality of life of patients with BC. As an essential chemotherapeutic agent, cisplatin has successfully enhanced the survival rate of patients with cancer, such as gastric (21), testicular cancer (22) and BC (23). However, lower cisplatin sensitivity still exists due to chemoresistance and leads to poor prognosis (24). Recent studies have indicated that altered of miRNAs act as a regulator of chemosensitivity in BC, which has become a research focus (25,26).
The present study investigated the potential role of miR-181a-5p in altering cisplatin resistance in BC (11,12). Five types of BC cell lines were chosen to identify a suitable cell line for subsequent experiments, which included HS578T, HCC70, MDA-MB-231, MDA-MB-468 and BT549 cells. miR-181a-5p expression and cisplatin IC50 in these cell lines were determined. Finally, HS578T chosen for subsequent experiments, due to the average levels of miR-181a-5p and cisplatin IC50 measured compared with other cells. HS578T cells were transfected of miR-181a-5p mimic, miR-181a-5p inhibitor or control and then treated with or without cisplatin. Compared with the control group, the cell viability and invasion abilities were significantly inhibited in the mimics-181a group, while viability and invasion was recovered to a certain extent in the inhibitor-181a group. It was found that cisplatin treatment in the mimics-181a group resulted in high levels of HS578T cell death. Compared with the control group, miR-181a-5p overexpression increased cisplatin-induced apoptosis, while in the inhibitor-181a group, apoptosis was significantly decreased. In conclusion, upregulation of miR-181a-5p resulted in an increased apoptosis ratio in HS578T cells, while proliferation and migration abilities were inhibited. Meanwhile, the sensitivity of HS578T cells to cisplatin increased.
TargetScan prediction revealed VDR as a potential target gene of miR-181a-5p.VDR is expressed in a wide variety of tissues. Vitamin D signals through VDR, where 1,25-dihydroxy vitamin D3 binds with VDR to modulate target gene transcription. In cancer cells, the modulation includes preventing cell differentiation and proliferation and the regulation of programmed cell death, such as apoptosis and autophagy (27,28). Studies have shown that VDR regulates autophagy in luminal BC cells (29) and other cells (16,30). To verify whether VDR can regulate autophagy as the target gene of miR-181a-5p, immunofluorescence and western blot assays were performed. The western blotting results showed that miR-181a-5p overexpression inhibited the expression of VDR levels in HS578T cells, and VDR expression significantly increased in the inhibitor-181a group compared with the control group. Immunofluorescence assay results were consistent with the western blotting. Due to time constraints, luciferase reporter assay was not performed to directly prove that miR-181a-5p directly suppresses VDR expression. However, both western blotting and immunofluorescence assays results showed that miR-181a-5p could negatively regulate VDR. A luciferase reporter assay will be performed in future studies. LC3 immunoblotting was performed to determine cell autophagy. It was shown that decreased expression of VDR resulted in the increased autophagy of HS578T cells. Additionally, the administration of cisplatin increased autophagy in cells. This indicated that VDR inhibits the occurrence of autophagy, and the administration of cisplatin can increase the levels of miR-181a-5p. Increasing miR-181a-5p expression can increase autophagy. As aforementioned, the synergy of miR-181a-5p and cisplatin can promote HS578T cell apoptosis and inhibit migration. Since all tests were performed under the same conditions, it can be concluded that the increase in autophagy could enhance cisplatin chemosensitivity in HS578T cells.
To conclude, miR-181a-5p upregulation could increase chemosensitivity to cisplatin, decrease VDR expression and result in increased autophagy in HS578T cells. These data indicated that there was an inverse association between VDR and autophagy. This was consistent with a previously published study showing that VDR can directly regulate autophagy in BC cells. miR-181a-5p increased the chemical sensitivity of HS578T cells to cisplatin by inhibiting VDR to promote autophagy. However, further research must be performed to determine the underlying mechanism. Furthermore, miR-181a-5p and VDR might be attractive candidates for a novel method for overcoming cisplatin resistance in BC.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Youth Project of Fujian Health Commission (grant no. 2018-2-71) and the Xiamen Youth Innovation Talents Project (grant no. 2015-A-03).
Funding
The present study was supported by the Youth Project of Fujian Health Commission (grant no. 2018-2-71) and the Xiamen Youth Innovation Talents Project (grant no. 2015-A-03).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HY, JL and ZS provided the study concept and design. JL, XC and MS wrote and revised the manuscript. JL, XC, MS, XQ and YW participated in discussion and made a significant contribution to the interpretation of the results. JL, CL, XL and LZ collected the data, and performed the experiments. XC and MS performed the statistical analyses. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 2.Macpherson IR, Spiliopoulou P, Rafii S, Saggese M, Baird RD, Garcia-Corbacho J, Italiano A, Bonneterre J, Campone M, Cresti N, et al. A phase I/II study of epertinib plus trastuzumab with or without chemotherapy in patients with HER2-positive metastatic breast cancer. Breast Cancer Res. 2019;22:1. doi: 10.1186/s13058-019-1178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin T, Li B, Feng X, Fan S, Liu L, Liu D, Mao J, Lu Y, Yang J, Yu X, et al. Abnormally elevated USP37 expression in breast cancer stem cells regulates stemness, epithelial-mesenchymal transition and cisplatin sensitivity. J Exp Clin Cancer Res. 2018;37:287. doi: 10.1186/s13046-018-0934-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Grady S, Finn SP, Cuffe S, Richard DJ, O'Byrne KJ, Barr MP. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev. 2014;40:1161–1170. doi: 10.1016/j.ctrv.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Wu G, Zhou W, Pan X, Sun Y, Xu H, Shi P, Li J, Gao L, Tian X. miR-100 reverses cisplatin resistance in breast cancer by suppressing HAX-1. Cell Physiol Biochem. 2018;47:2077–2087. doi: 10.1159/000491476. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Kwon Y, Kim Y, Jung HS, Jeoung D. Role of HDAC3-miRNA-CAGE network in anti-cancer drug-resistance. Int J Mol Sci. 2018;20:51. doi: 10.3390/ijms20010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, Shen H. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer. 2017;16:148. doi: 10.1186/s12943-017-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y, Fu X, Li Q, Wang Y, Fan D, Zhou Q, Kuang W, Shen L. MicroRNA-181 serves an oncogenic role in breast cancer via the inhibition of SPRY4. Mol Med Rep. 2018;18:5603–5613. doi: 10.3892/mmr.2018.9572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Strotbek M, Schmid S, Sánchez-González I, Boerries M, Busch H, Olayioye MA. miR-181 elevates Akt signaling by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int J Cancer. 2017;140:2310–2320. doi: 10.1002/ijc.30661. [DOI] [PubMed] [Google Scholar]
- 13.Kim KH, Lee MS. Autophagy-a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10:322–337. doi: 10.1038/nrendo.2014.35. [DOI] [PubMed] [Google Scholar]
- 14.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo XL, Hu F, Wang H, Fang JM, Zhu ZΖ, Wei LΧ, Xu Q. Inhibition of autophagy in hepatocarcinoma cells promotes chemotherapeutic agent-induced apoptosis during nutrient deprivation. Oncol Rep. 2018;39:773–783. doi: 10.3892/or.2017.6115. [DOI] [PubMed] [Google Scholar]
- 16.Sun J. VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy. 2016;12:1057–1058. doi: 10.1080/15548627.2015.1072670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhu J, DeLuca HF. Where is the vitamin D receptor? Arch Biochem Biophys. 2012;523:123–133. doi: 10.1016/j.abb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, Collisson EA, Connor F, Van Dyke T, Kozlov S, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulgakova O, Zhabayeva D, Kussainova A, Pulliero A, Izzotti A, Bersimbaev R. miR-19 in blood plasma reflects lung cancer occurrence but is not specifically associated with radon exposure. Oncol Lett. 2018;15:8816–8824. doi: 10.3892/ol.2018.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014;14:342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 21.Mahlberg R, Lorenzen S, Thuss-Patience P, Heinemann V, Pfeiffer P, Möhler M. New perspectives in the treatment of advanced gastric cancer: S-1 as a novel oral 5-FU therapy in combination with cisplatin. Chemotherapy. 2017;62:62–70. doi: 10.1159/000443984. [DOI] [PubMed] [Google Scholar]
- 22.Albrecht W. Long-term effects of cisplatin-based chemotherapy in testicular cancer patients-what is important? Urologe A. 2019;58:1212–1216. doi: 10.1007/s00120-019-1000-8. (In German) [DOI] [PubMed] [Google Scholar]
- 23.Al-Bahlani S, Al-Lawati H, Al-Adawi M, Al-Abri N, Al-Dhahli B, Al-Adawi K. Fatty acid synthase regulates the chemosensitivity of breast cancer cells to cisplatin-induced apoptosis. Apoptosis. 2017;22:865–876. doi: 10.1007/s10495-017-1366-2. [DOI] [PubMed] [Google Scholar]
- 24.Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol Res. 2016;106:27–36. doi: 10.1016/j.phrs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Adhami M, Haghdoost AA, Sadeghi B, Malekpour Afshar R. Candidate miRNAs in human breast cancer biomarkers: A systematic review. Breast Cancer. 2018;25:198–205. doi: 10.1007/s12282-017-0814-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang YW, Zhang W, Ma R. Bioinformatic identification of chemoresistance-associated microRNAs in breast cancer based on microarray data. Oncol Rep. 2018;39:1003–1010. doi: 10.3892/or.2018.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Høyer-Hansen M, Nordbrandt SP, Jäättelä M. Autophagy as a basis for the health-promoting effects of vitamin D. Trends Mol Med. 2010;16:295–302. doi: 10.1016/j.molmed.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Welsh J. Vitamin D and cancer: Integration of cellular biology, molecular mechanisms and animal models. Scand J Clin Lab Invest Suppl. 2012;243:103–111. doi: 10.3109/00365513.2012.682870. [DOI] [PubMed] [Google Scholar]
- 29.Tavera-Mendoza LE, Westerling T, Libby E, Marusyk A, Cato L, Cassani R, Cameron LA, Ficarro SB, Marto JA, Klawitter J, Brown M. Vitamin D receptor regulates autophagy in the normal mammary gland and in luminal breast cancer cells. Proc Natl Acad Sci USA. 2017;114:E2186–E2194. doi: 10.1073/pnas.1615015114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paik S, Kim JK, Chung C, Jo EK. Autophagy: A new strategy for host-directed therapy of tuberculosis. Virulence. 2019;10:448–459. doi: 10.1080/21505594.2018.1536598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.