Abstract
Liver cancer ranks in the top 10 most common malignancies for both mortality rate and incidence worldwide. Hepatocellular carcinoma (HCC) is the most common subtype of liver cancer. It has been reported that long non-coding RNA GABPB1 intronic transcript 1 (IT1) is downregulated in lung cancer and predicts poor survival. However, its role in live cancer remains unclear. Therefore, the present study aimed to investigate the role of GABPB1-IT1 in HCC. A total of 64 patients with HCC (40 males and 24 females; range, 43–67 years old; mean age=55.1±5.1 years) were enrolled at the 96604 Military Hospital of the Chinese People's Liberation Army between May 2012 and May 2014. The expression levels of GABPB1-IT1 and microRNA (miR)-93 in tumor and adjacent normal tissues were measured using quantitative PCR. A dual luciferase activity assay was performed to analyze the interaction between miR-93 and GABPB1-IT1. A Cell Counting Kit-8 assay was used to analyze the effect of miR transfection on the proliferation of SNU-398 cells. It was demonstrated that GABPB1-IT1 can interact with miR-93 in HCC cells, while overexpression of GABPB1-IT1 and miR-93 in HCC cells did not affect the expression of each other. GABPB1-IT1 was downregulated in HCC tissues compared with paired non-tumor tissues and predicted poor survival. Notably, overexpression of GABPB1-IT1 in HCC cells led to upregulation of pigment epithelium-derived factor (PEDF), a target of miR-93. In addition, overexpression of GABPB1-IT1 reduced the enhancing effects of miR-93 on HCC cell proliferation. Therefore, GABPB1-IT1 may upregulate PEDF through miR-93 to suppress cell proliferation in HCC.
Keywords: GABPB1 intronic transcript 1, hepatocellular carcinoma, miR-93, pigment epithelium-derived factor
Introduction
Liver cancer ranks in the top 10 most common malignancies for both mortality rate and incidence (1). In 2018, there were 841,080 new cases of liver cases (4.7% of all new cancer cases) and caused 781,631 deaths (8.2% of all cancer-associated deaths) worldwide (2). The number of deaths is close to the number of new cases. Hepatocellular carcinoma (HCC) is the most common subtype of liver cancer (3). Hepatitis B virus (HBV) or C virus (HCV) infections, as well as alcohol abuse, are major risk factors for HCC (4). With the improved understanding of HBV and HCV infections, the molecular pathogenesis of HCC has also been extensively studied (5,6). However, the precise molecular pathways underlying HCC development have not been fully elucidated.
The majority of studies on the molecular pathogenesis of HCC focus on the regulation of protein-coding genes (7–9), while the roles of most non-coding (nc)RNAs, such as micro (mi)RNAs (miRs) and long non-coding (lnc)RNAs, remain unclear in HCC. Although ncRNAs do not encode proteins, they participate in the regulation of gene expression at multiple levels to promote or suppress cancer development (10–12). Therefore, functional characterization of critical ncRNAs in cancer biology provides novel insights into the development of novel targeted therapies (13). In a recent study, lncRNA GABPB1 intronic transcript 1 (IT1) was reported to be downregulated in non-small cell lung cancer (NSCLC) and predicts poor survival, indicating its potential tumor suppressive role (14). However, the involvement of GABPB1-IT1 in other cancer types remains unknown. miR-93, whose sequence is located in the intron of the MCM7 gene at chromosome 7q22.1, has been found to be an oncogenic micro RNA in several cancer types, such as liver and bladder cancer (15,16). Moreover, it has been reported that miR-93 promotes the proliferation and invasion of bladder cancer cells by targeting pigment epithelium-derived factor (PEDF), a glycoprotein from the family of non-inhibitory serpins (16). Considering that the interaction between GABPB1-IT1 and miR-93 was predicted by IntaRNA 2.0, the present study aimed to investigate the role of GABPB1-IT1 in HCC, and whether it functions by regulating miR-93/PEDF axis.
Materials and methods
Patients with HCC and follow-up
The present study was approved by The Ethics Committee of 96604 Military Hospital of the Chinese People's Liberation Army (Lanzhou, China). A total of 64 patients with HCC (40 males and 24 females; age range, 43–67 years; mean age ± SEM=55.1±5.1 years) were enrolled between May 2012 and May 2014. All patients were subjected to histopathological examination to confirm HCC. No previous history or family history of malignancies were observed and patients with other clinical disorders were excluded. No therapy was initiated prior to the study. All patients provided informed written consent. Biopsies were performed on all patients using fine needle to collect paired HCC and adjacent (3-cm away from tumor) tissue samples from each patient. Tissue samples were frozen in liquid nitrogen and stored at −80°C before use. There were 12, 14, 17 and 21 cases at American Joint Committee on Cancer stage I, II, III and IV, respectively (17). From the day of admission, all patients were followed-up for 5 years in a monthly manner through telephone to record their survival. The survival of patients was recorded and all patients completed the follow-up. Overall survival was calculated from the day of diagnosis to the day of death or last follow-up.
HCC cells and transfection
Human HCC cell lines SNU-398 and C3A (American Type Culture Collection) were used. SNU-398 and C3A cells were cultivated in a cell culture medium composed of 10% FBS and 90% RPMI-1640 (both Gibco; Thermo Fisher Scientific, Inc.). Cell culture conditions were 5% CO2 at 37°C and 95% humidity. Cells were collected at ~80% confluence to perform subsequent transfections. The backbone to construct the expression vectors of GABPB1-IT1 and pigment epithelium-derived factor (PEDF). miR-93 mimic and negative control (NC) miRNA (scrambled) were purchased from Sigma-Aldrich; Merck KGaA. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect miRNA (45 nM) or vector (12 nM) into SNU-398 cells with a cell density of 106 cells/ml at 37°C for 24 h. Untransfected cells were used as control (C) cells. NC cells were cells transfected with empty pcDNA 3.1 vector or NC miRNA. Subsequent experiments were carried out using cells collected 48 h after transfection at room temperature. IntaRNA 2.0 predicted that GABPB1-IT1 could bind to the 3′UTR region of miR-93 and then a dual luciferase activity assay was performed to verify their interaction. The GABPB1-IT1 luciferase reporter vector was constructed using the pGL3 vector (Promega Corporation). SNU-398 cells were co-transfected with miR-93 mimic + GABPB1-IT1 vector (miR-93 group) or NC miRNA + GABPB1-IT1 vector (NC miRNA group) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). In addition, SNU-398 cells were also co-transfected with miRNA and GABPB1-IT1 inhibitor (Beyotime Institute of Technology) using Lipofectamine 2000. Luciferase activity was measured 48 h post-transfection by a Dual-Luciferase Reporter Assay kit (Promega Corporation). The firefly luciferase activity was normalized to the Renilla luciferase activity.
RNA extraction and reverse transcription-quantitative (RT-q)PCR assays
Ribozol (Sigma-Aldrich; Merck KGaA) was used to isolate total RNAs from tissues and cells. miRNAs were harvested by precipitating RNAs with 85% ethanol. RNA concentrations were measured using NanoDrop 2000 (Thermo Fisher Scientific. Inc.) and genomic DNAs were digested using gDNA eraser (Takara Bio, Inc.). RNA samples with an OD 260/280 ratio ~2.0 (pure RNA) were reverse transcribed using the PrimeScript RT Reagent kit (Takara Bio, Inc.). The detailed workflow was as follows: 37°C For 15 min and 85°C for 5 sec then maintained at 4°C. Then the qPCR assays performed using SYBR-Green RT-PCR kit (Vazyme Biotech Co., Ltd.) to measure the expression levels of GABPB1-IT1 and PEDF mRNA. The detailed workflow was as follows: 95°C For 10 min, 40 cycles of 95°C for 15 sec and 60°C for 45 sec, final annealing at 72°C for 10 min. The expression levels of mature miR-93 were measured using the All-in-One™ miRNA RT-qPCR Detection kit (GeneCopoeia, Inc.). U6 and GAPDH were used as the endogenous controls of miR-93 and PEDF, respectively. PCR reactions were performed in triplicate and Cq values were processed using the Δ-Δq (ΔΔCq) method (18).
Western-blot assay
Total proteins were isolated from SNU-398 cells using RIPA lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.). Protein concentrations were measured using a BCA assay (Sigma-Aldrich; Merck KGaA). All protein samples were denatured at 95°C for 10 min, followed by loading 25 µg protein/lane onto 8% gels and resolving using SDS-PAGE. Then these samples were transferred to the polyvinylidene fluoride (PVDF) membranes. All membranes were blocked with PBS containing 5% non-fat milk at room temperature for 2 h. Primary rabbit antibody of GAPDH (1:1,000; cat. no. ab9485) or PTEN (1:500; cat. no. ab14993) (both Abcam) was used to incubate with the membranes at 4°C for 12 h, followed by incubation with HRP Goat Anti-Rabbit (IgG) (1:1,000; ab97051; Abcam) secondary antibody at room temperature for 2 h. Enhanced chemiluminescence kit (Sigma-Aldrich; Merck KGaA) was used to visualize the signals, which were quantified by Quantity One software version 4.6 (Bio-Rad Laboratories, Inc.).
Cell proliferation assay
A Cell counting Kit (CCK)-8 (Dojindo Molecular Technologies, Inc.) was used to investigate the effect of transfection on the proliferation of SNU-398 and C3A cells. Each well of a 96-well plate was seeded with 3,000 cells in 0.1 ml cell culture medium, followed by cell culture under the aforementioned conditions. Cells were collected every 24 h for 4 days. At 4 h before cell collection, 10 µl of CCK-8 solution was added to each well. OD values were measured at 450 nM using a microplate reader.
In vivo animal study
In total, 20 female nude mice (4–6 weeks old, 20–22 g) were used. These mice were raised in a specific pathogen-free room under standardized housing conditions (temperature 22–25°C, relative humidity 40–50%, 12 h artificial light daily) with free access to food and water. Mice were observed daily and humanely euthanized by injection of sodium pentobarbital with humane endpoint criteria as previously described, including severe tumor burden (>20 mm in diameter), labored breathing, prostration, unresponsive to external stimuli, obviously body weight loss and body temperature drop (19,20). For investigating the role of GABPB1-IT1, 106 C3A cells stably overexpressing GABPB1-IT1 were suspended in mixture of PBS and Matrigel (1:1) and subcutaneously injected into the flank of nude mice. Tumor sizes were measured every week. The tumor size was measured using a Vernier caliper using the formula V=1/2 (LxW2), where L represents the length (longest dimension), and W represents the width (shortest dimension). After 6 weeks, all mice were euthanized by injection of sodium pentobarbital (100 mg/kg) followed by cervical dislocation. Finally, the xenograft tissues were used for qPCR to analyze the expression of GABPB1-IT1. The animal experiments were approved by The Ethics Committee of 96604 Military Hospital of the Chinese People's Liberation Army (Lanzhou, China; approval no. 202065433).
Statistical analysis
Data from three biological replicates were expressed as mean values ± SEM, unless otherwise shown. Unpaired t-tests were used to compare differences between two groups. Paired t-tests were used to compare HCC and adjacent normal tissues. ANOVA (one-way) and Tukey's post hoc test were used to compare differences among multiple groups. With the median expression levels of GABPB1-IT1 (1.96) in HCC tissues as cut-off value, the 64 patients were grouped into high and low GABPB1-IT1 level groups (both n=32). Survival curves were plotted for both groups and the log-rank test was used to compare survival curves. Linear regression was used for correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
GABPB1-IT1 and miR-93 interact with each other but do not regulate the expression of each other
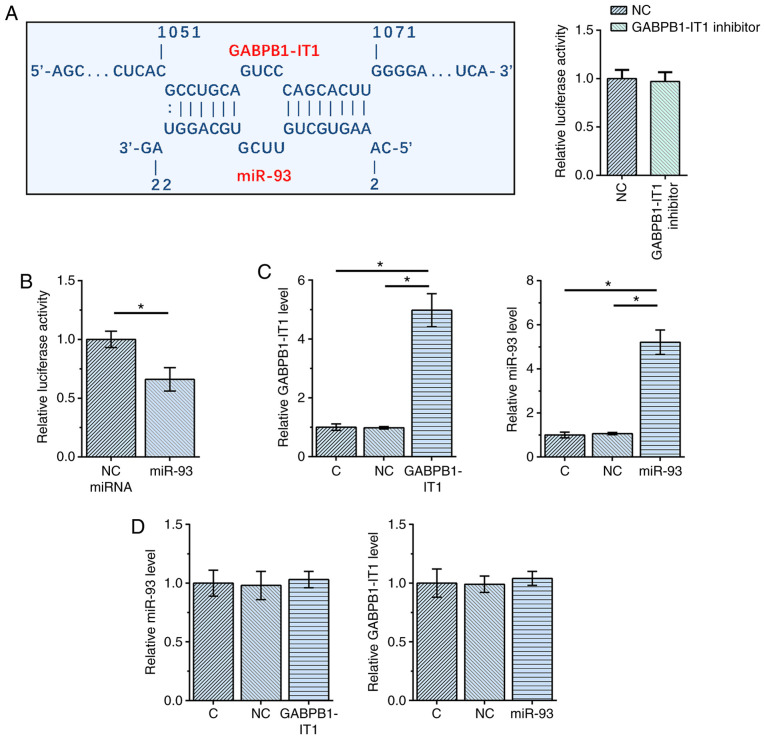
The interaction between GABPB1-IT1 and miR-93 was first predicted using IntaRNA 2.0 (21). It was observed that GABPB1-IT1 and miR-93 could form multiple base pairs. The relative activity of luciferase was not significantly changed by co-transfection of miRNA and GABPB1-IT1 inhibitor (Fig. 1A). A dual luciferase activity assay was performed by co-transfecting SNU-398 cells with miR-93 mimic + GABPB1-IT1 vector (miR-93 group) or NC miRNA + GABPB1-IT1 vector (NC miRNA group). Compared with NC miRNA group, the relative luciferase activity was significantly lower in miR-93 group (P<0.05; Fig. 1B), indicating direct interaction. SNU-398 cells were transfected with GABPB1-IT1 expression vector or miR-93 mimic, followed by confirmation of the overexpression of GABPB1-IT1 and miR-93 using RT-qPCR at 48 h post-transfection (both P<0.05 vs. NC and C; Fig. 1C). Notably, overexpression of GABPB1-IT1 and miR-93 did not affect the expression of each other (Fig. 1D). Taken together, our data suggest that GABPB1-IT1 and miR-93 interact with each other but do not regulate the expression of each other.
Figure 1.
GABPB1-IT1 and miR-93 interact with each other but do not regulate the expression of each other. (A) Interaction between GABPB1-IT1 and miR-93 was predicted using IntaRNA 2.0 and (B) a dual luciferase activity assay was performed. SNU-398 cells were transfected with (C) GABPB1-IT1 expression vector or (D) miR-93 mimic, followed by the confirmation of the overexpression of GABPB1-IT1 and miR-93 using RT-qPCR at 48 h post-transfection *P<0.05 vs. respective control. RT-q, reverse transcription-quantitative; NC, negative control; miR, microRNA.
Downregulation of GABPB1-IT1 predicts poor survival for patients with HCC
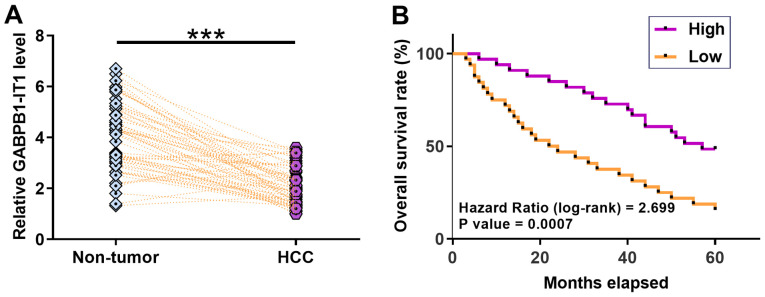
According to patients' medical records, 23 patients were positive for HBV only, 18 were positive for HCV only, eight were HCV and HBV positive and 15 were HCV and HBV negative. The expression levels of GABPB1-IT1 in paired HCC and non-tumor tissues from 64 patients with HCC were measured using RT-qPCR. It was observed that GABPB1-IT1 was significantly downregulated in HCC tissues compared with that in non-tumor tissues (P<0.001; Fig. 2A). Survival curve analysis showed that patients in the low GABPB1-IT1 level group had a lower overall survival rate during the 5-year follow-up compared with patients in the high GABPB1-IT1 level group (P=0.0007; Fig. 2B). miR-93 was significantly upregulated in HCC (Fig. S1A), and PEDF was significantly downregulated in HCC (Fig. 1SB) compared with in non-tumor tissues (both P<0.001). No significant differences in the expression levels of GABPB1-IT1 were observed among patients at different clinical stages (Fig. S2). Linear regression analysis showed that the expression of GABPB1-IT1 and miR-93 were not significantly correlated with each other in HCC tissues (R2=0.02 and P=0.223), while the expression of GABPB1-I T1 and PEDF were positively correlated (R2=0.41 and P<0.001), and miR-93 and PEDF (R2=0.67 and P<0.001) were negatively correlated in HCC tissues (Fig. S3). Taken together, our data suggest that decreased GABPB1-IT1 predicts a poor survival for patients with HCC.
Figure 2.
Downregulation of GABPB1-IT1 predicts the poor survival of patients with HCC. (A) Expression levels of GABPB1-IT1 in paired HCC and non-tumor tissues from 64 patients were measured using reverse transcription-quantitative PCR. (B) Survival curves were plotted for both groups and log-rank test was used to compare survival curves. ***P<0.001. HCC, hepatocellular carcinoma.
Overexpression of GABPB1-IT1 upregulates PEDF
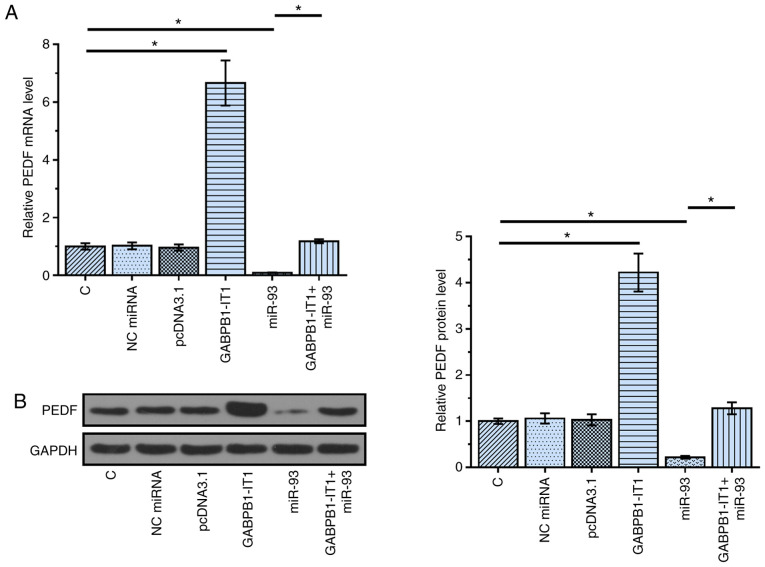
PEDF is a direct target of miR-93 (16). The function of miRNA sponges is to suppress the function of miRNAs, such as targeting downstream genes (22). To test the possibility that GABPB1-IT1 can sponge miR-93, the effects of overexpression of GABPB1-IT1 and miR-93 on the expression of PEDF were analyzed using RT-qPCR (Fig. 3A) and western blotting (Fig. 3B). Compared with the C group, overexpression of miR-93 resulted in the downregulation of PEDF (P<0.05). In contrast, compared with the C group, overexpression of GABPB1-IT1 in GABPB1-IT1 group led to the upregulation of PEDF (P<0.05). Compared with the miR-93 group, overexpression of GABPB1-IT1 in GABPB1-IT1+miR-93 group reduced the effects of miR-93 on the expression of PEDF (P<0.05). Taken together, our data suggest that GABPB1-IT1 may sponge miR-93 to up-regulate PEDF.
Figure 3.
Overexpression of GABPB1-IT1 upregulates PEDF. PEDF is a direct target of miR93. To test the possibility that GABPB1-IT1 can sponge miR-93, the effects of overexpression of GABPB1-IT1 and miR-93 on the expression of PEDF were analyzed using (A) reverse transcription-quantitative PCR and (B) western blotting. *P<0.05 vs. C or miR-93. C, control; NC, negative control; miR, microRNA; PEDF, pigment epithelium-derived factor.
GABPB1-IT1 suppresses the proliferation of SNU-398 and C3A cells through the miR-93/PEDF axis
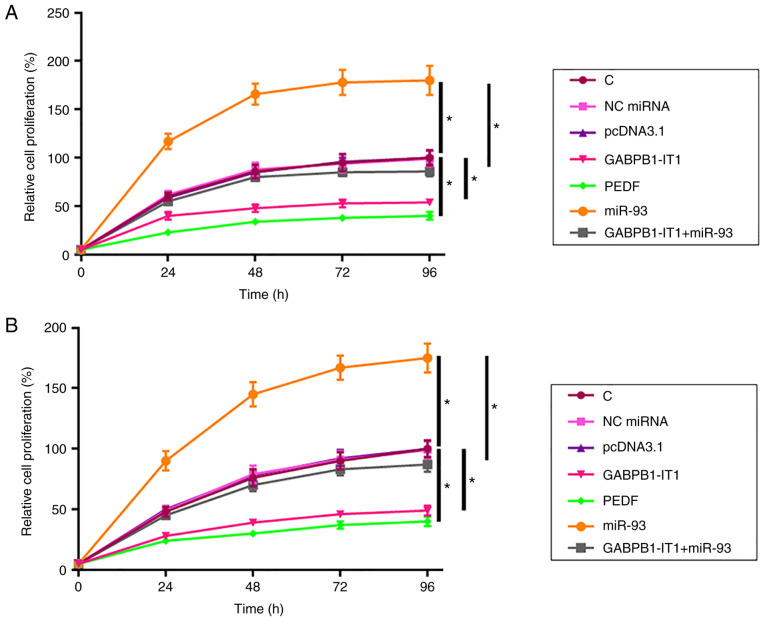
The effects of transfection on the proliferation of SNU-398 cells were analyzed using the CCK-8 assay. Compared with the C group, overexpression of GABPB1-IT1 and PEDF led to decreased proliferation rate of SNU-398 cells (both P<0.05; Fig. 4A). In addition, overexpression of GABPB1-IT1 led to decreased miR-93-mediated promotion of HCC cell proliferation (P<0.05). C3A cells were used to repeat the cell proliferation assay and similar results were obtained (all P<0.05; Fig. 4B). Taken together, our data suggest that GABPB1-IT1 inhibited HCC cell proliferation by regulating miR-93/PEDF axis.
Figure 4.
GABPB1-IT1 suppresses the proliferation of SNU-398 and C3A cells through the miR-93/PEDF axis. Effects of transfections on the proliferation of (A) SNU-398 and (B) C3A cells were analyzed using a Cell Counting Kit-8 assay. *P<0.05 vs. C or pcDNA3.1. miR, microRNA; PEDF, pigment epithelium-derived factor; C, control; NC, negative control.
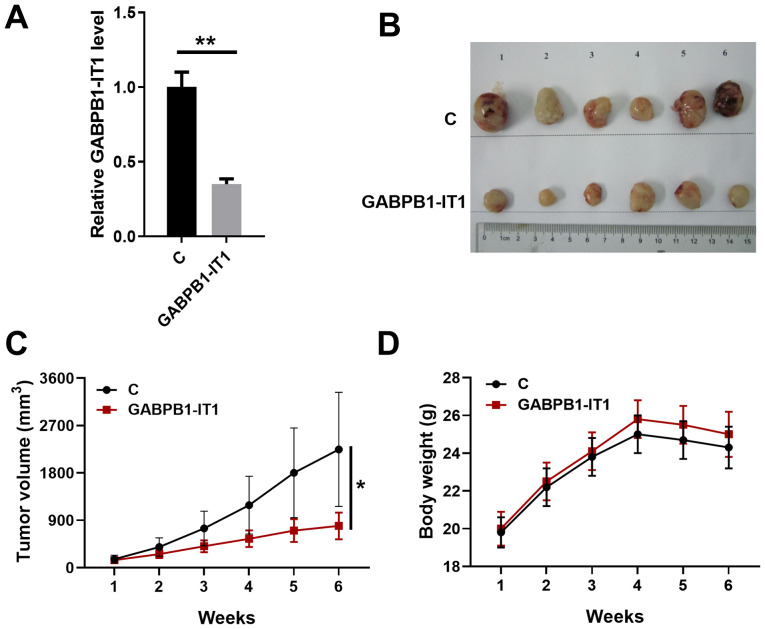
Overexpression of GABPB1-IT1 inhibits HCC tumor growth in vivo
The effect of GABPB1-IT1 on tumorigenesis in a xenograft model was examined. C3A cells stably overexpressing GABPB1-IT1 were subcutaneously injected into the flank of the nude mice. Tumor sizes were measured every week. As shown Fig. 5A, GABPB1-IT1 was significantly downregulated in the xenografts formed by the overexpression of GABPB1-IT1 transfected cells (P<0.01). As shown in Fig. 5B and C, the xenograft formed by overexpression of GABPB1-IT1 had a significantly smaller tumor volume compared with that in the control group (P<0.05). As shown in Fig. 5D, compared with control group, no significant change in bodyweight was observed in mice injected with C3A cells stably overexpressing GABPB1-IT1 compared with the C group. Taken together, our data suggest that GABPB1-IT1 overexpression represses HCC tumor growth in vivo.
Figure 5.
Overexpression of GABPB1-IT1 inhibits HCC tumor growth in vivo. HCC cells infected with GABPB1-IT1 or control were injected into nude mice. (A) Quantitative PCR analysis of the expression of GABPB1-IT1 in xenograft tumors. (B) Images of the tumors extracted from the nude mice. (C) Tumor volumes were measured after injection every week. (D) Body weights were measured after injection every week. *P<0.05, **P<0.01 vs. control. C, control; HCC, hepatocellular carcinoma.
Discussion
The present study investigated the role of GABPB1-IT1 in HCC. It was demonstrated that downregulation of GABPB1-IT1 in HCC was associated with poor survival. In addition, GABPB1-IT1 may upregulate PEDF by sponging miR-93 to suppress cancer cell proliferation.
A recent study reported that GABPB1-IT1 was downregulated in NSCLC and predicted poor survival (14). However, the function of GABPB1-IT1 in cancer biology, such as the regulation of cancer cell behaviors, remains unknown. The current study demonstrated the downregulation of GABPB1-IT1 in HCC. In addition, it was reported that GABPB1-IT1 inhibited the proliferation of HCC cells. Therefore, GABPB1-IT1 is likely a tumor suppressor in HCC.
Due to the lack of early diagnostic markers, most patients with HCC are diagnosed at advanced stages, and the overall survival rate is generally poor (23,24). For example, the 5-year overall survival rate of HCC is only 30–40% (25). The present study showed that the low expression levels of GABPB1-IT1 in HCC tissues before therapy could be used to predict the survival of patients with HCC patients. Therefore, GABPB1-IT1 may serve as a prognostic marker for HCC to improve survival by guiding treatment decisions and the development of personalized care programs. However, the accuracy GABPB1-IT1 as a prognostic remains to be further confirmed.
It has been reported that miR-93 can target PEDF to promote the proliferation of bladder cancer cells (16). Moreover, miR-93-3p is a likely clear cell renal cell carcinoma oncogene that acts by regulating PEDF (26). The present study showed that miR-93 could also target PEDF and increased the proliferation rate of HCC cells. Therefore, different cancer types may share similar molecular pathogenesis and miR-93 may serve as a target for the treatment of multiple types. The present study demonstrated that miR-93 and GABPB1-IT1 could directly interact with each other; however, the expression levels of each were not affected. Instead, overexpression of GABPB1-IT1 upregulated the expression of PEDF, a characterized target of miR-93 (16). It has been well established that miRNA sponging suppresses the function of miRNAs but may not regulate the expression of the target miRNA (22). Therefore, the current data suggested that GABPB1-IT1 could serve as an endogenous sponge of miR-93 to suppress its function. However, other mechanisms may also exist. For instance, GABPB1-IT1 may form certain tertiary structure to participate in the migration and invasion of cancer cells (27).
There were some limitations to the present study. Firstly, the sample size of patients with HCC was small and more samples are needed to confirm the current findings. In addition, the study only investigated the effect of GABPB1-IT1 on HCC cell proliferation. Future studies should investigate the involvement of GABPB1-IT1 in other cellular functions, such as cell migration and invasion.
In conclusion, GABPB1-IT1 is downregulated in HCC and may regulate the miR-93/PEDF axis to suppress cancer cell proliferation.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BL and YW performed the experiments, analyzed the data, interpreted the results and wrote the manuscript. QG and YD performed the experiments. LG designed the study and revise the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval was obtained from The 96604 Military Hospital of The Chinese People's Liberation Army Medical Research Ethics Committee (approval no. 2012-06-027), and informed written consent was obtained from all patients. The animal experiments were approved by The Ethics Committee of 96604 Military Hospital of the Chinese People's Liberation Army (approval no. 202065433).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM, et al. Annual report to the nation on the status of cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312–1337. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 4.Janevska D, Chaloska-Ivanova V, Janevski V. Hepatocellular carcinoma: Risk factors, diagnosis and treatment. Open Access Maced J Med Sci. 2015;3:732–736. doi: 10.3889/oamjms.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Negash AA, Olson RM, Griffin S, Gale M., Jr Modulation of calcium signaling pathway by hepatitis C virus core protein stimulates NLRP3 inflammasome activation. PLoS Pathog. 2019;15:e1007593. doi: 10.1371/journal.ppat.1007593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang F, Yu X, Zhou C, Mao R, Zhu M, Zhu H, Ma Z, Mitra B, Zhao G, Huang Y, et al. Hepatitis B e antigen induces the expansion of monocytic myeloid-derived suppressor cells to dampen T-cell function in chronic hepatitis B virus infection. PLoS Pathog. 2019;15:e1007690. doi: 10.1371/journal.ppat.1007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L, Zhang W, Zou F, Mei L, Wu G, Teng Y. KLHL21, a novel gene that contributes to the progression of hepatocellular carcinoma. BMC Cancer. 2016;16:815. doi: 10.1186/s12885-016-2851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen HC, Jeng YM, Yuan RH, Hsu HC, Chen YL. SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann Surg Oncol. 2012;19:2011–2019. doi: 10.1245/s10434-011-2159-4. [DOI] [PubMed] [Google Scholar]
- 9.Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 10.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. Apr 15; doi: 10.1093/hmg/ddl046. 200 (Epub ahead of print). doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 11.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 14.Xie J, Xie G, Chen Q, Xu Z, Bai W, Chen M. Identification of a novel lncRNA GABPB1-IT1 that is downregulated and predicts a poor prognosis in non-small cell lung cancer. Oncol Lett. 2019;18:838–845. doi: 10.3892/ol.2019.10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, Huang SK, Colquhoun S, Hoon DS. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6:3211–3224. doi: 10.18632/oncotarget.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Bu Q, Zeng M, Xia D, Wu A. MicroRNA-93 promotes bladder cancer proliferation and invasion by targeting PEDF. Urol Oncol. 2019;37:150–157. doi: 10.1016/j.urolonc.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Lei HJ, Chau GY, Lui WY, Tsay SH, King KL, Loong CC, Wu CW. Prognostic value and clinical relevance of the 6th Edition 2002 American Joint Committee on Cancer staging system in patients with resectable hepatocellular carcinoma. J Am Coll Surg. 2006;203:426–435. doi: 10.1016/j.jamcollsurg.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Higuchi T, Oshiro H, Zhang Z, Miyake K, Sugisawa N, Katsuya Y, Yamamoto N, Hayashi K, Kimura H, Miwa S, et al. Osimertinib regresses an EGFR-mutant cisplatinum-resistant lung adenocarcinoma growing in the brain in nude mice. Transl Oncol. 2019;12:640–645. doi: 10.1016/j.tranon.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X, Zheng G, Li C, Liu C. Long non-coding RNA Fer-1-like family member 4 suppresses hepatocellular carcinoma cell proliferation by regulating PTEN in vitro and in vivo. Mol Med Rep. 2019;19:685–692. doi: 10.3892/mmr.2018.9629. [DOI] [PubMed] [Google Scholar]
- 21.Mann M, Wright PR, Backofen R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3827. [DOI] [PubMed] [Google Scholar]
- 23.Forner A, Bruix J. Biomarkers for early diagnosis of hepatocellular carcinoma. Lancet Oncol. 2012;13:750–751. doi: 10.1016/S1470-2045(12)70271-1. [DOI] [PubMed] [Google Scholar]
- 24.Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109:542–553. doi: 10.1038/ajg.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, Chu R, Song H, Xie D, Jiang X, Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2015;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Yang G, Zhu X, Wang Z, Wang H, Bai Y, Sun P, Peng L, Wei W, Chen G, et al. miR-93-3p inhibition suppresses clear cell renal cell carcinoma proliferation, metastasis and invasion. Oncotarget. 2017;8:82824–82834. doi: 10.18632/oncotarget.20458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Leontis NB, Zirbel CL, Bisaro DM, Ding B. A three-dimensional RNA motif mediates directional trafficking of Potato spindle tuber viroid from epidermal to palisade mesophyll cells in Nicotiana benthamiana. PLoS Pathog. 2019;15:e1008147. doi: 10.1371/journal.ppat.1008147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.