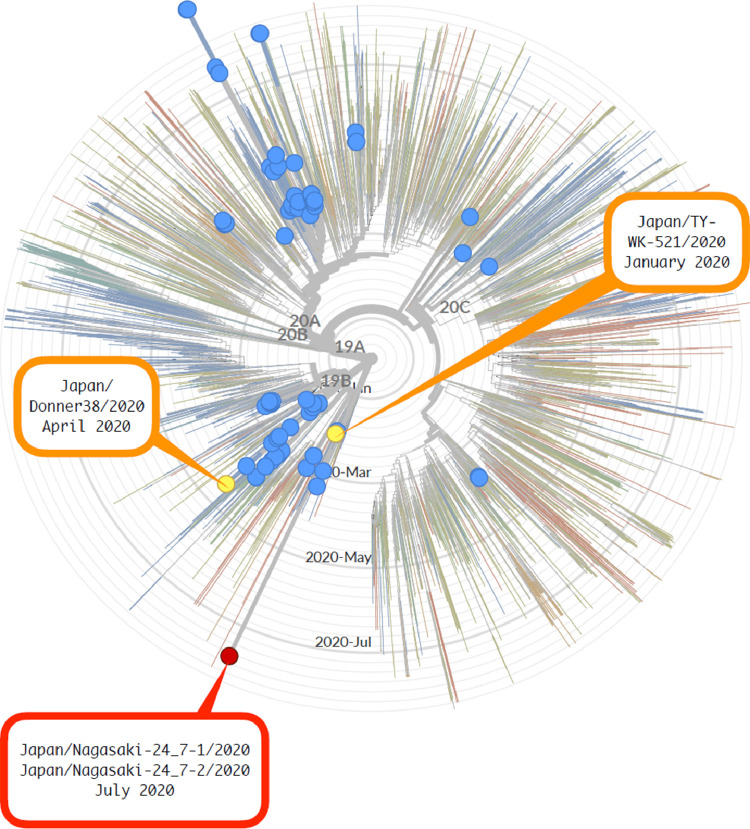
We read with interest on the case of SARS-CoV-2 re-infection on the importance of genomic information in understanding the true burden of COVID-19 [1]. After the emergence of SARS-CoV-2 in China, initial cases in Japan were travel-associated cases from affected areas, as determined by the viral sequence analyses. In concordance with global COVID-19 epidemic, the Asian SARS-CoV-2 clades were dominant until the beginning of March, 2020 [2]. However, as of April, with the emergence of European clades (20A, 20B, 20C) as the dominant clade in Japan, there were no reports on local transmission of the Asian clades (19A and 19B) within the following weeks (figure). Our analysis however indicates the Asian clade remains in circulation, with at least two strains confirmed locally. Both strains (Japan/Nagasaki-24_7-1/2020 and Japan/Nagasaki-24_7-2/2020), isolated in July 2020, belonged to the “diminished” Asian clade, 19B (figure). In addition to the 628 isolates from Japan, and the 4981 globally isolated strains, we observed that both of the sampled strains were of an orphaned monophyletic clade, in which the next closest strain of this clade was a locally strain that was isolated in January (Japan/TY-WK-521/2020) (Fig. 1). While there may be limited sampling in certain geographical areas, we are confident that this distinct clade represents initial introduction from China, followed by basal amplification and persistent community maintenance of transmission cycle. While outbreaks may derive from local spread after introduction events, these events can generally be traced by using phylogenetically ancestral viruses of outbreak clades. In this context, the detection of persistent transmission of an orphan clade that is unique to Japan indicates local transmission that may remain undetected by the Japanese surveillance mechanism. While mortality rate is low with advanced healthcare in Japan, there remains major challenges in Japan's response to COVID-19: publicly available SARS-CoV-2 genomes were not sampled in strict proportion to the reflect the national burden of infection, and genetic information processing and there may be time-lag between information sharing. Although both traditional public health surveillance and genomic epidemiology has provided insights into outbreak events [4], in this instance, blackouts in information sharing and transparency can potentially lead to information disparity. Additionally, genetic information may offer crucial evidence for re-infection and elucidation of disease burden. As genetic relationships can shed light on the underlying transmission patterns of spread in Japan and, considering the rapid global expansion of COVID-19 epidemic, our results highlight that policies to include real-time public-sharing, transparency and empowering of new systems for infectious disease surveillance would be critical in the control of SARS-CoV-2 transmission.
Fig. 1.
Maximum-likelihood phylogeny of 682 SARS-CoV-2 viruses collected from Japan (blue closed circles) on a background of 4981 globally collected virus strains from GISAID. Maximum-likelihood phylogeny of SARS-CoV-2 viruses collected from Japan (blue closed circles) on a background of 4981 globally collected virus strains from GISAID[3]. Nextstrain[5] was used to conduct phylogenetic analysis. Using Augur's subsampling process, 4981 records were selected from the GISAID records. The phylogenetic tree was constructed with the addition of strains isolated from Nagasaki. At the time point of September 2020, Nextstrain defined 5 major clades. Up to March 2020, the clades 19A and 19B (correspond to clades L+V and S in the classification of GISAID) were predominantly circulating in Asia. The clades 20A, 20B and 20C (G, GR and GH in GISAID) appeared in April 2020 and spread across Europe. In turn, the European clades became global dominant clade. The first case of COVID-19 was confirmed in January 2020 in Japan. Nagasaki prefecture is part of the Kyushu region of Japan and is located on the western edge of mainland Japan. The first case of COVID-19 in Nagasaki was confirmed in March 2020. Between March to April 2020, 16 cases were reported in Nagasaki prefecture, with no cases reported between May and June. Both strains of the clade 19B were isolated from patients in Nagasaki whom had returned from a neighboring prefecture.
We declare no competing interests. We thank health care practitioners of the clinics and hospitals in Japan who supported the study. This research was supported by Japan Agency for Medical Research and Development (AMED) under Grant Number JP20wm0125006 (Japan Program for Infectious Diseases Research and Infrastructure), and the Japan Society for the Promotion of Science KAKENHI (JP19K24679).
WHO Global Reference Laboratory for COVID-19, WHO Collaborating Center for Reference and Research on Tropical and Emerging Virus Diseases, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan (TN, SI, MMNT, MLM, KM), Department of Respiratory Medicine, Nagasaki University Hospital, Nagasaki, Japan, Department of Infectious Diseases (TT, NA, TM, HM), Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan (TT, TM, KI), Infection Control and Education Center, Nagasaki University Hospital, Nagasaki, Japan (KI).
Contributor Information
Meng Ling Moi, Email: sherry@nagasaki-u.ac.jp.
Kouichi Morita, Email: moritak@nagasaki-u.ac.jp.
References
- 1.Prado-Vivar B, Becerra-Wong M, Guadalupe JJ. A case of SARS-CoV-2 reinfection in Ecuador. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse Y, Ko YK, Saito M. Epidemiology of COVID-19 outbreak in Japan, from January-March 2020. Jpn J Infect Dis. 2020;73(5):391–393. doi: 10.7883/yoken.JJID.2020.271. [DOI] [PubMed] [Google Scholar]
- 3.Shu Y, McCauley J.GISAID. Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabata S, Imai K, Kawano S. Clinical characteristics of COVID-19 in 104 people with SARS-CoV-2 infection on the Diamond Princess cruise ship: a retrospective analysis. Lancet Infect Dis. 2020;20(9):1043–1050. doi: 10.1016/S1473-3099(20)30482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadfield James, Megill Colin, Bell Sidney M. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]