To the Editor,
Nursing homes (NH) have been severely affected by the coronavirus disease 2019 (COVID-19) pandemic, largely due to their congregate nature and the vulnerability of residents [1]. Advanced age, frailty and concurrence of underlying chronic health conditions place NH residents at high risk for developing severe forms of COVID-19 and for death. Long-lasting virus shedding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in urine and faeces has been documented in both symptomatic and asymptomatic infected adults [2]. As a result, near-source tracking in the sewers serving particular buildings (i.e. campus dormitories, workplaces, correctional facilities, schools) has emerged as an appealing non-invasive tool that, when combined with subsequent targeted population screening when SARS-CoV-2 is detected, may enable rapid identification and control of facility outbreaks [3]. In this pilot study, we provide evidence demonstrating the feasibility and utility of this wastewater-based epidemiological approach for early identification of isolated cases or outbreaks of SARS-CoV-2 infection in NH.
This study involved five NH facilities (listed as A to E) located in northeast Valencia (Spain), affiliated to the Clínico-Malvarrosa Health Department. These are nursing or mixed nursing/care homes, altogether providing care for 472 residents attended by 309 staff (Table 1 ). Selection from among the 17 NH supported by the Clínico-Malvarrosa Health Department was based upon two criteria: (a) existence of one (NH A,B,C and D) or more (NH E, n = 4) sewer manholes allegedly not shared with nearby buildings and (b) personal autonomy of most residents. Permission to analyse the wastewater was granted by the NH operator and the local authority responsible for the sewer system. NH sewage drain(s) were monitored for presence of SARS-CoV-2 RNA by testing near-source wastewater samples at least 5 days per week from 7 October to 28 December 2020. Grab samples were collected on site from water outlets at each facility. All samples were taken early in the morning, collecting 1 L of water in sterile plastic containers with sodium thiosulphate (VWR International, Radnor, PA, USA). Refrigerated water samples were transferred to the Global Omnium laboratory (Valencia, Spain), and concentrated within 24 h using the aluminium adsorption–precipitation method. Spiked Mengovirus (vMCO CECT 100000) acted as an internal control. Viral extraction from wastewater concentrates and RT-qPCR were performed as previously described [4,5]. The results are reported as genome copies (GC)/L. The limit of detection of the RT-qPCR assay was 670 GC/L.
Table 1.
Detection of SARS-CoV-2 in wastewater, residents and staff at nursing homes included in the study
| Nursing home (no. of residents/no. of staff) | Surveillance period | Date of first detection of SARS-CoV-2 RNA in wastewatera/day at which peak RNA levels (log10 GC/L) was reached | Date of first reported case of SARS-CoV-2 infection at the nursing home | No. of residents testing positive for SARS-CoV-2b,c | No. of staff testing positive for SARS-CoV-2b,c | Last SARS-CoV-2 infection case documented among residents or staff | Previous outbreaks |
|---|---|---|---|---|---|---|---|
| A (103/58) | 14 October to 28 December | 21 October/29 October (4.5) | 9 November | 1d | — | 9 November | Yes (16 June) |
| A (103/58) | 14 October to 28 December | 10 December/28 December (8.6) | 17 December | 25 | 13 | Outbreak ongoing | Yes (16 June and 21 October) |
| B (105/60) | 6 November to 28 December | 6 November/19 November (4.5) | 11 November | — | 1e | 11 November | Yes (17 June and 5 October) |
| C (48/25) | 6 November to 28 December | ND | NR | — | — | — | No |
| D (101/81) | 7 October to 28 December | 7 October/12 November (6.9) | NRf | — | — | — | Yes (9 July) |
| E (115/85) | 7 October to 28 December | 26 October/30 October (8.3)g | 17 October | 14 | 10 | 16 November | Yes (17 June and 13 July) |
ND, not detected; NR, not reported.
RNA extraction from sewage material was carried out using the NucleoSpin RNA virus Kit (Macherey-Nagel, Düren, Germany). SARS-CoV-2 RNA detection was performed by RT-qPCR using One-Step PrimeScript™ RT-PCR Kit (Perfect Real Time) (Takara Bio, Mountain View, CA, USA), targeting the nucleoprotein (N), N1 and N2 fragments, and envelope protein (E) gene [4,5]. RNA samples were analysed in duplicate. Each RT-qPCR run included negative (nuclease-free water) and positive controls. RT-qPCR targets were quantified by plotting the quantification cycles (CT) to an external standard curve built with ten-fold serial dilution of the 2019-nCoV_N_Positive Control and 2019-nCoV_E_Positive Control (IDT). Mengovirus RNA recovery rates were calculated and used as quality assurance parameters according to ISO 15216-1:2017.
Nasopharyngeal swabs (NP) for RT-PCR testing were collected by experienced nurses at the Nursing Home sites and immediately placed in 3 mL of Universal Transport Medium (UTM, Becton Dickinson, Sparks, MD, USA). RT-qPCRs were conducted within 24 h of specimen collection at the Microbiology Service of Hospital Clínico Universitario (Valencia, Spain) with the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA was extracted using the Applied Biosystems MagMAX™ Viral/Pathogen II Nucleic Acid Isolation Kits coupled with KingFisher Flex automated instrument (Thermo Fisher Scientific).
No reinfections were documented among residents and staff.
Resident tested for SARS-CoV-2 infection because appearance of symptoms compatible with COVID-19 (fever and cough).
Staff (asymptomatic) tested as a close household contact of a COVID-19 case.
Residents and Staff members were screened for SARS-CoV-2 infection by RT-PCR on 29 October.
Two of the four sewage draining sites were not tested until 26 October.
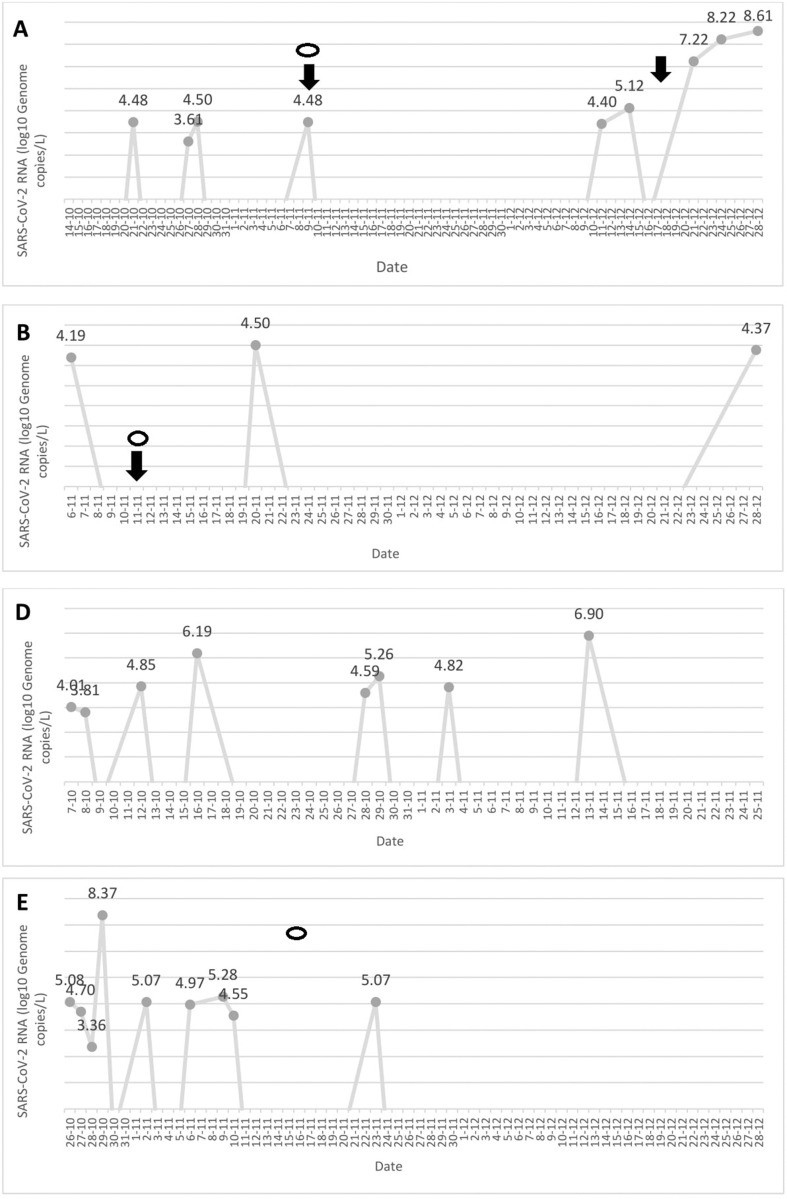
As shown in Table 1, SARS-CoV-2 RNA was detected in wastewater samples collected from four out of the five NH (A, B, D, E). The dynamics of SARS-CoV-2 RNA detection and viral loads measured at each NH are depicted in the Supplementary material (Fig. S1). SARS-CoV-2 infection cases among residents or staff, either asymptomatic or symptomatic, were documented in three of the four NH (A, B, E) (Table 1). According to public health policies in place in our Health Department within the study period, systematic testing of residents and staff at NH A and NH E was triggered by the occurrence of symptomatic cases and conducted within 48 h. Index cases were diagnosed within 24 h after symptom onset. At NH B, testing of all residents and staff was driven by detection of a single asymptomatic case among staff workers; this individual had been identified as a household contact of a COVID-19 case. In NH B, first detection of SARS-CoV-2 RNA in wastewater occurred approximately 1 month after an outbreak declaration involving three residents, who were promptly isolated following diagnosis, and 5 days before detection of the staff case. Hence, the possibility exists that detection of viral RNA in wastewater resulted from long-term shedding by infected residents.
No cases were identified at NH D within the study period, despite repeated detection of SARS-CoV-2 RNA in the sewage drain. Of note, as a ‘proof of concept’ approach, residents and staff at NH D were screened by RT-PCR in nasopharyngeal swabs on 29 October, 12 days after SARS-CoV-2 was first detected in sewage—all yielded negative results. A thorough investigation conducted afterwards revealed the existence of cross-contamination between sewage drains of this NH and that of an adjacent building that had gone unnoticed. SARS-CoV-2 RNA was not detected in samples collected from NH C and no cases were documented throughout the follow-up period.
Presence of SARS-CoV-2 RNA in sewage preceded identification of isolated cases among residents or staff (in both cases symptomatic) or outbreak declaration in two NH (NH A on two different occasions, and NH B), with lag times ranging from 5 to 19 days. Interestingly, by using an epidemic transmission model, Kaplan et al. [6] demonstrated that tracking SARS-CoV-2 RNA concentration in sewage sludge provides a 3- to 5-day lead time over tracking hospital admissions.
Repeated detection of SARS-CoV-2 RNA in sewage was not documented until after outbreak declaration in NH E, although it should be noted that between 7 October and the first case detection on 17 October only two of the four sewage drains at NH E had been sampled.
SARS-CoV-2 RNA levels in wastewater samples increased exponentially over the course of NH outbreaks (NH A and NH E), reaching peak levels above 8.0 log10 GC/L (see Supplementary material, Fig. S1).
Finally, disappearance of SARS-CoV-2 RNA from sewers was associated with control of outbreaks or absence of new case documentation following implementation of adequate measures (isolation of positive case and quarantining of close contacts) (see Supplementary material, Fig. S1). The SARS-CoV-2 outbreak at NH A was currently still active as of January 12.
The present study has several limitations that must be acknowledged. First, no sequencing data were available for the clinical and environmental SARS-CoV-2 variants detected, so sequence-matching analyses could not be performed. Second, cell cultures from sewage specimens were not performed.
In conclusion, results from this study suggested that SARS-CoV-2 RNA surveillance of sewage drains may serve as an early warning system for isolated cases or outbreak declaration of SARS-CoV-2 infection in NH. Frequent SARS-CoV-2 RT-qPCR sewage testing coupled with targeted screening of residents and staff may prove useful for early blunting of virus transmission and spread at NH. Further studies with a larger site sample are warranted to confirm this assumption.
Transparency declaration
The authors declare that they have no conflicts of interest.
This work received no public or private funds.
Author contributions
LD, RS, EA and IT contributed to methodology and data validation. LA, RS, JFM, GS and DN contributed to formal analysis. LD, RS, JFM, GS and DN contributed to conceptualization and supervision. PB, MJB, PL-F and RO contributed to the supervision of RT-PCR testing at NH facilities. LD, GS and DN wrote the original draft. All authors reviewed and approved the original draft.
Acknowledgements
We thank all personnel working in the nursing homes and the Hospital Clínico Universitario of Valencia for their unwavering commitment in the fight against COVID-19.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.02.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
Figure S1. Dynamics of SARS-CoV-2 RNA detection in sewage drains of nursing homes.
References
- 1.Grabowski D.C., Mor V. Nursing home care in crisis in the wake of COVID-19. JAMA. 2020;324:23–24. doi: 10.1001/jama.2020.8524. [DOI] [PubMed] [Google Scholar]
- 2.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassard F., Lundy L., Singer A.C., Grimsley J., Di Cesare M. Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools. Lancet Microb. 2020 doi: 10.1016/S2666-5247(20)30193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int J Hyg Environ Health. 2020;230:11362. doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag Sci. 2020 doi: 10.1007/s10729-020-09525-1. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]