Abstract
The Quercus species serve as a powerful model for studying introgression in relation to species boundaries and adaptive processes. Coexistence of distant relatives, or lack of coexistence of closely relative oak species, introgression may play a role. In the current study, four closely related oak species were found in Zijinshan, China. We generated a comprehensive genome size (GS) database for 120 individuals of four species using flow cytometry‐based approaches. We examined GS variability within and among the species and hybridization events among the four species. The mean GSs of Q. acutissima, Q. variabilis, Q. fabri, and Q. serrata var. brevipetiolata were estimated to be 1.87, 1.92, 1.97, and 1.97 pg, respectively. The intraspecific and interspecific variations of GS observed among the four oak species indicated adaptation to the environment. Hybridization occurred both within and between the sections. A hybrid offspring was produced from Q. fabri and Q. variabilis, which belonged to different sections. The GS evolutionary pattern for hybrid species was expansion. Hybridization between the sections may be affected by habitat disturbance. This study increases our understanding of the evolution of GS in Quercus and will help establish guidelines for the ecological protection of oak trees.
Keywords: flow cytometry, genome size variation, hybridization, Quercus
We examined GS variability within and among the species and present hybridization among four species. It occurred hybridization both within the section and between the sections. A hybrid offspring produced from Q. fabri and Q. variabilis, which belong to a different section. The pattern of GS evolution for hybrids species is GS expansion. Hybridization between the sections may be affected by habitat disturbance.

1. INTRODUCTION
Nuclear genome size (GS), a critical organismal characteristic and structural component, has high practical and predictive values for studying adaptive or stochastic processes in evolutionary biology (Talla et al., 2017). The GS varies considerably among organisms, with a greater than 2,360‐fold variation existing among different angiosperms (Greilhuber et al., 2006; Leitch et al., 2019). This variation is found within both species and genera (Castro et al., 2013; Denaeghel et al., 2017; Li et al., 2017; Nowicka et al., 2016; Prancl et al., 2014; Qiu et al., 2018). GS evolves in both directions, expanding as well as shrinking. Polyploidization events and transposable element (TE) amplification in plants lead to genome expansion (Hawkins et al., 2008; Kumar & Bennetzen, 1999; Wood et al., 2009), while recombination processes may lead to genome contraction (Devos et al., 2002; Hawkins et al., 2009; Schubert & Vu, 2016). Natural selection exerts a force on the genome; consequently, GS variation is usually related to the living environment, including the altitude, latitude, temperature, and precipitation level (Bennett et al., 2000; Hidalgo et al., 2015; Knight & Ackerly, 2002; Knight et al., 2005; Li et al., 2017; Zhang et al., 2019). GS allows the detection of interspecific hybrids and/or backcrosses (Vit et al., 2014; Yan et al., 2016) and has been widely applied to various plants, such as Sarcococca (Denaeghel et al., 2017; Prancl et al., 2014; Tlili et al., 2020).
Flow cytometry (FCM), which is a fast and effective tool to estimate GS, has been successfully applied to ploidy identification, cell cycle analysis, and species identification, including hybrids, rarely occurring cytotypes, and aneuploids (Francis et al., 2008; Hanusova et al., 2014; Vit et al., 2014; Zhang et al., 2019). It has been used successfully in plant genetic variation studies, genetic analyses, and breeding, as well as in studies of reproductive ecology, evolution, and plant system classification (Bilinski et al., 2018; Galbraith, 2004; Sharma et al., 2019; Spaniel et al., 2019). In the 1980s, Galbraith et al. (1983) developed a fast, efficient, and convenient method for isolating plant nuclei, which meant that FCM could be more widely used in botanical research, especially to determine plant GS.
Quercus L. is a genus that contains many economically and ecologically important tree species found in the northern hemisphere (Aldrich & Cavender‐Bares, 2011). From an evolutionary point of view, Quercus is a good material for studying the species boundaries and adaptive evolution (Porth et al., 2016; Yuan et al., 2018; Manuel et al., 2020; Petersson et al., 2020). Quercus is famous for its remarkable natural hybridization, which makes it difficult to identify species owing to the formation of hybrids (Song et al., 2015). Additionally, the frequent hybridization events of oak trees lead to shaping the community assembly and structure, as well as to the evolution of species and the generation of new species (Cannon & Scher, 2017; Wetherbee et al., 2020). Closely related species rarely co‐exist in the same forest land, because hybridization and introgression lead to species merging over time, eliminating their co‐existence (Cavender‐Bares & Pahlich, 2009; Pollock et al., 2015). Even when oaks occur in sympatry, there is significant gene flow. These may include the introgression events that lead to adaptation.
In this study, we found four closely related species of Quercus in Zijinshan, China that allowed us to study whether hybridization occurs between sympatric Quercus. The four oaks belonged to two sections in the genus Quercus. Quercus acutissima and Quercus variabilis belong to the Section Cerris, which is called the Old world clade, while Quercus fabri, and Quercus serrata var. brevipetiolata belong to the Section Quercus, which is called the New world clade (Manos et al., 2001). Members of the same sections easily form hybrids (Cottam et al., 1982), and there is an especially high frequency of hybrid formation within the Quercus section. We investigated whether hybridization occurs between sections. First, we determine the GSs of the four oak species, and then we determined whether there were variations in the GSs of these oak species. Finally, we analyzed the species hybridization within and between the two sections and determined the evolution of GS in these four oak species.
2. MATERIALS AND METHODS
2.1. Research site overview and field sampling
A total of 120 samples of Q. acutissima, Q. variabilis, Q. fabri, and Q. serrata var. brevipetiolata were collected in Zijin Mountains. Zijin Mountains is located to the east of Nanjing, Jiangsu Province (118°48′24″–118°53′04″ E, 32°01′57″–32°06′15″ E) and has an area of 3,008.8 ha, with a forest area of 2,107.6 ha, which has a forest canopy coverage of 0.75–0.80. At present, the vegetation of Zijin Mountains is mainly artificial and natural secondary forests, which are transitional between subtropical evergreen broad‐leaved forests and warm temperate deciduous broad‐leaved forests. The zonal vegetation is a mixture of deciduous evergreen and broad‐leaved forests that is rich in various plant resources. Q. acutissima and Q. variabilis are important deciduous tree species in this area that are widely distributed and occupy dominant positions. The samples were prepared from the fresh mature leaves of natural wild plants growing in the Zijin Mountains, Nanjing, Jiangsu Province, China, during late October 2011 (Figure 1). The sampling site is a mixed plot with four deciduous oaks, Q. acutissima and Q. variabilis, Q. fabri, Q. serrata var. glandulifera. 120 samples of four species of oak including seedlings (DBH ≤ 1), saplings (1 < DBH≤10), and trees (DBH > 10) were collected randomly and evenly in this forest stand.
FIGURE 1.
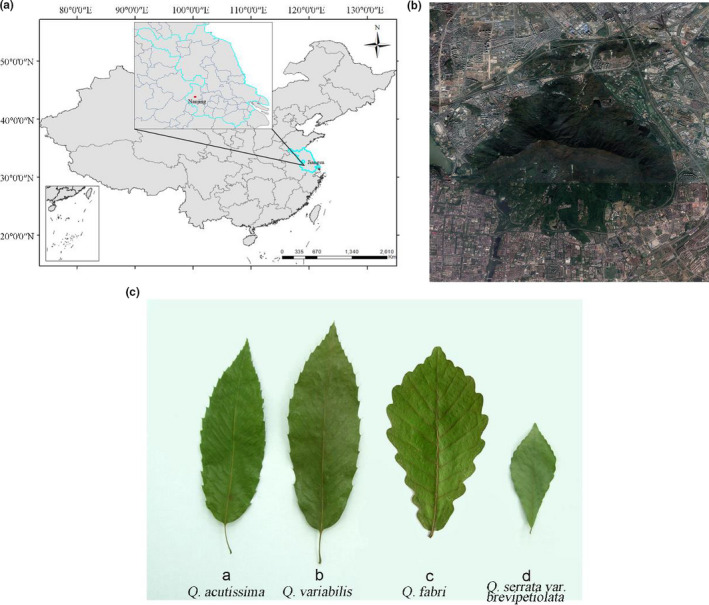
The geographical location of the study area and plant materials (a) Location of the research area in China; (b) Google Earth high‐resolution image of Zijin Mountain; (c) Features of four Quercus leaves
2.2. FCM measurement of the nuclear DNA content
Approximately 200 mg of oak leaves was used for the FCM analysis, and Petunia hybrida (2C = 2.85 pg) from the Nanjing Forestry University nursery was included as the internal standard plant (Marie & Brown, 1993). We used the Marie's Buffer (50 mM C6H12O6, 15 mM NaCl, 15 mM KC1, 5 mM EDTA Na2, 50 mM Na3C6H5O7.2H2O, 0.5%(v/v) Tween 20, and 50 mM Hepes, pH 7.2) and 2.2 μl β‐mercaptoethanol should be added to each ml of buffer before use (Favre & Brown, 1996; Marie & Brown, 1993). Owing to the collection of mature leaves, we modified Marie's Buffer to obtain the maximum nuclear yield. The most suitable conventional of β‐mercaptoethanol and Tween 20 for Q. acutissima, Q. variabilis, Q. fabri, and Q. serrata var. brevipetiolata was 45 mM, 2.5%; 15 mM, 2.0%; 60 mM, 1.0%; 30 mM, 1.0%, respectively (Wei & Fang, 2015). Pipettes were used to immerse 20 mg leaves in 1 ml of optimized Marie's Buffer. The leaves were then quickly chopped in Petri dishes. The dishes were gently shaken so that the chopped leaves and buffer completely mixed. A 25‐μm nylon mesh was used to remove any fragments and large debris. The nuclei were stained with propidium iodide (PI, Fluka, Buchs, Switzerland), and RNase (Fluka) was added to a final concentration of 50 μg/mL. The samples were treated at room temperature for 15 min in a dark environment before being analyzed using a flow cytometer (FACSCalibur, BD USA).
2.3. Data analyses
The 2C DNA content was calculated using the following formula:
The conversion ratio between the DNA mass and Base logarithm was 1 pg = 978 Mb (Doležel et al., 2003).
The DNA content data for 120 individuals were subjected to standardized processing. The average value and coefficient of variation (CV) were calculated and the variance in GS variation was analyzed using SPSS 19.0 software (SPSS, Inc.) and R studio 3.6.2. We set the GSs of the individual to be a variable, calculated the Euclidean genetic distances among the 120 samples and set 4 clustering categories. Finally, the hierarchical clustering was selected to realize the analysis with R studio 3.6.2.
3. RESULTS
3.1. GSs of the four oak species
Owing to the low nuclear yields of mature leaves, we optimized Marie's Buffer for each of the four oak species. The CV values of the four oak species were all within the normal range, and the GSs of the individual species overlapped (Table S1). The results for the mixed samples prepared using oak and P. hybrida are shown in Figure 2. The oak and petunia formed narrow and high DNA peaks, respectively, and the petunia DNA peaks were generally lower than those of the oak peaks (Table 1).
FIGURE 2.
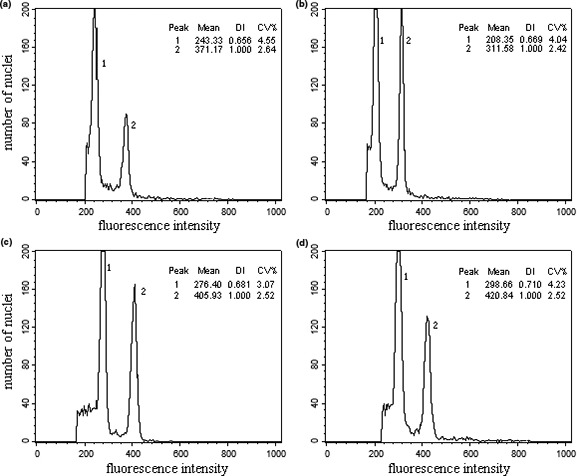
Quercus and Petunia hybrida fluorescence intensities. (a) Q. acutissim; (b) Q. variabilis; (c) Q. Fabri; (d). Q. serrata var. brevipetiolata. Peak 1: Oak nuclei at the G0/G1 phase; peak 2, P. hybrida nuclei at the G0/G1 phase. DI, mean nuclear DNA fluorescence index (oak/P. hybrida)
TABLE 1.
The 2C nuclear DNA contents for the four Quercus species measured in this study
| Species | N | Mean 2C‐value (pg) | SD | MIN | MAX |
|---|---|---|---|---|---|
| Q. acutissima | 30 | 1.87 | 0.02 | 1.83 | 1.91 |
| Q. variabilis | 30 | 1.92 | 0.31 | 1.87 | 1.99 |
| Q. fabri | 33 | 1.97 | 0.27 | 1.91 | 2.06 |
| Q. serratav ar. brevipetiolata | 27 | 1.97 | 0.34 | 1.93 | 2.06 |
| Average | 120 | 1.93 | 0.05 |
GS estimates varied 1.04‐fold for Q. acutissima, ranging from 1.83 to 1.91 pg (CV value range, 3.96–6.64), with a mean of 1.87 pg. In Q. variabilis individuals, estimates varied 1.06‐fold, ranging from 1.87 to 1.99 pg (CV value range, 4.26–6.78), with a mean of 1.92 pg. Estimates varied 1.05‐fold in Q. fabri individuals, ranging from 1.91 to 2.02 pg (CV value range, 3.11–6.71), with a mean of 1.97 pg. The GS of Q. serrata var. brevipetiolata varied 1.06‐fold, ranging from 1.93 to 2.06 pg (CV value range, 3.82–6.51), with a mean of 1.97 pg. Quercus fabri and Q. serrata var. brevipetiolata had the same mean values (1.97 pg). The average GSs for Q. fabri and Q. serrata var. brevipetiolata were the largest, and the average GS for Q. variabilis was only slightly larger than that for Q. acutissima. As shown in Figure 3, the GSs of most Q. acutissima individuals were less than the mean GS (1.82 pg), the GSs of most Q. variabilis individuals were greater than the mean GS (1.92 pg), and the GSs of most Q. serrata var. brevipetiolata and Q. fabri individuals were less than the mean GS (1.97 pg).
FIGURE 3.
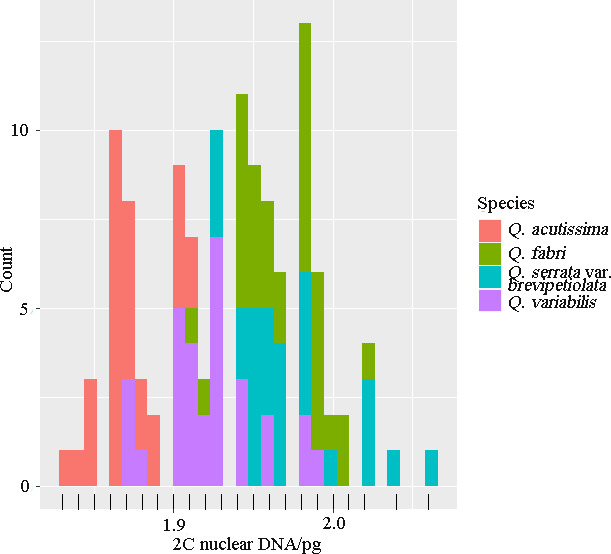
Distributions of the 2C nuclear DNA contents of 120 individuals
3.2. GS variation among the four oak species' populations
To assess the GS variation among the four oak species' populations, repeated‐measures ANOVAs were used (Table 2). There were significant differences in the GS variations among the populations, except those of Q. serrata var. brevipetiolata and Q. fabri. Additionally, variations between the populations were greater than variations within the populations. The intraspecific variation decreased in the following order: Q. serrata var. brevipetiolata > Q. variabilis > Q. fabri > Q. acutissima (Figure 4).
TABLE 2.
An ANOVA of 2C DNA values among populations of the four Quercus species
| pops | Source of difference | SS | df | MS | F | p‐value | ||
|---|---|---|---|---|---|---|---|---|
| ZJ_A × ZJ_V | Between pops | 0.04 | 1 | 0.04 | 55.91 | 4.63E−10** | ||
| Within pops | 0.04 | 58 | 0.71 × 10–3 | |||||
| ZJ_V × ZJ_F | Between pops | 0.03 | 1 | 0.03 | 38.92 | 4.62E−08** | ||
| Within pops | 0.05 | 61 | 0.84 × 10–3 | |||||
| ZJ_F × ZJ_G | Between pops | 0.58 × 10–3 | 1 | 0.58 × 10–3 | 0.62 | 0.43 | ||
| Within pops | 0.05 | 56 | 0.93 × 10–3 | |||||
| ZJ_A × ZJ_F | Between pops | 0.15 | 1 | 0.15 | 256.89 | 1.53E−23** | ||
| Within pops | 0.04 | 61 | 0.57 × 10–3 | |||||
| ZJ_A × ZJ_G | Between pops | 0.15 | 1 | 0.15 | 187.65 | 2.27E−19** | ||
| Within pops | 0.04 | 55 | 0.78 × 10–3 | |||||
| ZJ_V × ZJ_G | Between pops | 0.04 | 1 | 0.04 | 33.43 | 3.61E−07** | ||
| Within pops | 0.06 | 55 | 0.11 × 10–2 | |||||
ZJ_A represents the Q. acutissima population, ZJ_V represents the Q. variabilis population, ZJ_F represents the Q. fabri population, and ZJ_G represents the Q. serrata var. brevipetiolata population.
Statistically different (p < .01).
FIGURE 4.
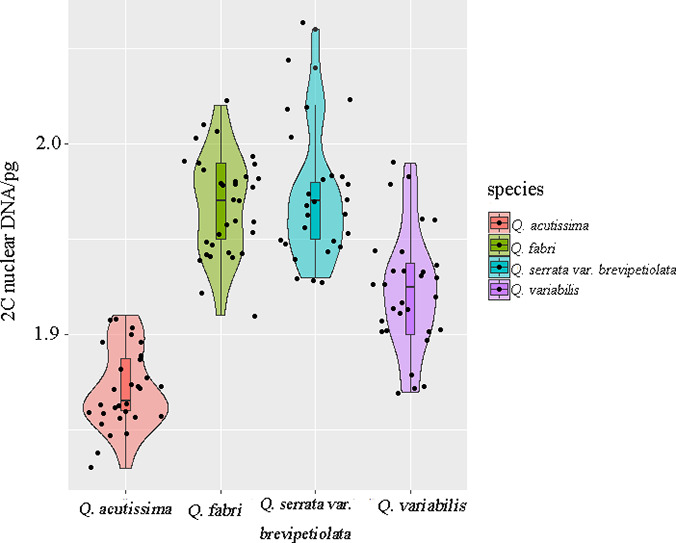
Genomic size variation in four oak species' populations
3.3. Clustering analysis of GS variation within and between the four Quercus species
In this study, only GS variation parameters were used for cluster analysis without other indicator (Figure 5). The results showed that all the samples clustered into three clades. The two individuals ZJ116 and ZJ105 formed a single clade in the clustering analysis, and their GSs 2.024 pg and 2.06 pg, respectively, which were greater than those of the other species. This indicated that the two Q. serrata var. brevipetiolata individuals (ZJ116 and ZJ105) may be hybrids. The clade II was mostly composed of Q. acutissima individuals and a few of Q. variabilis individuals. The members of clade III are relatively complex, mainly including four oak species. Firstly, a small number of Q. acutissima individuals; secondly, Q. variabilis and Q. fabri individuals make up a large proportion of this clade, lastly the Q. serrata var. brevipetiolata individuals were scattered, without specific rules in this clade. The discontinuous cross‐distribution of four species of oaks in the clade III indicated that the GSs of these individuals had interspecific transition values. Moreover, Q. acutissima individuals did not appear in the clade which composed of Q. fabri and Q. serrata var. brevipetiolata. This confirmed that there were no interspecific hybridization between Q. acutissima and either Q. fabri or Q. serrata var. brevipetiolata.
FIGURE 5.
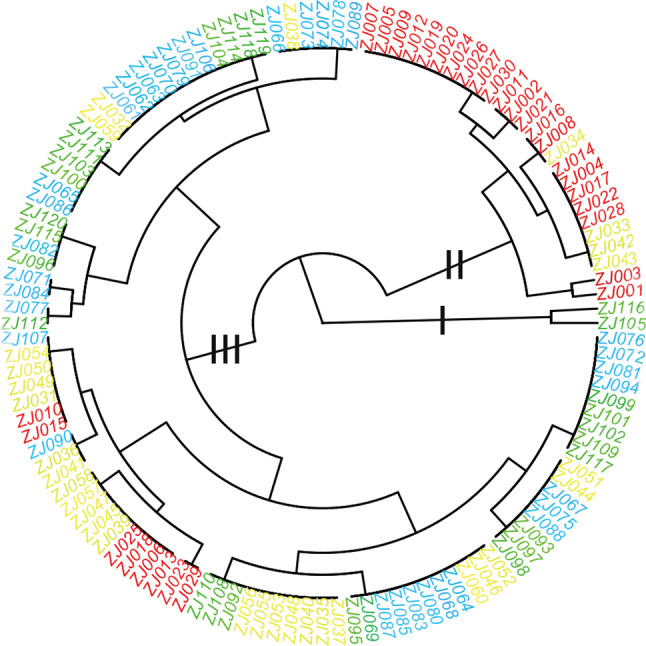
Cluster analysis results for 120 Quercus individuals based on nuclear 2C DNA content variation. Red, Q. acutissima; Yellow, Q. variabilis; Blue, Q. fabri; Green: Q. serrata var. Brevipetiolata
3.4. GS variation in different evolutionary branches of angiosperms
It has been reported that the GSs of angiosperms are significantly non‐normal, and some lineages have extremely large GSs (Leitch & Leitch, 2013). In order to explore the characteristics of GSs of Fagaceae in angiosperms, we used the angiosperm DNA C‐value database (Leitch et al., 2019) and counted the GS variations of five groups: base taxa of angiosperm, monocots, base taxa of eudicots, rosids, and Fagaceae. Figure 6 shows the total numbers of samples and random samples in each group. The monocots had the largest variation in GS (0.76–141.92 pg). As shown in Figure 7, the mean GS of the monocots was greater than the mean of the other groups (16.96 pg). The mean GS of species in Fagaceae was small (2.04 pg).
FIGURE 6.
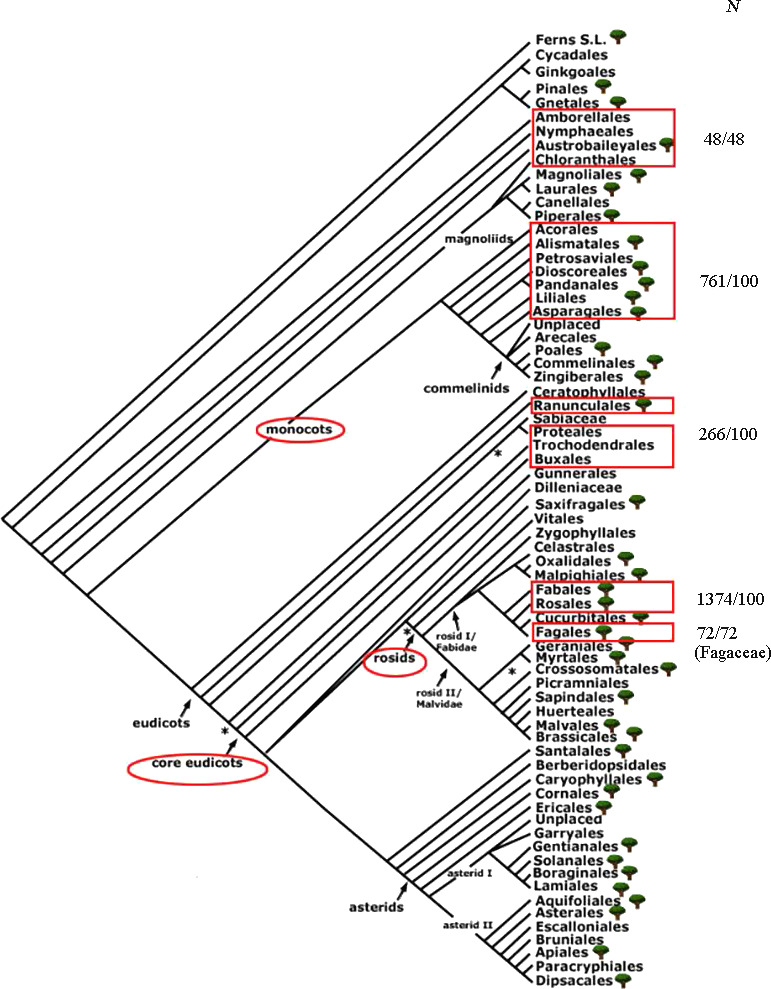
The total numbers of samples and random samples in each group. The phylogeny is adapted from Angiosperm Phylogeny Group IV
FIGURE 7.
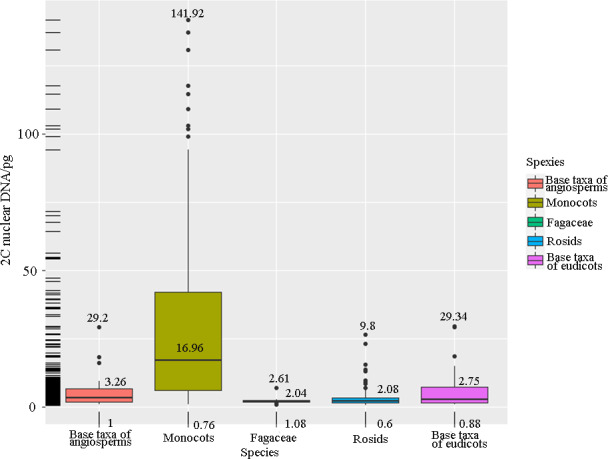
Box patterns of genomic size variation in five groups of angiosperms
4. DISCUSSION
4.1. The GS evolution of four oak species
The GSs in 10,770 angiosperms have been estimated and ranged between 0.062 pg (Genlisea tuberosa, Lentibulariaceae) and 304.46 pg (Paris japonica, Melanthiaceae) (Fleischmann et al., 2014; Leitch et al., 2019). The ancestral genomes of most major clades of core eudicots, such as Caryophyllales, Saxifragales, and Asterids, are very small (Soltis et al., 2003). Additionally, some genera (e.g., some Brassicaceae, Rutaceae, and Onagraceae) are clearly large genome plants. By comparing the GSs of five groups of species, we found that the GSs of Fagaceae species belong to a smaller group and the GS varies narrowly in angiosperms. This finding is consistent with that of Chen et al. (2014) who analyzed the GS variation in the Fagaceae. The Soltis division of GS was used in this study and is as follows: ≤1.4 pg, very small; >1.4 pg ≤ 3.5 pg, small; >3.5 pg ≤ 14.0 pg, intermediate; >14 pg < 35 pg, large; ≥35 pg, very large. In this study, the GS range for the four oak species was 1.83–2.06 pg. Therefore, the four oak species all had small GSs. The genomes in Sections Quercus and Cerris are about five times larger than the Arabidopsis genome, which has a GS that ranges from 1.84 to 2.00 pg (Aldrich & Cavender‐Bares, 2011). Our results corroborate those observed in earlier studies (Kremer et al., 2007; Leitch et al., 2019). However, the estimates for deciduous species calculated in this study were larger (1.93 vs. 1.64 pg/2C) than those calculated previously (Kremer et al., 2007). A possible explanation was that GS evolution was unidirectional, resulting in a model for overall growth (Bennetzen & Kellogg, 1997).
4.2. Intraspecific and interspecific variation in GS
In this study, considerable GS variability was found both within and among the four oak species. The GS estimate within the species of Section Cerris (1.08‐fold) was greater than that of section Quercus (1.07‐fold). This observation may be correlated with environmental parameters. According to the study of Zhang and Li on the prediction of the habitable zone of Q. acutissima and Q. variabilis in China, we found that optimal distribution regions for two species were the Yunnan‐Guizhou Plateau and Qin‐Ba Mountains having both a high altitude and latitude (Li et al., 2014; Zhang et al., 2014). However, the two oaks in this study came from Zijin Mountain, and their areas and altitudes are quite different compared with the most suitable areas. The Zijin Mountain is located in the plain area with an average altitude of 20–30 m, while the Yunnan‐Guizhou Plateau has an average altitude of 2,000–4,000 m, and the Qin‐Ba Mountains with altitude of 1,500–2,500 m. Species may experience natural selection when adapting to such an environment, resulting in large GS variability (Bilinski et al., 2018; Li et al., 2017). In addition, a comparison of mean GS estimates across two species in Section Cerris showed that the mean GS in Q. acutissima (1.87pg) is smaller than in Q. variabilis (1.92 pg). Some of the phenotypic traits, such as plant height, seed mass, cell size, and cell cycle time, may also facilitate GS variability (Benor et al., 2011; Kang et al., 2014; Knight et al., 2005). The differences in these phenotypic features may be related to growth rate, leaf anatomy, and photosynthesis. Previous continuous observations of the seedlings of biennial Q. acutissima and Q. variabilis revealed that the growth rate of the latter is less than that of the former (Li et al., 2020). This is in line with the theory that plans with larger genomes have lower growth rates (Kang et al., 2014; Knight et al., 2005).
The use of GS may not be very useful for classification at higher taxonomic levels, but it is particularly valuable at the species level (Liu et al., 2020; Qiu et al., 2018; Zonneveld, 2008; Zonneveld et al., 2005). The GS variation is approximately 20% across species in a single genus. In woody plants, GS and chromosomal structure are highly conserved. Therefore, the interspecific variation between genomes is greater than the intraspecies variation (Chen et al., 2014). In this study, an analysis of variance showed that most of the variation is interspecific variation. The GS variation among populations, except for those of Q. serrata var. brevipetiolata and Q. fabri, showed significant differences. Several explanations for the interspecific variation have been proposed, such as repeated cycles of polyploidy, which is supported by genomic and isozyme evidence (Bowers et al., 2003; Otto & Whitton, 2000; Wendel, 2000). Earlier studies indicated that the interspecific variation in GS results partly from the appearance of extra B chromosomes, which are caused by the irregular segregation of additional chromosomes during mitosis (Piscor & Parise‐Maltempi, 2015; Zoldos et al., 1998). However, recent technological advances have shown that the presence of B chromosomes generally increases the size of an individual genome, but it does not affect the extent of the variation within the population, regardless of whether it includes individuals having satellite chromosomes. The new findings also suggest that the GS variation may be related to the A chromosome (Chumová et al., 2016). These hypotheses are all possible because the chromosome and ploidy numbers in the Fagaceae remain stable (2n = 24) in most genera, except for extra chromosomes in some Quercus populations (Dzialuk et al., 2007; Zoldos et al., 1998). When the number of chromosome changes results from fission and fusion, then the evolution of the chromosomes may result in recombination between populations. Here, we speculated that the variation in GS was owing to hybridization. We suspect that ZJ116 and ZJ105 were hybrid individuals using a clustering analysis with GS expansion (2.024 pg and 2.06 pg). They may be hybrids resulting from crosses between two species in Section Quercus. The white oaks are wind‐pollinated and unable to discriminate pollen from other species in the same section. In addition, we speculated that hybrid offspring having expanded GSs were produced from Section Quercus and Section Cerris. Cluster analysis showed that there were always Q. fabri and Q. serrata var. brevipetiolata individuals interspersed in the Q. variabilis population. The phenomenon of hybridization within different groups is rare and blocked by reproductive isolation, but it exists and has been reported (Burgarella et al., 2009). In general, a GS increases after polyploidization, but it may undergo a decrease in noncoding DNA sequences, leading to a reduced GS after polyploidization (Li et al., 2013). Furthermore, an increase or decrease in DNA repeat sequences during oak hybridization leads to variations in GS and is the main reason for GS changes in angiosperms.
4.3. Ecological protection proposals for oak trees
Interspecific hybridization and the introgression of Quercus leads to a series of systematic evolutionary and ecological results, such as community recombination and structural adjustment (Aldrich & Cavender‐Bares, 2011; McVay et al., 2017; Song et al., 2015). In addition to the impact on community succession, the hybridization and introgression of oaks are conducive to increasing the genetic diversity and the rapid transformation and fixation of adaptive genes among species. This is conducive to the better survival of species in new environments (Ramirez‐Valiente & Cavender‐Bares, 2017; Ramirez‐Valiente et al., 2018). However, closely related species seldom coexist owing to functional divergences that allow them to occupy different habitats (Klein et al., 2016). In our study, the four oak species were found coexisting on the same land. Their coexistence may result from the vegetation in Zijin Mountains being mainly artificial and natural secondary forests. In the 1950s and 1960s, the inbreeding decline of related species was not considered in afforestation. The GS variations in the four species of Quercus may have been required to adapt to such an environment. In addition, the hybridization in different sections may have resulted from habitat disturbance. As a tourist attraction, Zijinshan has year‐round human activities that change the diffusion patterns of seeds and pollens, disturbing the habitat to a certain extent. Thus, local authorities should guide tourists, strengthen the ecological management, perform appropriate thinning activities, and reduce inbreeding.
5. CONCLUSIONS
In this study, the mean GSs of Q. acutissima, Q. variabilis, Q. fabri, and Q. serrata var. brevipetiolata were 1.87, 1.92, 1.97, and 1.97 pg, respectively, which were within a reasonable range. Thus, there was a low level of intraspecific variation in GSs among Q. acutissima, but it was relatively high among individuals of the other three oak species. Furthermore, there was a high level of interspecific variation among the four oak populations. The oaks in the same section produced hybrid introgression. Additionally, a hybrid offspring was produced from Q. fabri and Q. variabilis, which belong to different sections. The pattern of GS evolution for hybrids species is expansion. This study on GS described a valuable complementary method for studying genetic variation in oak species and has significance in guiding the ecological protection of oaks.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
GaoMing Wei: Data curation (lead); formal analysis (lead); methodology (lead); resources (equal); writing–original draft (equal). Xuan Li: Formal analysis (supporting); methodology (supporting); software (lead); writing–original draft (equal); writing–review and editing (lead). YanMing Fang: Resources (equal); supervision (lead); validation (equal); writing–review and editing (supporting).
Supporting information
Table S1
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation in China (31770699 and 31370666), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Forestry Innovation Project of Jiangsu [LYSX(2016)49], the Nanjing Forestry University Excellent Doctoral Thesis Fund, and the Jiangsu Innovation Engineering Fund. Thanks to Lei Xie for his guidance in drawing.
Wei G, Li X, Fang Y. Sympatric genome size variation and hybridization of four oak species as determined by flow cytometry genome size variation and hybridization. Ecol Evol. 2021;11:1729–1740. 10.1002/ece3.7163
Co‐first author: GaoMing Wei, Xuan Li.
DATA AVAILABILITY STATEMENT
Genome size (GS) database for 120 individuals of four Querucs species is publicly available at Dryad repository: https://doi.org/10.5061/dryad.hx3ffbgcv.
REFERENCES
- Aldrich, P. R. , & Cavender‐Bares, J. (2011). Genomics and breeding of oaks and their slightly less‐domesticated wild oak relatives In Kole C. (Ed.), Wild crop relatives: Genomic and breeding resources (pp. 89–129). Springer. [Google Scholar]
- Bennett, M. D. , Bhandol, P. , & Leitch, I. J. (2000). Nuclear DNA amounts in angiosperms and their modern uses‐807 new estimates. Annals of Botany, 86, 859–909. [Google Scholar]
- Bennetzen, J. L. , & Kellogg, E. A. (1997). Do plants have a one‐way ticket to genomic obesity? The Plant Cell, 9(9), 1509–1514. 10.2307/3870439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benor, S. , Fuchs, J. , & Blattner, F. R. (2011). Genome size variation in Corchorus olitorius (Malvaceaes.l.) and its correlation with elevation and phenotypic traits. Genome, 54(7), 575–585. [DOI] [PubMed] [Google Scholar]
- Bilinski, P. , Albert, P. S. , Berg, J. J. , Birchler, J. A. , Grote, M. N. , Lorant, A. , Quezada, J. U. , Swarts, K. , Yang, J. L. , & Ross‐Ibarra, J. (2018). Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLoS Genetics, 14(5), e1007162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J. E. , Chapman, B. A. , Rong, J. K. , & Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature, 422(6930), 433–438. [DOI] [PubMed] [Google Scholar]
- Burgarella, C. , Lorenzo, Z. , Jabbour‐Zahab, R. , Lumaret, R. , Guichoux, E. , Petit, R. J. , Soto, A. , Gil, L. (2009). Detection of hybrids in nature: Application to oaks (Quercus suber and Q. ilex). Heredity, 102(5), 442–452. [DOI] [PubMed] [Google Scholar]
- Cannon, C. H. , & Scher, C. L. (2017). Exploring the potential of gametic reconstruction of parental genotypes by F1 hybrids as a bridge for rapid introgression. Genome, 9(60), 713–719. [DOI] [PubMed] [Google Scholar]
- Castro, S. , Romeiras, M. M. , Castro, M. , Duarte, M. C. , & Loureiro, J. (2013). Hidden diversity in wild Beta taxa from Portugal: Insights from genome size and ploidy level estimations using flow cytometry. Plant Science, 207(3), 72–78. [DOI] [PubMed] [Google Scholar]
- Cavender‐Bares, J. , & Pahlich, A. (2009). Molecular, morphological, and ecological niche differentiation of sympatric sister oak species, Quercus virginiana and Q. geminate (Fagaceae). American Journal of Botany, 96(9), 1690–1702. [DOI] [PubMed] [Google Scholar]
- Chen, S. C. , Cannon, C. H. , Kua, C. S. , Liu, J. J. , & Galbraith, D. W. (2014). Genome size variation in the Fagaceae and its implications for trees. Tree Genetics & Genomes, 10(4), 977–988. [Google Scholar]
- Chumová, Z. , Mandáková, T. , & Trávníček, P. (2016). Are B‐chromosomes responsible for the extraordinary genome size variation in selected Anthoxanthum annuals? Plant Systematics & Evolution, 302(6), 731–738. 10.1007/s00606-016-1295-5 [DOI] [Google Scholar]
- Cottam, W. P. , Tucker, J. M. , & Santamour, F. S. (1982). Oak Hybridization at the University of Utah, Salt Lake City: State Arboretum of Utah. [Google Scholar]
- Denaeghel, H. , Van Laere, K. , Leus, L. , Van Huylenbroeck, J. , & Van Labeke, M. C. (2017). Interspecific hybridization in Sarcococca supported by analysis of ploidy level, genome size and genetic relationships. Euphytica, 213(7), 149 10.1007/s10681-017-1934-0 [DOI] [Google Scholar]
- Devos, K. M. , Brown, J. K. , & Bennetzen, J. L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis . Genome Research, 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel, J. , Bartos, J. , Voglmayr, H. , & Greilhuber, J. (2003). Nuclear DNA content and genome size of trout and human. Cytometry, 51A, 127–128. [DOI] [PubMed] [Google Scholar]
- Dzialuk, A. , Chybicki, I. , Welc, M. , Sliwinska, E. , & Burczyk, J. (2007). Presence of triploids among oak species. Annals of Bbotany, 99(5), 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre, J. M. , & Brown, S. (1996). A flow cytometric evaluation of the nuclear DNA content and GC percent in genomes of European oak species. Annales Des Sciences Forestières. EDP Sciences, 53(4), 915–917. [Google Scholar]
- Fleischmann, A. , Michael, T. P. , Rivadavia, F. , Sousa, A. , Wang, W. Q. , Temsch, E. , Greilhuber, J. , Muller, K. F. , & Heubl, G. (2014). Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Annals of Botany, 114(8), 1651–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, D. , Davies, M. S. , & Barlow, P. W. (2008). A strong nucleotypic effect on the cellcycle regardless of ploidy level. Annals of Botany, 101(6), 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, D. W. (2004). Cytometry and plant sciences: a personal retrospective. Cytometry Part A, 58A(1), 37–44. [DOI] [PubMed] [Google Scholar]
- Galbraith, D. W. , Harkins, K. R. , Maddox, J. M. , Ayres, N. M. , Sharma, D. P. , & Firoozabady, E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science, 220(4601), 1049–1051. [DOI] [PubMed] [Google Scholar]
- Greilhuber, J. , Borsch, T. , Muller, K. , Worberg, A. , Porembski, S. , & Barthlott, W. (2006). Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biology, 8(6), 770–777. [DOI] [PubMed] [Google Scholar]
- Hanusova, K. , Ekrt, L. , Vit, P. , Kolar, F. , & Urfus, T. (2014). Continuous morphological variation correlated with genome size indicates frequent introgressive hybridization among Diphasiastrum species (Lycopodiaceae) in Central Europe. PLoS One, 9(6), e99552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, J. S. , Grover, C. E. , & Wendel, J. F. (2008). Repeated big bangs and the expanding universe: Directionality in plant genome size evolution. Plant Science, 174, 557–562. [Google Scholar]
- Hawkins, J. S. , Proulx, S. R. , Rapp, R. A. , & Wendel, J. F. (2009). Rapid DNA loss as acounter balance to genome expansion through retrotransposon proliferation in plants. Proceedings of the National Academy of Sciences, USA, 106(42), 17811–17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo, O. , Garcia, S. , Garnatje, T. , Mumbru, M. , Patterson, A. , Vigo, J. , & Valles, J. (2015). Genome size in aquatic and wetland plants: Fitting with the large genome constraint hypothesis with a few relevant exceptions. Plant Systematics & Evolution, 301(7), 1927–1936. [Google Scholar]
- Kang, M. , Tao, J. J. , Wang, J. , Ren, C. , Qi, Q. W. , Xiang, Q. Y. , & Huang, H. W. (2014). Adaptive and nonadaptive genome size evolution in karst endemic flora of China. New Phytologist, 202(4), 1371–1381. [DOI] [PubMed] [Google Scholar]
- Klein, E. K. , Lagache‐Navarro, L. , & Petit, R. J. (2016). Demographic and spatial determinants of hybridization rate. Journal of Ecology, 105(1), 29–38. [Google Scholar]
- Knight, C. A. , & Ackerly, D. D. (2002). Variation in nuclear DNA content across environmental gradients: A quantile regression analysis. Ecology Letters, 5(1), 66–76. 10.1046/j.1461-0248.2002.00283.x [DOI] [Google Scholar]
- Knight, C. A. , Molinari, N. A. , & Petrov, D. A. (2005). The large genome constraint hypothesis: Evolution, ecology and phenotype. Annals of Botany, 95(1), 177–190. 10.1093/aob/mci011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, A. , Casasoli, M. , Barreneche, T. , Bodénès, C. , Sisco, P. , Kubisiak, T. , Scalfi, M. , Leonardi, S. , Bakker, E. , Buiteveld, J. , Romero‐Severson, J. , Arumuganathan, K. , Derory, J. , Scotti‐Saintagne, C. , Roussel, J. , Bertocchi, M. E. , Lexer, C. , Porth, I. , Hebard, F. , … Villani, F. (2007). Fagaceae trees (pp. 161–187). Springer, Berlin Heidelberg. [Google Scholar]
- Kumar, A. , & Bennetzen, J. L. (1999). Plant retrotransposons. Annual Review of Genetics, 33(1), 479–532. [DOI] [PubMed] [Google Scholar]
- Leitch, I. J. , Johnston, E. , Pellicer, J. , Hidalgo, O. , & Bennett, M. D. (2019). Angiosperm DNA C‐values database (release 9.0, Apr 2019). Retrieved from https://cvalues.science.kew.org/ [Google Scholar]
- Leitch, I. J. , & Leitch, A. R. (2013). Genome size diversity and evolution in land plants In Leitch I. J. (Ed.), Plant genome diversity (Vol. 2, pp. 307–322). Springer. [Google Scholar]
- Li, D. D. , Lu, Y. L. , Guo, S. L. , Yin, L. P. , Zhou, P. , & Lou, Y. X. (2017). Nuclear DNA contents of Echinchloa crus‐galli and its Gaussian relationships with environments. Acta Oecologica, 79, 36–47. 10.1016/j.actao.2017.01.002 [DOI] [Google Scholar]
- Li, X. , Hu, D. , Luo, M. M. , Zhu, M. , Li, X. W. , Luo, F. , Li, J. Q. , & Yan, J. (2013). Nuclear DNA content variation of three Miscanthus species in china. Genes & Genomics, 35(1), 13–20. 10.1007/s13258-013-0063-y [DOI] [Google Scholar]
- Li, X. , Xue, M. L. , Hu, R. , Yang, J. D. , & Fang, Y. M. (2020). Analysis and evaluation of seedling growth difference between Quercus acutissima and Quercus variabilis, China: (under review). Journal of Northeast Forestry University. [Google Scholar]
- Li, Y. , Zhang, X. W. , & Fang, Y. M. (2014). Predicting the impact of global warming on the geographical distribution pattern of Quercus variabilis in China. Chinese Journal of Plant Ecology, 12, 3381–3389. [PubMed] [Google Scholar]
- Liu, G. C. , Chang, Z. , Chen, L. , He, J. W. , Dong, Z. W. , Yang, J. , Lu, S. H. , Zhao, R. P. , Wan, W. T. , Ma, G. L. , Li, J. , Zhang, R. , Wang, W. , & Li, X. Y. (2020). Genome size variation in butterflies (Insecta, Lepidotera, Papilionoidea): A thorough phylogenetic comparison. Systematic Entomology, 45(3), 571–582. 10.1111/syen.12417 [DOI] [Google Scholar]
- Manos, P. S. , Zhou, Z. K. , & Cannon, C. H. (2001). Systematics of Fagaceae: Phylogenetic tests of reproductive trait evolution. International Journal of Plant Sciences, 162(6), 1361–1379. 10.1086/322949 [DOI] [Google Scholar]
- Manuel, R. P. J. , Hernando, R. C. , Antonio, G. R. , Victor, R. R. , & Ken, O. (2020). High genetic diversity and stable Pleistocene distributional ranges in the widespread Mexican red oak Quercus castanea Née (1801) (Fagaceae). Ecology and Evolution, 10(10), 4204–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie, D. , & Brown, S. C. (1993). A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biology of the Cell, 78(1–2), 41–51. [DOI] [PubMed] [Google Scholar]
- Mcvay, J. D. , Hauser, D. , Hipp, A. L. , & Manos, P. (2017). Phylogenomics reveals a complex evolutionary history of lobed‐leaf white oaks in western north America. Genome, 60(9), 733–742. [DOI] [PubMed] [Google Scholar]
- Nowicka, A. , Sliwinska, E. , Grzebelus, D. , Baranski, R. , & Grzebelus, E. (2016). Nuclear DNA content variation within the genus Daucus (Apiaceae) determined by flow cytometry. Scientia Horticulturae, 209, 132–138. [Google Scholar]
- Otto, S. P. , & Whitton, J. (2000). Polyploid incidence and evolution. Annual Review of Genetics, 34(1), 401–437. [DOI] [PubMed] [Google Scholar]
- Petersson, L. K. , Dey, D. C. , Felton, A. M. , Gardiner, E. S. , & Löf, M. (2020). Influence of canopy openness, ungulate exclosure, and low‐intensity fire for improved oak regeneration in temperate Europe. Ecology and Evolution, 10(5), 2626–2637. 10.1002/ece3.6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscor, D. , & Parise‐Maltempi, P. P. (2015). First description of B chromosomes in the Hyphessobrycon (Characiformes, Characidae) genus: A hypothesis for the extra element of Hyphessobrycon eques Steindachner, 1882. Comparative Cytogenetics, 9(3), 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, L. J. , Bayly, M. J. , & Vesk, P. A. (2015). The roles of ecological and evolutionary processes in plant community assembly: The environment, hybridization, and introgression influence co‐occurrence of Eucalyptus. The American Naturalist, 185(6), 784–796. [DOI] [PubMed] [Google Scholar]
- Porth, I. , Garnier‐Gere, P. , Klapste, J. , Scotti‐Saintagne, C. , El‐Kassaby, Y. A. , Burg, K. , & Kremer, A. (2016). Species‐specific alleles at a beta‐tubulin gene show significant associations with leaf morphological variation within Quercus petraea and Q. robur populations. Tree Genetics & Genomes, 12(4), 81. [Google Scholar]
- Prancl, J. , Kaplan, Z. , Travnicek, P. , & Jarolimova, V. (2014). Genome size as a key to evolutionary complex aquatic plants: Polyploidy and hybridization in Callitriche (Plantaginaceae). PLoS One, 9(9), e105997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, F. , Baack, E. J. , Whitney, K. D. , Bock, D. G. , Tetreault, H. M. , Rieseberg, L. H. , & Ungerer, M. C. (2018). Phylogenetic trends and environmental correlates of nuclear genome size variation in Helianthus sunflowers. New Phytologist, 221(3), 1609–1618. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Valiente, J. A. , & Cavender‐Bares, J. (2017). Evolutionary trade‐offs between drought resistance mechanisms across a precipitation gradient in a seasonally dry tropical oak (Quercus oleoides). Tree Physiology, 37(7), 889–901. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Valiente, J. A. , Deacon, N. J. , Etterson, J. , Center, A. , Sparks, J. P. , Sparks, K. L. , Longwell, T. , Pilz, G. , & Cavender‐Bares, J. (2018). Natural selection and neutral evolutionary processes contribute to genetic divergence in leaf traits across a precipitation gradient in the tropical oak Quercus oleoides . Molecular Ecology, 27(9), 2176–2192. [DOI] [PubMed] [Google Scholar]
- Schubert, I. , & Vu, G. T. H. (2016). Genome stability and evolution: Attempting a holistic view. Trends in Plant Science, 21(9), 749–757. [DOI] [PubMed] [Google Scholar]
- Sharma, S. , Kaushik, S. , & Raina, S. N. (2019). Estimation of nuclear DNA content and its variation among Indian Tea accessions by flow cytometry. Physiology and Molecular Biology of Plants, 25(2), 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis, D. E. , Soltis, P. S. , Bennett, M. D. , & Leitch, I. J. (2003). Evolution of genome size in the angiosperms. American Journal of Botany, 90(11), 1596–1630. [DOI] [PubMed] [Google Scholar]
- Song, Y. G. , Deng, M. , Hipp, A. L. , & Li, Q. S. (2015). Leaf morphological evidence of natural hybridization between two oak species (Quercus austrocochinchinensis and Q. kerrii) and its implications for conservation management. European Journal of Forest Research, 134(1), 139–151. [Google Scholar]
- Spaniel, S. , Marhold, K. , & Zozomova‐Lihova, J. (2019). Polyphyletic Alyssum cuneifolium (Brassicaceae) revisited: Morphological and genome size differentiation of recently recognized allopatric taxa. Journal of Systematics and Evolution, 57(3), 287–301. [Google Scholar]
- Talla, V. , Suh, A. , & Kalsoom, F. , Dinca, V. , Vila, R. , Friberg, M. , Wiklund, C. , & Backström, N. (2017). Rapid increase in genome size as a consequence of transposable element hyperactivity in wood‐white (leptidea) butterflies. Genome Biology & Evolution, 9(10), 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlili, A. , Gouja, H. , Vallès, J. , Garnatje, T. , Buhagia, J. , & Neffati, M. (2020). Chromosome number and genome size in Atriplex mollis from southern Tunisia and Atriplex lanfrancoi from Malta (Amaranthaceae). Plant Systematics & Evolution, 306, 11 10.1007/s00606-020-01643-1 [DOI] [Google Scholar]
- Vit, P. , Wolfova, K. , Urfus, T. , Tajek, P. , & Suda, J. (2014). Interspecific hybridization between rare and common plant congeners inferred from genome size data: Assessing the threat to the Czech serpentine endemic Cerastiumalsinifolium . Preslia, 86(1), 95–117. [Google Scholar]
- Wei, G. M. , & Fang, Y. M. (2015). Establishment and optimization of an experiment system for flowcytometry in Quercus . Journal of Nanjing Forestory University (Natural Sciences Edition), 39(1), 167–172. [Google Scholar]
- Wendel, J. F. (2000). Genome evolution in polyploids. Plant Molecular Biology, 42(1), 225–249. [PubMed] [Google Scholar]
- Wetherbee, R. , Birkemoe, T. , Skarpaas, O. , & Sverdrup‐Thygeson, A. (2020). Hollow oaks and beetle functional diversity: Significance of surroundings extends beyond taxonomy. Ecology and Evolution, 10(2), 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, T. E. , Takebayashi, N. , Barker, M. S. , Mayrose, I. , Greenspoon, P. B. , & Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascularplants. Proceedings of the National Academy of Sciences, USA, 106(33), 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. H. , Martin, S. L. , Bekele, W. A. , Latta, R. G. , Diederichsen, A. , Peng, Y. Y. , & Tinker, N. A. (2016). Genome size variation in the genus Avena . Genome, 59(3), 209–220. [DOI] [PubMed] [Google Scholar]
- Yuan, Z. L. , Wei, B. L. , Chen, Y. , Jia, H. R. , Wei, Q. N. , & Ye, Y. Z. (2018). How do similarities in spatial distributions and interspecific associations affect the coexistence of Quercus species in the Baotianman National Nature Reserve, Henan. China. Ecology and Evolution, 8(5), 2580–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Wang, M. , Guo, Z. , Guan, Y. , & Yan, X. (2019). Variation in ploidy level and genome size of Cynodon dactylon (L.) Pers. along a latitudinal gradient. Folia Geobotanica, 54(3), 267–278. [Google Scholar]
- Zhang, X. W. , Li, Y. , & Fang, Y. M. (2014). Geographical distribution and prediction of potential ranges of Quercus acutissima in China. Acta Botanica Sinica, 34(8), 1685–1692. [Google Scholar]
- Zoldos, V. , Papes, D. , Brown, S. C. , Panaud, O. , & Siljak‐Yakovlev, S. (1998). Genome size and base composition of seven Quercus species: inter‐ and intra‐population variation. Genome, 41, 162–168. [Google Scholar]
- Zonneveld, B. J. M. (2008). The systematic value of nuclear DNA content for all species of Narcissus L. (Amaryllidaceae). Plant Systematics & Evolution, 275(1–2), 109–132. [Google Scholar]
- Zonneveld, B. J. M. , Leitch, I. J. , & Bennett, M. D. (2005). First Nuclear DNA Amounts in more than 300 Angiosperms. Annals of Botany, 96(2), 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
Genome size (GS) database for 120 individuals of four Querucs species is publicly available at Dryad repository: https://doi.org/10.5061/dryad.hx3ffbgcv.


