Abstract
Objectives
Coronavirus disease‐19 (COVID‐19) is associated with various clinical manifestations, ranging from asymptomatic infection to critical illness. The aim of this study is to evaluate the clinical and laboratory characteristics of hospitalised COVID‐19 patients and construct a predictive model for the discrimination of patients at risk of disease progression.
Methods
A single‐centre cohort study was conducted including consecutively patients with COVID‐19. Demographic, clinical and laboratory findings were prospectively collected at admission. The primary outcome of interest was the intensive care unit admission. A risk model was constructed by applying a Cox's proportional hazard's model with elastic net penalty. Its diagnostic performance was assessed by receiver operating characteristic analysis and was compared with conventional pneumonia severity scores.
Results
From a total of 67 patients 15 progressed to critical illness. The risk score included patients’ gender, presence of hypertension and diabetes mellitus, fever, shortness of breath, serum glucose, aspartate aminotransferase, lactate dehydrogenase, C‐reactive protein and fibrinogen. Its predictive accuracy was estimated to be high (area under the curve: 97.1%), performing better than CURB‐65, CRB‐65 and PSI/PORT scores. Its sensitivity and specificity were estimated to be 92.3% and 93.3%, respectively, at the optimal threshold of 1.6.
Conclusions
A10‐variable risk score was constructed based on clinical and laboratory characteristics in order to predict critical illness amongst hospitalised COVID‐19 patients, achieving better discrimination compared with traditional pneumonia severity scores. The proposed risk model should be externally validated in independent cohorts in order to ensure its prognostic efficacy.
What's known
Several risk factors have been proposed to predispose for the development of critical illness amongst COVID‐19 patients, although the optimal screening model to be widely applied at admission remains unclear.
What's new
A 10‐variable model including demographical and laboratory parameters has been developed by elastic net regularisation, outperforming the conventional pneumonia severity scores.
1. INTRODUCTION
Coronavirus disease‐19 (COVID‐19) represents an emerging life‐threatening respiratory infection, caused by the beta coronavirus SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus‐2), first identified in Wuhan, China. 1 Cell entry depends on the expression of angiotensin‐converting enzyme 2 (ACE‐2) and thus lung and vascular epithelium, as well as macrophages and monocytes represent the main targets of the virus. 2 The pathophysiology of the disease has been suggested to be linked to a hyperinflammatory state, which in its severe form may lead to diffuse alveolar damage and the clinical development of acute respiratory distress syndrome (ARDS). 3 Coagulopathy has been also recognised as a critical aspect of the disease pathophysiology, associated with thromboembolic events, endothelial dysfunction and small vessel thromboses. 4 COVID‐19 is associated with a broad spectrum of clinical presentations, ranging from asymptomatic disease to acute lung injury with severe hypoxemia requiring mechanical ventilation, although the subgroup of patients prone to develop rapid respiratory deterioration remains still a matter of debate.
Early risk stratification is essential to guide decision making regarding clinical management and patient allocation, especially in resource‐limited settings. To this end, significant research effort has been devoted to the identification of factors associated with disease progression and the construction of predictive models, including clinical, laboratory or radiological parameters. 5 Nonetheless, inconsistent results have been reported leading to remarkable heterogeneity of the existing risk scores. In addition, the risk of bias and concerns about overfitting often exist, rendering the proposed models overoptimistic and limiting their direct clinical applicability. 6 As a result, the optimal screening model for the prediction of critical illness remains still under investigation.
The present study aims to comprehensively assess the clinical and laboratory characteristics at the admission of patients with COVID‐19 and identify those linked to worse prognosis and disease progression. A novel risk score is constructed by applying machine learning methodology in order to improve variable selection and effectively recognise patients at higher risk of severe disease. At the same time, traditional pneumonia severity scores are estimated and their predictive performance is compared with the proposed risk model.
2. MATERIALS AND METHODS
2.1. Study design
All consecutive adult patients with COVID‐19 admitted to our department in “Sotiria” General and Chest Diseases Hospital of Athens from 11 March to 1 June 2020 were prospectively enrolled. The diagnosis was based on the detection of SARS‐CoV‐2 by real‐time polymerase chain reaction (RT‐PCT) analysis of nasopharyngeal swabs. Non‐laboratory‐confirmed cases were excluded. The study was approved by the institutional review board of the hospital and all patients or their next of kin gave written informed consent.
Reporting of outcomes was performed in accordance with the TRIPOD (Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis) guidelines. 7
2.2. Procedures
Pre‐specified variables about clinical history, comorbidities, symptoms, laboratory tests on admission and treatment were registered in a comprehensive database. Variables of interest were baseline characteristics, comorbidities (diabetes mellitus, hypertension, coronary artery disease, heart failure, chronic obstructive pulmonary disease, asthma, liver disease, cancer, hematologic malignancy and immunodeficiency), clinical symptoms (fever, chills, cough, shortness of breath, sputum production, hemoptysis, fatigue, headache, diarrhoea and nausea/vomiting) and laboratory tests (complete blood count, coagulation tests, renal and liver function tests, serum electrolytes, glucose, lactate dehydrogenase, amylase, troponin, triglycerides, total cholesterol, C‐reactive protein, procalcitonin, ferritin and blood lactate levels). Derived neutrophil‐to‐lymphocyte ratio (dNLR) was defined as: dNLR = Neutrophil count/(White blood cell count − Lymphocyte count)). 8 Arterial blood gases were assessed on admission, calculating the PaO2/FiO2 ratio.
Furthermore, three clinical scores were measured on admission: CURB‐65 (Confusion, Urea, Respiratory rate, Blood pressure, age ≥ 65 years), 9 CRB‐65 (Confusion, Respiratory rate, Blood pressure, age ≥ 65 years) 10 and PSI/PORT (Pneumonia Severity Index/Pneumonia Outcome Research Trial‐PSI/PORT). 11 The main outcome of interest was set to be the transfer from the isolation ward to the intensive care unit (ICU). Secondary outcomes included the occurrence of acute respiratory distress syndrome (ARDS), 12 systemic inflammatory response syndrome (SIRS) 13 and acute kidney injury. 14 In addition, the maximum values of SOFA (Sequential Organ Failure Assessment), 15 APACHE II (Acute Physiology and Chronic Health Evaluation II) 16 and MEWS (Modified Early Warning Score) 17 scores during hospital stay were calculated.
2.3. Data analysis
Statistical analysis was conducted in R‐3.6.3 (“survival,” 18 “glmnet,” 19 and “pROC” 20 packages). Statistical significance was defined as P value < .05. Normality of continuous variables was tested by the Shapiro‐Wilk test because of the moderate sample size. 21 The possible linear correlation amongst laboratory variables was assessed by the Spearman correlation coefficient, because of the presence of skewed distributions. Strong correlations were detected by Spearman ρ < −0.6 or > 0.6. 22 Potential missing values were planned to be statistically imputed by the k‐nearest neighbour method. 23 Survival analysis was performed aiming to identify clinical and laboratory factors on admission associated with subsequent ICU admission. To avoid overfitting, the Cox's proportional hazard's model with elastic net penalty was implemented. The elastic net penalty was defined by the following equation:
| (1) |
where β represents the regression coefficient, λ the shrinkage parameter and α the elastic net mixing parameter, with 0 ≤ α ≤1. Values of α equal to 0 and 1 correspond to the ridge and the Least Absolute Shrinkage and Selection Operator (LASSO) regression models, respectively. 24 The values of α and λ were selected by 10‐fold cross‐validation. Specifically, the λ value providing minimum deviance (λ min) was chosen for the analysis and the α value providing the lowest mean cross‐validation error at λ min was selected. A risk prediction model was constructed by including the parameters with nonzero coefficients; hence, a risk score was calculated according to the following equation:
| (2) |
where βi refers to the regression coefficient and χi the value of the parameter. The Harrell concordance C‐index was used to evaluate the discrimination of the model. The estimated risk score was compared between patient subgroups based on age (≤65/>65 years), day from symptom onset (≤7/>7 days) and PaO2/FiO2 ratio on admission (≤200, 201‐300, >300) using the Mann‐Whitney U test and was incorporated in a multivariate Cox's proportional hazard's model including the above parameters. Moreover, the diagnostic accuracy of the risk score was tested by plotting the receiver operating characteristics (ROC) curve and calculating the area under the curve (AUC). The optimal threshold was specified by estimating the Youden index 25 and the corresponding sensitivity and specificity were reported. The diagnostic accuracy of the risk score was compared with those of CURB‐65, CRB‐65 and PSI/PORT scores. Kaplan‐Meier survival curves stratified by the outcomes of the four clinical scores were also constructed, using the log‐rank test to evaluate statistically significant differences.
Concerning secondary outcomes, the accuracy of the risk score in predicting the occurrence of ARDS, SIRS and acute kidney injury was estimated by performing ROC analysis and calculating the respective AUCs. In addition, the potential correlation of the risk score with the worst SOFA, APACHE II and MEWS scores was assessed by the Spearman rank correlation test.
3. RESULTS
3.1. Clinical characteristics
A total of 67 patients were included in the present study. Fifteen of them were transferred to the ICU, needing intubation and mechanical ventilation, while three of them subsequently died. Their demographic and clinical characteristics are summarised in Table 1. The source of exposure to SARS‐CoV‐2 was known for 27 cases (40.3%), while seven patients were healthcare workers. The mean age of patients was 59.04 years (standard deviation: 17.45, range: 19 to 92) and 44 of them (65.7%) were male. Fifteen patients (22.4%) were obese (body mass index ≥ 25 kg/m2), while 18 patients (26.9%) reported that they were ever‐smokers. The most common comorbidity was hypertension (29.9%), followed by coronary artery disease (8.9%), heart failure (8.9%) and history of malignant disease (8.9%). The majority of patients presented with cough (62.7%), fever (59.7%), or fatigue (55.2%), while approximately half of them (52.2%) reported shortness of breath. Diarrhoea was reported by 26.9% of patients, but nausea and vomiting were rare (7.5%).
TABLE 1.
Demographic and clinical characteristics of the included patients
| Clinical characteristics | Patients (N = 67) |
|---|---|
| Demographics | |
| Age (years) | 59.04 ± 17.45 |
| ≤65 | 46 (68.7%) |
| >65 | 21 (31.3%) |
| Male gender | 44 (65.7%) |
| Obesity | 15 (22.4%) |
| History of smoking | 18 (26.9%) |
| Known source of exposure | 27 (40.3%) |
| Day of symptoms on admission | 8 [5‐10] |
| Comorbidities | |
| Hypertension | 20 (29.9%) |
| Coronary artery disease | 6 (8.9%) |
| Heart failure | 6 (8.9%) |
| Cancer | 6 (8.9%) |
| Asthma | 5 (7.5%) |
| Diabetes mellitus | 3 (4.5%) |
| Immunodeficiency | 3 (4.5%) |
| Chronic obstructive pulmonary disease | 3 (4.5%) |
| Hematologic malignancy | 2 (3.0%) |
| Chronic liver disease | 0 (0%) |
| Symptoms at admission | |
| Cough | 42 (62.7%) |
| Fever | 40 (59.7%) |
| Fatigue | 37 (55.2%) |
| Shortness of breath | 35 (52.2%) |
| Chills | 19 (28.4%) |
| Diarrhoea | 18 (26.9%) |
| Headache | 13 (19.4%) |
| Sputum production | 10 (14.9%) |
| Nausea and vomiting | 5 (7.5%) |
| Hemoptysis | 1 (1.5%) |
| Vital signs – ABGs at admission | |
| Mean arterial pressure (mm Hg) | 90 [83.33‐96.67] |
| Heart rate (beats/minute) | 82.90 ± 13.84 |
| PaO2/FiO2 (mm Hg) | 338.1 [266.3‐370.7] |
| PaCO2 (mm Hg) | 34 [31‐35] |
| HCO3 (mmol/L) | 24.07 ± 2.12 |
| Arterial‐alveolar gradient (mm Hg) | 38.93 [31.33‐80.80] |
| Treatment | |
| Antimicrobial | 50 (74.6%) |
| Azithromycin | 43 (64.2%) |
| Hydroxychloroquine | 39 (58.2%) |
| Oseltamivir | 26 (38.8%) |
| Glucocorticoids | 9 (13.4%) |
| Tocilizumab | 1 (1.5%) |
| Convalescent plasma therapy | 1 (1.5%) |
| Complications | |
| Bacterial pneumonia | 23 (34.3%) |
| Acute respiratory distress syndrome | 17 (25.4%) |
| Systemic inflammatory response syndrome | 16 (23.9%) |
| Acute kidney injury | 15 (22.4%) |
| Pulmonary embolism | 3 (4.5%) |
| Acute coronary syndrome | 1 (1.5%) |
| Cerebrovascular accident | 1 (1.5%) |
| Admission to ICU | 15 (22.4%) |
| Death | 3 (4.5%) |
Treatment mainly included the administration of azithromycin (64.2%) and hydroxychloroquine (58.2%), while various antimicrobial agents (74.6%) were administered during the course of their illness because of suspected bacterial superinfection. In addition, oseltamivir therapy (38.8%) was initiated in patients because of suspected influenza co‐infection, but was subsequently discontinued in most cases, as influenza co‐infection was confirmed only in two patients. Bacterial pneumonia was assumed to complicate the clinical course of 23 patients based on clinical, microbiological and/or radiological criteria. 26 Moreover, ARDS complicated the clinical course of 17 cases (25.4%), SIRS of 16 (23.9%), acute kidney injury of 15 (22.4%), pulmonary embolism of 3 (4.5%) cases, while ischemic stroke was diagnosed in 1 patient.
3.2. Laboratory findings
The outcomes of laboratory tests are presented in Table 2. The most common abnormality was the elevation of inflammation markers, especially C‐reactive protein (91%), ferritin (76.1%) and fibrinogen (85.1%). Coagulation disorders were observed as increased activated partial thromboplastin time and D‐dimer levels occurred in 38.8% and 58.2% of patients, respectively. Furthermore, lactate dehydrogenase levels were above the normal range in 64.2% of patients, creatinine phosphokinase in 25.4% and troponin in 19.4% of patients. Liver damage was present in 25 (37.3%) patients, as reflected by increased aspartate aminotransferase values. Lymphopenia was detected in 22 (32.8%) cases, while the median neutrophil‐to‐lymphocyte ratio was estimated to be 3.20 (interquartile range: 2.43 to 4.91). The coefficients of correlation amongst laboratory variables are illustrated in Figure 1. The Spearman rank test indicated strong correlation of procalcitonin with C‐reactive protein (ρ = 0.67, P value < .001), ferritin (ρ = 0.64, P value < .001) and troponin (ρ = 0.66, P value < .001), of C‐reactive protein with ferritin (ρ = 0.74, P value < .001) and fibrinogen (ρ = 0.69, P value < .001), as well as of lactate dehydrogenase with ferritin (ρ = 0.66, P value < .001), aspartate aminotransferase (ρ = 0.65, P value < .001) and D‐dimers (ρ = 0.64, P value < .001).
TABLE 2.
Laboratory findings of the included patients
| Laboratory findings | Normal values | Patients (N = 67) | ||
|---|---|---|---|---|
| Total | Increased | Decreased | ||
| Complete blood count | ||||
| Hemoglobin (g/dL) | 13.0‐17.5 | 13.71 ± 1.41 | – | 19 (28.4%) |
| Mean Corpuscular volume (fL) | 80‐100 | 87.6 [85.5‐89.75] | – | 3 (4.5%) |
| White blood cells (/μL) | 4500‐10500 | 6470 [4685‐7710] | 6 (8.9%) | 15 (22.4%) |
| Neutrophil count (/μL) | 1500‐8000 | 4170 [3160‐5583] | 4 (6.0%) | 5 (7.5%) |
| Lymphocyte count (/μL) | 1000‐4800 | 1290 [926‐1745] | – | 22 (32.8%) |
| Platelet count (/μL) | 150‐450 | 200.16 ± 63.00 | – | 15 (22.4%) |
| Neutrophil‐to‐lymphocyte ratio | – | 3.20 [2.43‐4.91] | N/A | N/A |
| Derived neutrophil‐to‐lymphocyte ratio | – | 0.88 [0.83‐0.92] | N/A | N/A |
| Platelet‐to‐lymphocyte ratio | – | 227 [161‐350.8] | N/A | N/A |
| Coagulation function | ||||
| Activated partial thromboplastin time (s) | 28‐40 | 38.10 [35.10‐42.35] | 26 (38.8%) | – |
| International normalised ratio | <1.1 | 1.04 [1.01‐1.13] | 21 (31.3%) | – |
| D‐dimer (µg/mL) | <0.5 | 0.56 [0.41‐1.19] | 39 (58.2%) | – |
| Fibrinogen (mg/dL) | 200‐400 | 553.21 ± 162.22 | 57 (85.1%) | 1 (1.5%) |
| Blood biochemistry | ||||
| Glucose (mg/dL) | 75‐115 | 105 [94‐116.5] | 18 (26.9%) | – |
| Urea (mg/dL) | 10‐40 | 30 [24‐40] | 16 (23.9%) | – |
| Creatinine (mg/dL) | 0.6‐1.2 | 0.90 [0.80‐1.00] | 8 (11.9%) | 3 (4.5%) |
| Aspartate aminotransferase (U/L) | 10‐40 | 32 [24‐45] | 25 (37.3%) | – |
| Alanine aminotransferase (U/L) | 10‐50 | 31 [19.5‐46] | 14 (20.9%) | – |
| Total bilirubin (mg/dL) | 0.2‐1.0 | 0.60 [0.45‐0.80] | 8 (11.9%) | – |
| Lactate dehydrogenase (mg/dL) | <250 | 268 [220‐364.5] | 43 (64.2%) | – |
| Amylase (U/L) | <90 | 61 [40.5‐77.5] | 7 (10.4%) | – |
| Creatine phosphokinase (U/L) | <170 | 99 [76.5‐165] | 17 (25.4%) | – |
| Uric acid (mg/dL) | 1.2‐6.0 | 4.9 [4.1‐5.9] | 12 (17.9%) | – |
| Troponin (ng/L) | <14 | 5 [2.7‐10.15] | 13 (19.4%) | – |
| Triglycerides (mg/dL) | <150 | 120.5 [92‐164.5] | 19 (28.4%) | – |
| Total cholesterol (mg/dL) | <200 | 145.5 [130.8‐174] | 7 (10.4%) | – |
| Sodium (mEq/L) | 135‐145 | 138 [135.5‐140] | – | 17 (25.4%) |
| Potassium (mEq/L) | 3.5‐5.5 | 4.1 ± 0.36 | – | 8 (11.9%) |
| Lactate (mmol/L) | <1.2 | 1.14 ± 0.36 | 23 (34.3%) | – |
| Inflammation‐related markers | ||||
| C‐reactive protein (mg/dL) | <0.3 | 6.55 [1.48‐12.64] | 61 (91.0%) | – |
| Procalcitonin (ng/mL) | <0.25 | 0.07 [0.04‐0.12] | 9 (13.4%) | – |
| Ferritin (ng/mL) | 12‐300 | 747 [309.3‐973.6] | 51 (76.1%) | – |
Data presented as mean ± standard deviation in normally distributed variables or otherwise as median [interquartile range].
Abbreviation: N/A, not applicable.
FIGURE 1.
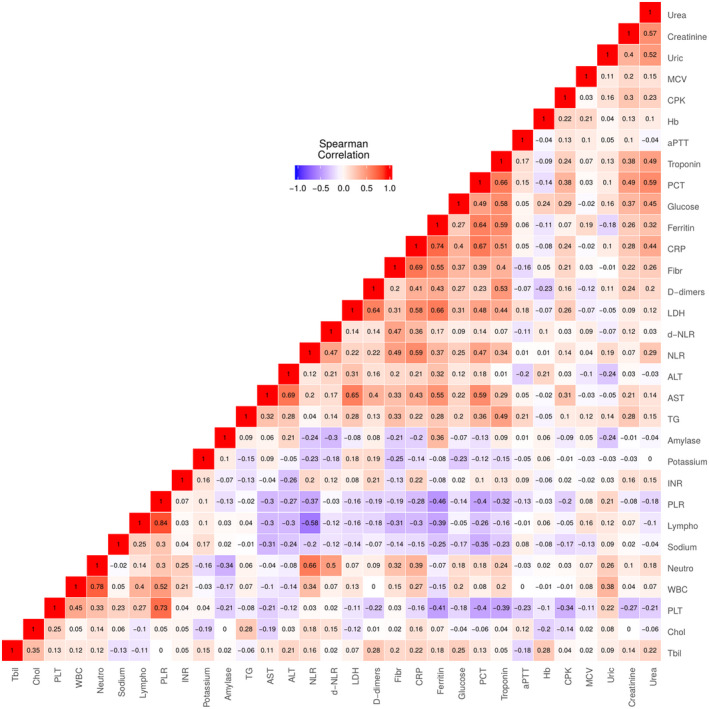
Correlation analysis of laboratory findings at admission
3.3. Risk score
Cross‐validation indicated an optimal value of α = 0.35 as it provided the lowest mean cross‐validation error at λ min = 0.34 (Figure S1). The elastic net model identified the following factors with nonzero coefficients for the construction of the risk model: female gender (β = −0.0086), hypertension (β = 0.0402), diabetes mellitus (β = 0.3269), fever at admission (β = 0.0458), shortness of breath at admission (β = 0.2060), serum C‐reactive protein (β = 0.0063), lactate dehydrogenase (β = 0.0008), aspartate aminotransferase (β = 0.0019), fibrinogen (β = 0.0005) and glucose (β = 0.0006) (Table 3). As a result, a risk score was calculated for each patient (median: 1.29, interquartile range: 0.94 to 1.61). The C‐index of the model was estimated to be 0.856 (standard error: 0.037). The risk score was significantly higher in patients that were subsequently admitted to ICU (P value < .0001), as well as to those developing ARDS (P value < .0001), SIRS (P value < .0001) and AKI (P value < .0001).
TABLE 3.
Risk model for the prediction of ICU admission
| Covariates | Regression coefficient (β) |
|---|---|
| Gender (0: male, 1: female) | −0.0086 |
| Hypertension (0: no, 1: yes) | 0.0402 |
| Diabetes mellitus (0: no, 1: yes) | 0.3269 |
| Fever at admission (0: no, 1: yes) | 0.0458 |
| Shortness of breath (0: no, 1: yes) | 0.2060 |
| Glucose (mg/dL) a | 0.0006 |
| C‐reactive protein (mg/dL) a | 0.0063 |
| Lactate dehydrogenase (mg/dL) a | 0.0008 |
| Aspartate aminotransferase (U/L) a | 0.0019 |
| Fibrinogen (mg/dL) a | 0.0005 |
per unit increase.
Subgrouping indicated that high‐risk score was significantly associated with ICU admission both in patients younger and older than 65 years (P value < .001), as well as in patients presenting both before and after the first week of symptoms (P value < .001). In addition, higher risk score was linked to ICU transfer both in patients with PaO2/FiO2 ≤ 200 mm Hg (P value < .05) and PaO2/FiO2 of 200‐300 mm Hg (P value < .01) at presentation, although no significant difference was observed for patients with initial PaO2/FiO2 > 300, as only one patient was subsequently admitted to ICU in this subgroup (Figure 2). The outcomes of the multivariate Cox regression model demonstrated that the association of risk score with disease progression remained significant after adjustment for age, day of symptom and PaO2/FiO2 ratio at admission (adjusted hazard ratio: 23.14, 95% confidence intervals: 2.43 to 220.37, P value: .006) (Table 4).
FIGURE 2.
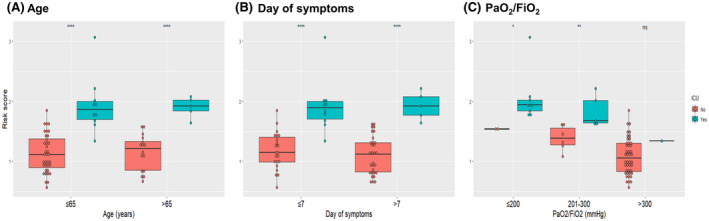
Boxplots of risk score of critically and noncritically ill patients categorised by age (A), day of symptoms (B) and oxygenation status (C). Asterisks denote the level of statistical significance
TABLE 4.
Outcomes of the multivariate Cox proportional hazard model
| Variable | Hazard ratio | 95% confidence intervals | P value |
|---|---|---|---|
| Age, years | |||
| ≤65 | reference | ||
| >65 | 1.46 | 0.39‐5.43 | .571 |
| Day of symptoms, days | |||
| ≤7 | reference | ||
| >7 days | 0.60 | 0.17‐2.13 | .428 |
| PaO2/FiO2, mm Hg | |||
| ≤200 | reference | ||
| 200‐300 | 0.49 | 0.15‐1.60 | .233 |
| >300 | 0.07 | 0.01‐0.66 | .021 |
| Risk score | |||
| ≤1.6 | reference | ||
| 1.6 | 23.14 | 2.43‐220.37 | .006 |
Bold text indicates statistical significance (P value < .05).
The AUC of the risk score for the prediction of ICU admission was calculated to be 97.1%, while it was estimated to provide a sensitivity of 92.3% and specificity of 93.3% at the threshold of 1.6. The diagnostic accuracy of the risk score was calculated to be higher than those of CURB‐65 (AUC: 86.6%), CRB‐65 (AUC: 86.3%) and PSI/PORT (AUC: 80.2%) (Figure 3). The Kaplan‐Meier survival curves stratified by the results of four clinical scores are depicted in Figure 4. Specifically, time‐to‐event analysis indicated that progression to critical‐illness was significantly higher in patients with risk score > 1.6 (P value < .0001), CURB‐65 ≥ 2 (P value < .0001), CRB‐65 ≥ 2 (P value: .00086) and PSI/PORT ≥ 90 (P value: .0018). Moreover, the risk score presented high diagnostic accuracy for the prediction of ARDS (AUC: 95.4%), SIRS (90.9%) and AKI (75.4%) (Figure S2), as well as significant correlation with worse SOFA (ρ = 0.52, P value < .001), APACHE II (ρ = 0.50, P value < .001) and MEWS score (ρ = 0.58, P value < .001) (Figures S3‐S5).
FIGURE 3.
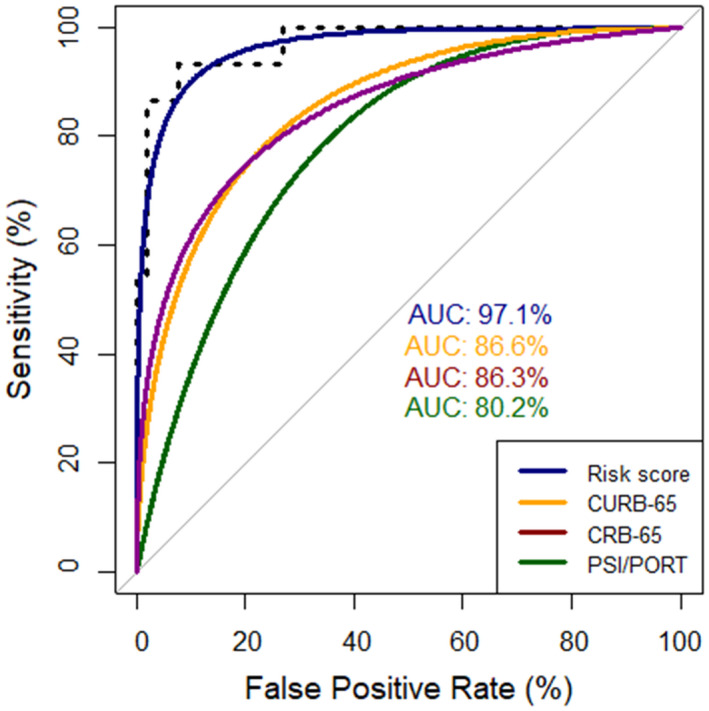
Receiver operating characteristic (ROC) curves of risk score, CURB‐65, CRB‐65 and PSI/PORT for the prediction of intensive care unit admission. AUC, area under the curve
FIGURE 4.
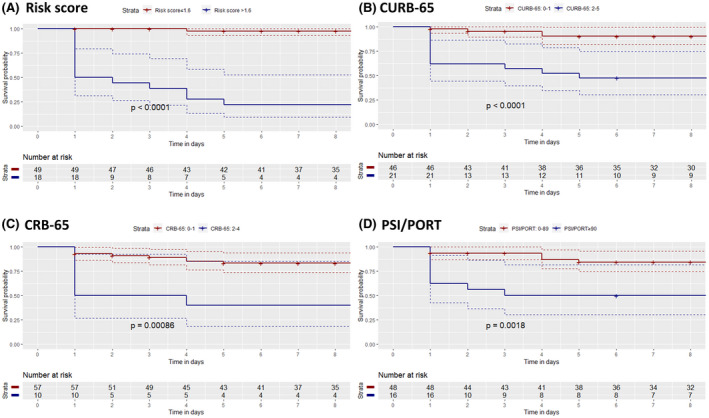
Kaplan‐Meier curves of time until intensive care unit admission for patients stratified by their risk score (A), CURB‐65 (B), CRB‐65 (C) and PSI/PORT score (D)
4. DISCUSSION
The present prospective study included a total of 67 patients, with 22.4% of them developing critical illness requiring intubation and mechanical ventilation. A risk score was developed aiming to optimise variable selection and minimise the risk of overfitting. Specifically, a model was constructed by taking into account the effects of gender, hypertension, diabetes mellitus, fever and shortness of breath at admission, initial serum glucose, lactate dehydrogenase, aspartate aminotransferase, C‐reactive protein and fibrinogen. The proposed risk model proved in our study population to be extremely accurate in the prediction of ICU admission, outperforming the conventional pneumonia severity scores, CURB‐65, CRB‐65 and PSI/PORT. Moreover, it was able to accurately predict disease complications, such as ARDS and SIRS and positively correlated with established scores of critical illness, especially SOFA, APACHE II and MEWS scores.
The findings of the present study are in accordance with other prediction models concerning the negative prognostic value of male gender, hypertension, elevated glucose, C‐reactive protein and lactate dehydrogenase. 27 , 28 , 29 Importantly, raised lactate dehydrogenase has been also recognised as a severity marker in patients with H1N1 30 and MERS‐CoV (Middle East respiratory syndrome coronavirus) infection, 31 reflecting subclinical tissue damage and activation of the cytokine cascade. Increased fibrinogen levels in patients at risk of critical illness may be explained by the COVID‐19‐induced coagulopathy, as well as by the hyperactivation of pro‐inflammatory pathways since recent studies have demonstrated a significant correlation of its values with interleukin‐6 levels. 32 Moreover, no prognostic role was proposed for lymphopenia, as suggested by previous reports 33 ; although neutrophil‐to‐lymphocyte‐ratio of the sample was estimated to be high (median: 3.20, interquartile range: 2.43‐4.91), no significant difference was observed amongst patients developing mild and severe disease.
Inspection of the Kaplan‐Meier curves indicated that most events (ie, transfer to ICU) occurred within the first 5 days of hospital stay, implying that patients tended to present at a late stage of the disease. For this reason, subgroup analysis was performed based on the day form the onset of symptoms and the presenting PaO2/FiO2 ratio, obtaining stable results and thus strengthening the generalisability of outcomes. Therefore, the proposed risk score may be applicable at the time of admission for all COVID‐19 patients requiring hospitalisation, allowing the early identification of those at high risk of disease progression and development of critical illness. Moreover, it should be noted that the internal validation of the model was performed by implementing cross‐validation techniques in order to account for potential optimism during variable selection and thus limit the overall risk of bias. 34
Nevertheless, the study presents several shortcomings. Specifically, the interpretation of outcomes is mainly limited by the available sample size, as well as by the lack of external validation. The study was a single‐centre one and thus generalisability to populations of other countries cannot be ascertained. In addition, the number of deaths in the present cohort was small; hence, mortality could not be assessed as an outcome of interest. The potential effects of different treatment options could also not be assessed since the majority of patients initially received similar therapeutic regimens, consisting mainly of azithromycin and hydroxychloroquine. Moreover, the present study adopted a prospective design with pre‐specified variables and end‐points, minimising thus the risk of selection bias. A variety of clinical and laboratory parameters was evaluated, allowing a comprehensive analysis of the potential prognostic factors amongst COVID‐19 patients. In addition, the selection of variables for the construction of the risk model was optimised by applying regularisation techniques, since standard regression methods may not converge in the setting of small sample size and a high dimensional dataset. 35
Several research questions need to be addressed by future studies in the field. Additional validation cohorts should evaluate the proposed risk model in order to test its efficacy in different populations and compare it to other published COVID‐19 prognostic scores. Clinical and laboratory findings may be combined with radiological features in order to construct models with improved discrimination. The potential value of viral load for the prediction of disease progression should be assessed by studies implementing droplet digital PCR allowing absolute quantification of SARS‐CoV‐2. 36 Finally, it is important to state that prediction models may serve as useful tools in the guidance of clinical practice regarding closer monitoring and treatment decisions, as well as during the selection of appropriate populations for inclusion in future clinical trials. In this context, their implementation may allow a precision medicine approach aiming to offer personalised therapeutic strategies depending on the accurate phenotypic recognition of COVID‐19 patients. 37
5. CONCLUSIONS
The present study developed a novel risk score based on clinical and laboratory characteristics in order to predict the occurrence of critical illness amongst patients hospitalised because of SARS‐CoV‐2 infection. A10‐variable model was constructed, achieving optimal discrimination of patients that were subsequently admitted to the intensive care unit. Further large‐scale studies are needed to validate the efficacy of the proposed risk factors and test whether early risk stratification is able to guide therapeutic decisions and optimise the clinical management of COVID‐19 patients.
DISCLOSURE
None declared.
AUTHOR CONTRIBUTIONS
Conception and design: I. Bellos, P. Lourida, A. Argyraki, E. Korompok
Analysis and interpretation of the data: I. Bellos, P. Lourida, E. Korompoki
Drafting of the article: I. Bellos, P. Lourida, E. Korompoki, C. Zirou, I. Kokkinaki
Critical revision for important intellectual content: A. Argyraki, A. Pefanis
Final approval of the article: I. Bellos, P. Lourida, A. Argyraki, E. Korompoki, C. Zirou, I. Kokkinaki, A. Pefanis
Statistical expertise: I. Bellos, E. Korompoki
Collection and assembly of data: I. Bellos, C. Zirou, I. Kokkinaki.
Funding information
This study was carried out as part of our routine work
Supporting information
Supplementary Material
Bellos I, Lourida P, Argyraki A, et al. Development of a novel risk score for the prediction of critical illness amongst COVID‐19 patients. Int J Clin Pract.2021;75:e13915. 10.1111/ijcp.13915
REFERENCES
- 1. Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A Novel coronavirus emerging in China—key questions for impact assessment. N Engl J Med. 2020;382:692‐694. 10.1056/NEJMp2000929 [DOI] [PubMed] [Google Scholar]
- 2. Yuki K, Fujiogi M, Koutsogiannaki S. COVID‐19 pathophysiology: a review. Clin Immunol. 2020;215:108427. 10.1016/J.CLIM.2020.108427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cao W, Li T. COVID‐19: towards understanding of pathogenesis. Cell Res. 2020;30:367‐369. 10.1038/s41422-020-0327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becker RC. COVID‐19 update: Covid‐19‐associated coagulopathy. J Thromb Thrombolysis. 2020;50:54‐67. 10.1007/s11239-020-02134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang JJY, Lee KS, Ang LW, Leo YS, Young BE. Risk factors of severe disease and efficacy of treatment in patients infected with COVID‐19: a systematic review, meta‐analysis and meta‐regression analysis. Clin Infect Dis. 2020;71:2199–2206. 10.1093/cid/ciaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid‐19: systematic review and critical appraisal. BMJ. 2020;369:1–24. 10.1136/BMJ.M1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Ann Intern Med. 2015;162:55. 10.7326/M14-0697 [DOI] [PubMed] [Google Scholar]
- 8. Capone M, Giannarelli D, Mallardo D, et al. Baseline neutrophil‐to‐lymphocyte ratio (NLR) and derived NLR could predict overall survival in patients with advanced melanoma treated with nivolumab. J Immunother Cancer. 2018;6:74. 10.1186/s40425-018-0383-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim W, van der Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377. 10.1136/THORAX.58.5.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McNally M, Curtain J, O’Brien KK, Dimitrov BD, Fahey T. Validity of British Thoracic Society guidance (the CRB‐65 rule) for predicting the severity of pneumonia in general practice: systematic review and meta‐analysis. Br J Gen Pract. 2010;60:e423‐e433. 10.3399/BJGP10X532422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fine MJ, Auble TE, Yealy DM, et al. A prediction rule to identify low‐risk patients with community‐acquired pneumonia. N Engl J Med. 1997;336:243‐250. 10.1056/NEJM199701233360402 [DOI] [PubMed] [Google Scholar]
- 12. The ARDS Definition Task Force . Acute respiratory distress syndrome. JAMA. 2012;307:2526‐2533. 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- 13. Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644‐1655. 10.1378/CHEST.101.6.1644 [DOI] [PubMed] [Google Scholar]
- 14. Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron. 2012;120:c179‐c184. 10.1159/000339789 [DOI] [PubMed] [Google Scholar]
- 15. Vincent J‐L, Moreno R, Takala J, et al. The SOFA (Sepsis‐related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis‐Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707‐710. 10.1007/BF01709751 [DOI] [PubMed] [Google Scholar]
- 16. Knaus W, Draper E, Wagner D, Zimmerman J. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818‐829. http://europepmc.org/article/MED/3928249. Accessed June 5, 2020. [PubMed] [Google Scholar]
- 17. Subbe CP, Kruger M, Rutherford P, Gemmel L. Validation of a modified early warning score in medical admissions. QJM. 2001;94(10):521‐526. 10.1093/qjmed/94.10.521 [DOI] [PubMed] [Google Scholar]
- 18. Therneau T. A Package for Survival Analysis in R. 2020. https://cran.r‐project.org/package=survival. Accessed May 10, 2020.
- 19. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1‐22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robin X, Turck N, Hainard A, et al. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mishra P, Pandey CM, Singh U, Gupta A, Sahu C, Keshri A. Descriptive statistics and normality tests for statistical data. Ann Card Anaesth. 2019;22:67‐72. 10.4103/aca.ACA_157_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akoglu H. User’s guide to correlation coefficients. Turkish J Emerg Med. 2018;18:91. 10.1016/J.TJEM.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Z. Introduction to machine learning: k‐nearest neighbors. Ann Transl Med. 2016;4:218. 10.21037/ATM.2016.03.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J, Liang G, Siegmund KD, Lewinger JP. Data integration by multi‐tuning parameter elastic net regression. BMC Bioinformatics. 2018;19:1–9. 10.1186/S12859-018-2401-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32‐35. [DOI] [PubMed] [Google Scholar]
- 26. Lansbury L, Lim B, Baskaran V, Lim WS. Co‐infections in people with COVID‐19: a systematic review and meta‐analysis. J Infect. 2020. May;81:266‐275. 10.1016/j.jinf.2020.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ji D, Zhang D, Xu J, et al. Prediction for progression risk in patients with COVID‐19 pneumonia: the CALL score. Clin Infect Dis. 2020. April;71:1393‐1399. 10.1093/cid/ciaa414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang D, Wang T, Chen Z, Yang H, Yao R, Liang Z. A novel risk score to predict diagnosis with Coronavirus Disease 2019 (COVID‐19) in suspected patients: a retrospective, multi‐center, observational study. J Med Virol. 2020. June:jmv.26143. 10.1002/jmv.26143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galloway JB, Norton S, Barker RD, et al. A clinical risk score to identify patients with COVID‐19 at high risk of critical care admission or death: an observational cohort study. J Infect. 2020;81:282‐288. 10.1016/j.jinf.2020.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xi X, Xu Y, Jiang L, Li A, Duan J, Du B. Hospitalized adult patients with 2009 influenza A(H1N1) in Beijing, China: risk factors for hospital mortality. BMC Infect Dis. 2010;10:256. 10.1186/1471-2334-10-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Assiri A, Al‐Tawfiq JA, Al‐Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752‐761. 10.1016/S1473-3099(13)70204-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ranucci M, Ballotta A, Di Dedda U, et al. The procoagulant pattern of patients with COVID‐19 acute respiratory distress syndrome. J Thromb Haemost. 2020;18:1747‐1751. 10.1111/jth.14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients With COVID‐19. JAMA Int Med. 2020;180:1081. 10.1001/jamainternmed.2020.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moons KGM, Wolff RF, Riley RD, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170:W1. 10.7326/M18-1377 [DOI] [PubMed] [Google Scholar]
- 35. Finch W, Finch M. Regularization methods for fitting linear models with small sample sizes: fitting the Lasso estimator using R. Pract Assessment Res Eval. 2019;21:1–13. 10.7275/jr3d-cq04 [DOI] [Google Scholar]
- 36. Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS‐CoV‐2 in infected patients. Clin Infect Dis. 2020;71:793‐798. 10.1093/cid/ciaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crisci CD, Ardusso LRF, Mossuz A, Müller L. A precision medicine approach to SARS‐CoV‐2 pandemic management. Curr Treat Options Allergy. 2020;7:422‐440. 10.1007/s40521-020-00258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


