Abstract
Thalidomide causes teratogenic effects by inducing protein degradation via cereblon (CRBN)‐containing ubiquitin ligase and modification of its substrate specificity. Human P450 cytochromes convert thalidomide into two monohydroxylated metabolites that are considered to contribute to thalidomide effects, through mechanisms that remain unclear. Here, we report that promyelocytic leukaemia zinc finger (PLZF)/ZBTB16 is a CRBN target protein whose degradation is involved in thalidomide‐ and 5‐hydroxythalidomide‐induced teratogenicity. Using a human transcription factor protein array produced in a wheat cell‐free protein synthesis system, PLZF was identified as a thalidomide‐dependent CRBN substrate. PLZF is degraded by the ubiquitin ligase CRL4CRBN in complex with thalidomide, its derivatives or 5‐hydroxythalidomide in a manner dependent on the conserved first and third zinc finger domains of PLZF. Surprisingly, thalidomide and 5‐hydroxythalidomide confer distinctly different substrate specificities to mouse and chicken CRBN, and both compounds cause teratogenic phenotypes in chicken embryos. Consistently, knockdown of Plzf induces short bone formation in chicken limbs. Most importantly, degradation of PLZF protein, but not of the known thalidomide‐dependent CRBN substrate SALL4, was induced by thalidomide or 5‐hydroxythalidomide treatment in chicken embryos. Furthermore, PLZF overexpression partially rescued the thalidomide‐induced phenotypes. Our findings implicate PLZF as an important thalidomide‐induced CRBN neosubstrate involved in thalidomide teratogenicity.
Keywords: CRBN, protein degradation, thalidomide metabolite, thalidomide teratogenicity, ubiquitin
Subject Categories: Development & Differentiation; Chemical Biology; Post-translational Modifications, Proteolysis & Proteomics
The vertebrate transcription factor PLZF/ZBTB16 emerges as potential key mediator of limb defects caused by CRBN ubiquitin ligase modulators, providing insight into the teratogenic contributions of thalidomide metabolites.
Introduction
In many countries, thalidomide (Fig 1A) was popular in the 1960s as a sedative/hypnotic for pregnant women. Its use caused embryopathies, such as limb defects, ear damage and congenital heart diseases (Smithells & Newman, 1992; Miller & Strömland, 1999). Thalidomide binds to the cereblon (CRBN) protein within the CRL4 E3 ubiquitin ligase complex. CRBN is a key molecule for thalidomide‐induced teratogenesis (Ito et al, 2010). Recently, many studies have reported that the interaction of CRBN with thalidomide or its derivatives, which include lenalidomide, pomalidomide (Fig 1A) and CC‐885, alters the binding specificity of CRBN to proteins. This alteration induces the ubiquitination and degradation of binding proteins, such as Ikaros (IKZF1) (Krönke et al, 2014; Lu et al, 2014), casein kinase I (Krönke et al, 2015), GSPT1 (Matyskiela et al, 2016) and SALL4 (Donovan et al, 2018; Matyskiela et al, 2018). In humans, mutations of SALL4 are found in Duane‐radial ray syndrome (DRRS, Okihiro syndrome). The phenotypic features of DRRS include limb deformities (Kohlhase et al, 2005). Hind limb defects have also been reported in Sall4‐conditional knockout (Sall4‐CKO) mice (Akiyama et al, 2015). These findings suggest that SALL4 is partially involved in teratogenesis. Thalidomide embryopathy has been demonstrated in chickens and zebrafish (Therapontos et al, 2009; Ito et al, 2010; Mahony et al, 2013). However, the use of zebrafish as a model system to study limb teratogenicity is controversial (Mahony et al, 2013). The sequence of thalidomide‐binding sites in these SALL4 proteins differs between humans and these animals (Donovan et al, 2018), suggesting the possibility of other target proteins for teratogenesis.
Figure 1. Identification of thalidomide‐dependent interactors of CRBN using a cell‐free‐based human TF protein array.
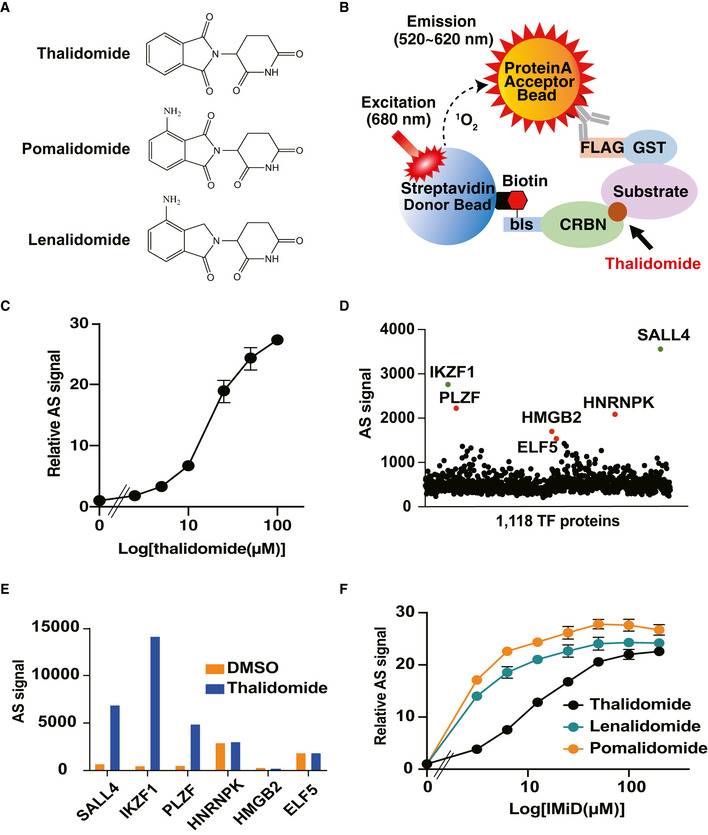
- Chemical structures of thalidomide, pomalidomide and lenalidomide.
- Schematic diagram of the thalidomide‐dependent in vitro binding assay between CRBN and substrates using AlphaScreen technology.
- Detection of luminescent signals of thalidomide‐dependent interactions between bls‐CRBN and FLAG‐GST‐IKZF1. Dose‐dependent signals (DMSO, 2.5, 5, 10, 25, 50 or 100 µM thalidomide) were analysed with an in vitro binding assay using the AlphaScreen technology.
- Results of in vitro high‐throughput screening targeting 1,118 human transcription factors. Green and red spots denote known neosubstrates and candidate clones, respectively.
- Confirmation of thalidomide dependency on six hit proteins using an in vitro binding assay. Interaction between bls‐CRBN and FLAG‐GST‐protein in the presence of DMSO or 50 µM thalidomide was detected using the AlphaScreen technology.
- In vitro binding assay for thalidomide, pomalidomide and lenalidomide. Interaction between bls‐CRBN and FLAG‐GST‐PLZF in the presence of (DMSO, 3.125, 6.25, 12.5, 25, 50, 100 or 200 µM) thalidomide, pomalidomide or lenalidomide was analysed using the AlphaScreen technology.
Data information: All relative AlphaScreen (AS) signals are expressed as relative luminescent signal with luminescent signal of DMSO as one. Error bars denote ± standard deviation (n = 3).
Promyelocytic leukaemia zinc finger (PLZF), also known as ZBTB16 or ZFP145, is a transcription factor (TF) with nine C2H2‐type zinc finger domains (ZNFs) (Suliman et al, 2012). It is involved in a broad range of developmental and biological processes, such as haematopoiesis, limb skeletal formation, spermatogenesis and immune regulation (Barna et al, 2000, 2005; Suliman et al, 2012). Loss of PLZF function in both human patients and mutant mice results in limb defects, which appear as an elongation defect of the zeugopod and thumb (Barna et al, 2000, 2005; Fischer et al, 2008). A recent study indicated that the 6th and 7th ZNFs in PLZF are not targets of CRBN (Sievers et al, 2018). However, it remains unknown whether the full‐length PLZF protein is a target for the binding of CRBN to thalidomide.
Thalidomide is metabolised by several isoforms of human cytochrome P450 (CYPs) into 5‐hydroxythalidomide and 5′‐hydroxythalidomide (Yamamoto et al, 2009; Chowdhury et al, 2010), involving oxidation of the phthalimido and glutarimide ring, respectively. CRBN mainly recognises the glutarimide ring in thalidomide (Fischer et al, 2014). Thus, the 5′‐hydroxythalidomide metabolite does not bind to CRBN and hence may not contribute to teratogenesis (Therapontos et al, 2009). Mouse embryos that express placental human CYP3A isoforms exhibited modest thalidomide‐induced limb defects in embryo culture compared to wild‐type (WT) controls, suggesting that placental CYP3A isoforms may convert thalidomide to an embryopathic reactive intermediate via the formation of 5‐hydroxythalidomide, although no limb defects were observed in vivo (Kazuki et al, 2016). However, there is no evidence that 5‐hydroxythalidomide is involved in CRBN‐dependent thalidomide teratogenicity.
We aimed to identify the protein required for the binding of thalidomide to CRBN. We constructed a human TF protein array (HuTFPA) consisting of 1,118 human recombinant proteins including mainly TFs and zinc finger proteins using a wheat cell‐free protein production system. Biochemical screening based on the interaction between TF and CRBN with thalidomide using the AlphaScreen system identified PLZF as a participant in the interaction between CRBN with thalidomide. PLZF was identified as a novel substrate of the CRL4CRBN E3 ubiquitin ligase complex with thalidomide, its derivatives and 5‐hydroxythalidomide. The amino acid sequences of PLZF were very similar among vertebrate species. In chick embryos, knockdown of Plzf/Zbtb16 induced abnormal limb development. More importantly, PLZF protein was decreased in the limb buds during thalidomide‐ and 5‐hydroxythalidomide‐induced teratogenicity, whereas the level of SALL4 protein did not change. PLZF overexpression in the chicken limb bud partially rescued the thalidomide‐induced phenotype, including reduction of Fgf10/Fgf8 expression.
Results
Screening for thalidomide‐dependent substrates of CRBN using a human TF protein array
Many TFs function as master regulators during the development and differentiation of embryos. In addition, many substrates of CRBN with thalidomide are ZNF‐type TFs. These include IKZF1 (Krönke et al, 2014; Lu et al, 2014), IKZF3 (Krönke et al, 2014; Lu et al, 2014) and SALL4 (Donovan et al, 2018; Matyskiela et al, 2018). We aimed to identify new substrates of CRBN with thalidomide from human TFs. Based on a wheat cell‐free protein synthesis system (Sawasaki et al, 2002), we previously developed a technology for the construction of a protein array that synthesises an individual protein in each well of a 96‐ or 384‐well plate. So far, we have identified substrate proteins from protein kinase (Tadokoro et al, 2010) and E3 ligase (Takahashi et al, 2016) protein arrays. The combination of our protein array and AlphaScreen technology is advantageous for the screening of protein–protein interactions. It can be used directly without protein purification, is sensitive and can be used as a high‐throughput system. A human TF protein array (HuTFPA) consisting mainly of human TFs (Table EV1), synthesised as N‐terminal FLAG‐glutathione S‐transferase (GST) fusions, produced by a wheat cell‐free system. CRBN was synthesised as an N‐terminal single biotin‐labelled form using the same system. The detection principle for this biochemical interaction is shown in Fig 1B. Using this cell‐free system, an interaction between FLAG‐GST‐IKZF1 and biotinylated CRBN was detected in a dose‐dependent manner using the AlphaScreen method (Fig 1C). As shown in the flowchart (Fig EV1A), screening of the substrate human TFs of CRBN with thalidomide (50 µM) on HuTFPA identified six TF proteins as CRBN binding proteins in the presence of thalidomide (Fig 1D). In contrast, several known substrates, including IKZF3 and CK1α, were not detected because these substrates were not included in the HuTFPA (Table EV1).
Figure EV1. Flowchart of in vitro high‐throughput screening and validation assay of candidate clones.
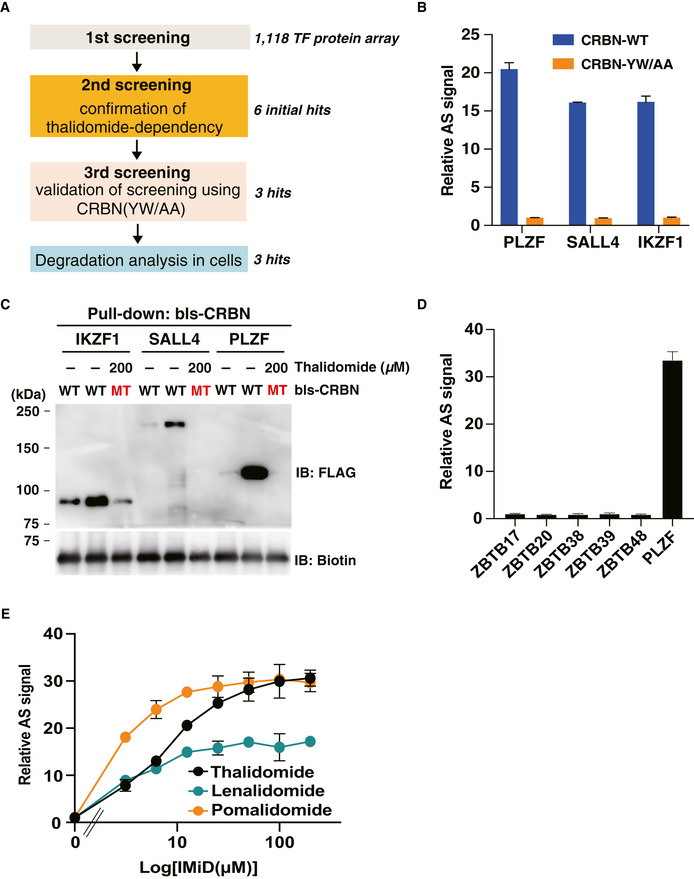
- Flowchart of in vitro high‐throughput screening.
- Validation of screening using CRBN mutant. Interaction between FLAG‐GST‐SALL4, FLAG‐PLZF or IKZF1 and bls‐CRB‐WT or bls‐CRBN‐YW/AA in the presence of DMSO or 50 µM thalidomide was analysed by in vitro binding assay using the AlphaScreen technology.
- In vitro binding assay using pull‐down and immunoblot analysis. bls‐CRBN‐WT or bls‐CRBN‐YW/AA was used as bait protein, and thalidomide‐dependent interactions between bls‐CRBN and FLAG‐GST‐SALL4, PLZF and IKZF were confirmed by immunoblot analysis after streptavidin pull‐down.
- In vitro binding assay for ZBTB family proteins using the AlphaScreen technology. FLAG‐GST‐ZBTB proteins (ZBTB17, ZBTB20, ZBTB38, ZBTB39, ZBTB48 and PLZF) were evaluated for thalidomide‐dependent interactions with bls‐CRBN by same procedures indicated in Fig EV1B.
- In vitro binding assay for thalidomide, pomalidomide and lenalidomide. Interaction between bls‐CRBN and FLAG‐GST‐SALL4 in the presence of (DMSO, 3.125, 6.25, 12.5, 25, 50, 100 or 200 µM) thalidomide, pomalidomide or lenalidomide was analysed using the AlphaScreen technology.
Data information: All relative AS (AlphaScreen) signals were expressed as relative luminescent signal with luminescent signal of DMSO as one. The error bars denote ± standard deviation (n = 3).
Source data are available online for this figure.
To investigate the thalidomide dependency of these proteins for CRBN binding, a biochemical assay was carried out with and without thalidomide. Three human TFs (IKZF1, SALL4 and PLZF) displayed the characteristics of thalidomide‐dependent binding to CRBN (Fig 1E), whereas HNRNPK, HMGB2 and ELF5 bound to CRBN in the absence of thalidomide. Furthermore, in the presence of thalidomide, these proteins did not bind to mutant CRBN‐YW/AA (Ito et al, 2010), which was unable to bind to thalidomide (Fig EV1B). Using recombinant proteins, an in vitro pull‐down assay confirmed the binding of PLZF to WT CRBN in the presence of thalidomide (Fig EV1C), as also observed for IKZF1 and SALL4. This binding was not observed using mutant CRBN‐YW/AA (MT) or in the absence of thalidomide (–). These results indicated the interaction of PLZF in the binding between CRBN and thalidomide.
PLZF binds to CRBN with thalidomide, pomalidomide and lenalidomide
The screening identified PLZF as a candidate substrate for CRBN with thalidomide. PLZF is a member of the zinc finger and bric à brac, tramtrack, and broad (ZBTB) protein family (Suliman et al, 2012). Since the other ZBTB proteins are included in the HuTFPA, these were analysed for interactions in the absence and presence of thalidomide. ZBTB proteins did not bind to CRBN with thalidomide (Fig EV1D), suggesting that CRBN with thalidomide recognises one or more specific region(s) in PLZF, but not a region common to the ZBTB family.
During the last two decades, two thalidomide derivatives, lenalidomide and pomalidomide (Fig 1A), have been developed for multiple myeloma or as immunomodulatory drugs (IMiDs) (Hideshima et al, 2000). Recent studies have reported that some proteins have different preferences between thalidomide and its derivatives for interaction with CRL4CRBN and/or protein degradation by CRL4CRBN (Krönke et al, 2015; Matyskiela et al, 2016). For example, the degradation of CK1α protein by CRL4CRBN was reportedly enabled by lenalidomide, but not by thalidomide or pomalidomide (Krönke et al, 2015), although the CK1α‐CRBN interaction was observed with thalidomide, lenalidomide and pomalidomide (Petzold et al, 2016). Therefore, we investigated the biochemical characteristics of the interactions between PLZF and CRBN with thalidomide and its two derivatives. Thalidomide, pomalidomide and lenalidomide induced a PLZF‐CRBN interaction, with a differential biochemical binding potency of pomalidomide > lenalidomide>thalidomide (Fig 1F). In a previous report (Donovan et al, 2018), time‐resolved fluorescence energy transfer (TR‐FRET) and cell‐based analyses showed that thalidomide and its two derivatives induced interaction of SALL4‐CRBN and protein degradation of SALL4. Consistent with a previous report, an in vitro binding assay using the AlphaScreen method confirmed that the order of binding potency of SALL4 was pomalidomide>thalidomide>lenalidomide (Fig EV1E). The similar binding potency of PLZF‐thalidomide and SALL4‐lenalidomide supported the prediction that PLZF and SALL4 are pan‐substrates for CRBN bound by thalidomide or either of its two derivatives.
PLZF is degraded by the CRL4CRBN complex with thalidomide and its derivatives
CRBN consists of a CRL4CRBN E3 ubiquitin ligase complex that includes DDB1, RBX1 and CUL4 (Ito et al, 2010). Therefore, the PLZF‐CRBN interaction with thalidomide and its two derivatives was expected to lead to the degradation of PLZF. For a cell‐based immunoblotting assay, we used the sensitive AGIA‐tag system on a rabbit monoclonal antibody (Yano et al, 2016). To investigate the stability of PLZF and SALL4, AGIA‐tagged PLZF or SALL4 was used to transfect HEK293T cells treated with thalidomide, pomalidomide or lenalidomide. All three compounds decreased the stability of PLZF and SALL4 (Fig EV2A). In addition, to explore whether PLZF is also a pan‐substrate like SALL4, endogenous PLZF or SALL4 protein levels were examined in HuH7 or HEK293T cells treated with thalidomide and its two derivatives. Immunoblot analyses revealed destabilisation of endogenous PLZF by thalidomide and the derivatives in both cell lines (Fig EV2B and C). The findings indicated that PLZF was also a pan‐substrate for CRBN bound by thalidomide or one of its two derivatives. To reduce experimental complexity, we focussed on thalidomide and lenalidomide in further analyses. A remarkable decrease in PLZF was observed 6 h after lenalidomide treatment (Fig EV2D). Degradation of IKZF1, IKZF3 and SALL4 was observed 3 h after the treatment (Krönke et al, 2014; Lu et al, 2014; Donovan et al, 2018). This time course analysis suggested the late reduction of PLZF compared to other CRBN substrates. The lenalidomide‐dependent loss of PLZF was completely inhibited by the proteasomal inhibitor MG132 and the NEDD8 inhibitor MLN4924 (Fig 2A). These findings suggested that PLZF is ubiquitinated by the CRL4 complex for degradation by the proteasome.
Figure EV2. Degradation of PLZF in IMiD‐treated cells.
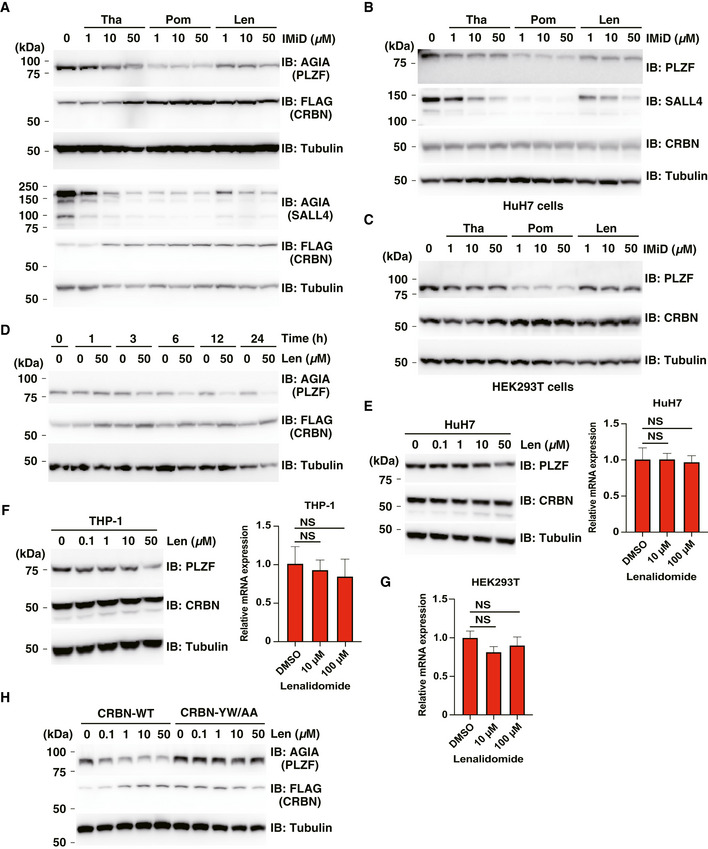
- Immunoblot analysis of AGIA‐PLZF or AGIA‐SALL4 protein levels in FLAG‐CRBN expressing HEK293T cells treated with DMSO, thalidomide (Tha), pomalidomide (Pom) or lenalidomide (Len) for 16 h.
- Immunoblot analysis of endogenous PLZF or SALL4 protein levels in HuH7 cells treated with DMSO, thalidomide (Tha), pomalidomide (Pom) or lenalidomide (Len) for 24 h.
- Immunoblot analysis of endogenous PLZF protein levels in HEK293T cells treated with DMSO, thalidomide (Tha), pomalidomide (Pom) or lenalidomide (Len) for 24 h.
- Time course of DMSO or lenalidomide (Len) treatment in AGIA‐PLZF and FLAG‐CRBN expressing HEK293T cells. AGIA‐PLZF protein levels were detected by immunoblot analysis.
- HuH7 cells were treated with the indicated concentrations of lenalidomide (Len) for 24 h. PLZF protein levels were analysed by immunoblot, and PLZF mRNA expression levels were measured by quantitative RT–PCR.
- THP‐1 cells were treated with the indicated concentrations of lenalidomide (Len) for 24 h. PLZF protein levels were analysed by immunoblot, and PLZF mRNA expression levels were measured by quantitative RT–PCR.
- HEK293T cells were treated with the indicated concentrations of lenalidomide (Len) for 24 h, and PLZF mRNA expression levels were measured by quantitative RT–PCR.
- Immunoblot analysis of AGIA‐PLZF protein levels in FLAG‐CRBN‐WT or FLAG‐CRBN‐YW/AA expressing CRBN−/− HEK293T cells treated with DMSO or lenalidomide (Len) for 16 h.
Data information: Relative mRNA expression used the expression level with DMSO treatment as one. Error bars denote ± standard deviation (n = 3), and P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (NS = not significant).
Source data are available online for this figure.
Figure 2. PLZF is a substrate of CRL4CRBN with thalidomide and lenalidomide for E3 ubiquitin ligase.
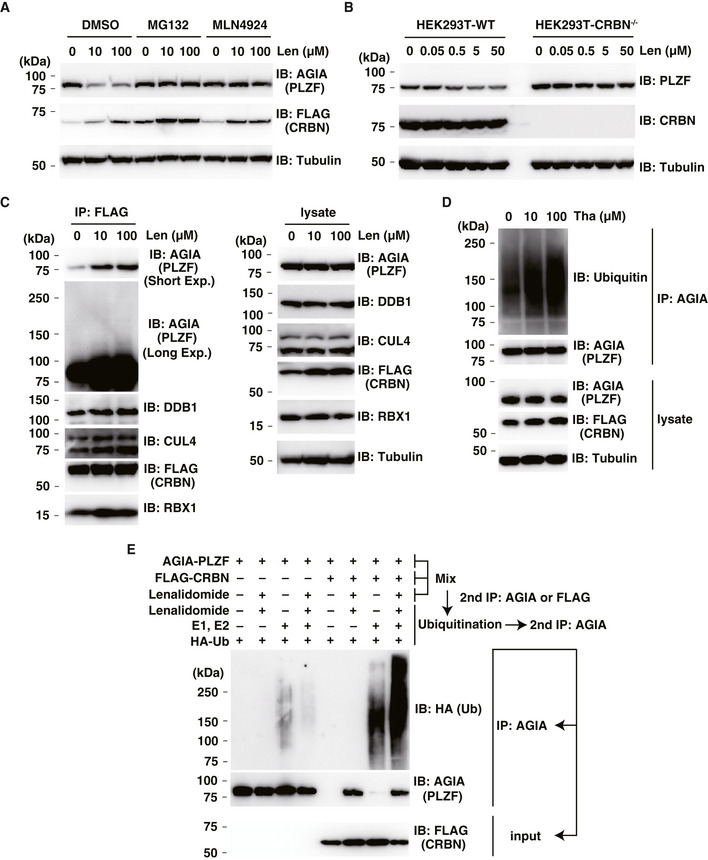
- Immunoblot analysis of AGIA‐PLZF protein levels in AGIA‐PLZF and FLAG‐CRBN expressing HEK293T cells treated with DMSO or lenalidomide (Len) in the presence of DMSO, MG132 or MLN4924 for 9 h.
- Immunoblot analysis of endogenous PLZF protein levels in HEK293T cells or CRBN−/− HEK293T cells treated with DMSO or lenalidomide (Len) for 24 h.
- Immunoprecipitation of FLAG‐CRBN in FLAG‐CRBN and AGIA‐PLZF expressing HEK293T cells treated with DMSO or lenalidomide (Len) in the presence of MG132 for 8 h. Components of CRL4FLAG‐CRBN and AGIA‐PLZF were detected using each specific antibody, as indicated.
- Ubiquitination of AGIA‐PLZF in AGIA‐PLZF and FLAG‐CRBN expressing CRBN−/− HEK293T cells treated with DMSO or thalidomide (Tha) in the presence of MG132 for 10 h. AGIA‐PLZF was immunoprecipitated using anti‐AGIA antibody, and the polyubiquitin chain on AGIA‐PLZF was analysed by immunoblot.
- In vitro binding and ubiquitination assay of AGIA‐PLZF. Empty vector, AGIA‐PLZF or FLAG‐CRBN expressing HEK293T cells were lysed and the lysates were mixed. The first immunoprecipitation with anti‐AGIA or anti‐FLAG antibodies was performed in the presence of DMSO or 200 µM lenalidomide. The purified AGIA‐PLZF or CRL4FLAG‐CRBN complex, including AGIA‐PLZF and FLAG‐CRBN, was incubated with recombinant E1, E2 and HA‐ubiquitin in the presence of DMSO or 200 µM lenalidomide. The second immunoprecipitation was performed using anti‐AGIA antibody. Ubiquitination of PLZF was analysed by immunoblot.
To investigate whether the loss of PLZF was dependent on CRBN, we generated CRBN‐deficient HEK293T cells using CRISPR/Cas9 (Yamanaka et al, 2020). In the presence of lenalidomide, degradation of endogenous PLZF was observed in normal HEK293T cells, whereas endogenous PLZF was not degraded in CRBN‐deficient cells (Fig 2B). Endogenous PLZF protein was also reduced by lenalidomide in HuH7 and THP‐1 cells (Fig EV2E and F). Expression of endogenous Plzf mRNA in these cells was unaffected by lenalidomide treatment (Fig EV2E–G). In addition, the FLAG‐CRBN mutant (YW/AA) did not degrade AGIA‐PLZF in the presence of lenalidomide (Fig EV2H), suggesting the CRBN‐dependent degradation of PLZF.
We next investigated whether PLZF is recruited to the CRL4CRBN complex and ubiquitinated. Immunoprecipitation of FLAG‐CRBN using an anti‐FLAG antibody pulled down the CRL4CRBN complex consisting of DDB1, RBX1 and CUL4. At the same time, AGIA‐tagged PLZF was also strongly associated with the complex after thalidomide or lenalidomide treatment (Fig 2C and Appendix Fig S1A), indicating the inclusion of PLZF in the CRL4CRBN complex in a thalidomide‐ or derivative‐dependent manner, depending on the binding of thalidomide or one of its two derivatives. In addition, to analyse the ubiquitination of PLZF, AGIA‐PLZF was transfected with FLAG‐CRBN and immunoprecipitated for immunoblotting. The smear band resulting from the immunoprecipitation of AGIA‐PLZF was increased by supplementation with thalidomide or lenalidomide, suggesting that PLZF ubiquitination was induced by treatment with thalidomide or one of its derivatives (Fig 2D and Appendix Fig S1B). Next, to investigate the in vitro ubiquitination of PLZF, CRL4FLAG‐CRBN and AGIA‐PLZF were coimmunoprecipitated with an anti‐FLAG antibody in the presence or absence of lenalidomide (Fig 2E, lanes 5–8). As a negative control, empty vector or AGIA‐PLZF expressing HEK293T cells were lysed and mixed, and the lysates were immunoprecipitated by anti‐AGIA antibody to demonstrate the ubiquitination of PLZF caused by CRL4FLAG‐CRBN (Fig 2E, lanes 1–4). When PLZF and CRBN plus exogenous E1 and E2 enzymes were coimmunoprecipitated, PLZF was ubiquitinated in the presence of lenalidomide (Fig 2E, lane 8), but not in its absence (Fig 2E, lane 7). The findings indicated that CRL4FLAG‐CRBN ubiquitinates PLZF in vitro. The collective results demonstrated that PLZF is a target of the CRL4CRBN complex for proteasomal degradation.
PLZF plays an important role in immune responses (Suliman et al, 2012). Hence, we investigated whether PLZF degradation also occurs in lymphoma cell lines. Immunoblot analyses showed that IMiD treatment induced protein degradation of PLZF in ABC‐DLBCL (TK), GCB‐DLBCL (BJAB and HT), adult T‐cell lymphoma/ATL (MT‐4) and Burkitt's lymphoma (Raji) cell lines (Fig EV3A and B). In contrast, in GCB‐DLBCL (SU‐DHL‐4) cells expressing CRBN at low protein levels (Fig EV3A), IMiD treatment minimally induced protein degradation of PLZF (Fig EV3A), suggesting that the strength of PLZF degradation depends on the CRBN protein level. The collective results demonstrated that IMiD‐dependent PLZF degradation occurs in various immune cells expressing CRBN.
Figure EV3. IMiD‐induced protein degradation of PLZF in B cell lymphomas.
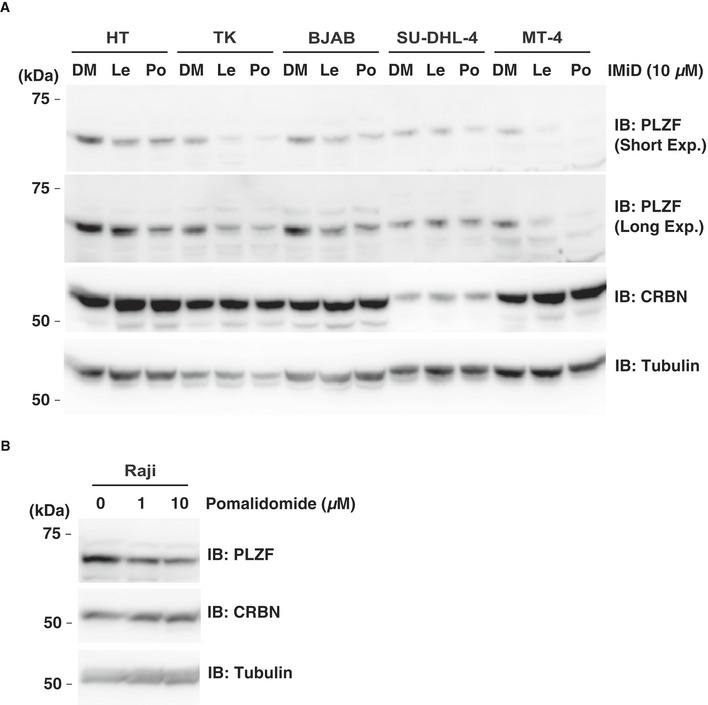
- HT, TK, BJAB, SU‐DHL‐4 and MT‐4 cells were treated with DMSO (DM), 10 µM lenalidomide (Le) or pomalidomide (Po) for 24 h. PLZF protein levels were analysed by immunoblot.
- Raji cells were treated with DMSO or pomalidomide for 24 h. PLZF protein levels were analysed by immunoblot.
Source data are available online for this figure.
Both ZNF1 and ZNF3 domains in PLZF are required for thalidomide‐dependent interaction with CRBN
We next attempted to determine the thalidomide‐dependent CRBN interaction region within PLZF. PLZF has a single BTB domain and nine ZNFs (Suliman et al, 2012) (Fig 3A). A single ZNF domain present in IKZF1 (Sievers et al, 2018) and SALL4 (Donovan et al, 2018; Matyskiela et al, 2018) is recognised by CRBN with thalidomide. As expected, the BTB domain alone in PLZF did not induce CRBN binding (Fig 3B). We therefore constructed a total of seven clones lacking different ZNFs by N‐terminal FLAG‐GST‐fusions (Fig 3A) and measured the interaction signals between each clone and CRBN with thalidomide. A ZNF1–4 clone displayed sufficient binding to CRBN with thalidomide but the ZNF1–3 clone did not (Fig 3B). We then investigated protein degradation of the truncated ZNF in CRBN‐deficient HEK293T cells. Surprisingly, immunoblot analysis revealed that all truncated ZNFs, including ZNF1–3, induced protein degradation after thalidomide treatment (Fig. 3C). Therefore, to refine the key domains on the native form of PLZF, each of the five ZNFs were individually interchanged with ZNF7 (Fig 3D), as this does not affect binding (Sievers et al, 2018). The binding assays revealed that the ZNF1 and ZNF3 domains were most important for binding (Fig 3E). In previous reports, a glycine residue in the ZNF domain of SALL4 and ZFP91 proteins was identified as a key amino acid for binding to CRBN with thalidomide (An et al, 2017; Donovan et al, 2018; Matyskiela et al, 2018). We constructed mutant clones (Gly to Ala shown in Fig 3F) with a single or double substitution in ZNF1 and ZNF3, which were then analysed in biochemical (Fig 3G) and cell‐based (Fig 3H) assays. Surprisingly, the double mutation completely abrogated binding and degradation. The binding ability of both single mutants was retained, but significantly reduced, and the degradation ability was retained (Fig 3G and H). Taken together, these results indicated that both ZNF1 and ZNF3 domains in PLZF are required for binding and degradation by CRBN with thalidomide.
Figure 3. Interaction regions in PLZF for binding to CRBN with thalidomide.
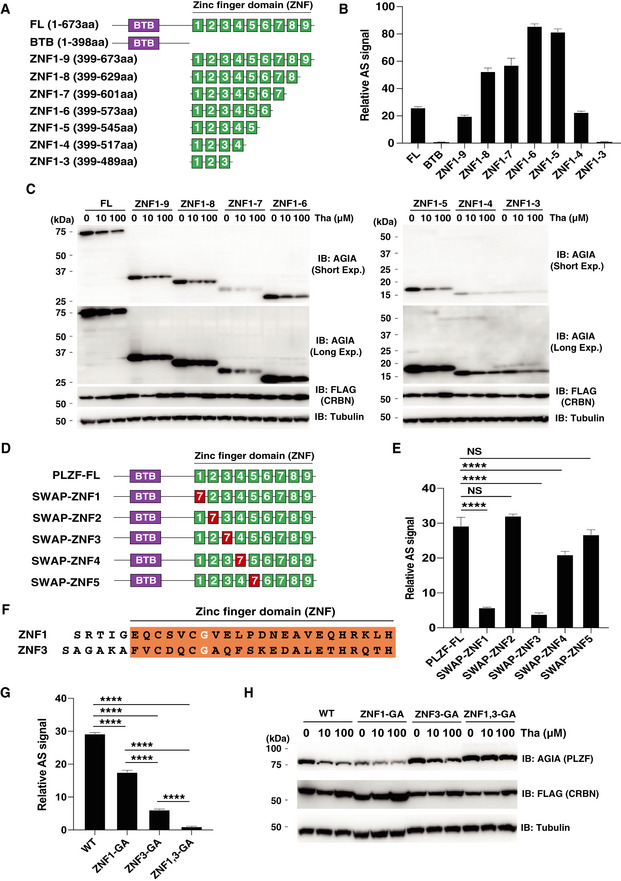
- Schematic diagram of PLZF and truncated PLZFs. FL; full length and BTB; BTB (broad‐complex, tramtrack, and bric à brac) domain.
- In vitro binding assay using truncated PLZF. Thalidomide‐dependent interaction between bls‐CRBN and FLAG‐GST‐PLZF‐full length (FL) or truncated FLAG‐GST‐PLZF was analysed in the presence of DMSO or 50 µM thalidomide using the AlphaScreen technology.
- Immunoblot analysis of AGIA‐PLZF protein levels in FLAG‐CRBN and PLZF‐full length (FL) or truncated FLAG‐GST‐PLZF expressing CRBN−/− HEK293T cells treated with DMSO or thalidomide (Tha) for 16 h.
- Schematic diagram of swapped PLZF mutants.
- In vitro binding assay using swapped PLZF mutants was performed using the same procedure as in Fig 3B.
- Amino acid sequences of ZNF1 and ZNF3 in PLZF. The white letters indicate critical Gly residue for the interaction between PLZF and CRBN with thalidomide or its derivatives.
- In vitro binding assay using point mutants of PLZF was performed using the same procedure as in Fig 3B.
- Immunoblot analysis of AGIA‐PLZF protein levels in FLAG‐CRBN and PLZF‐WT, PLZF‐ZNF1‐GA, PLZF‐ZNF3‐GA, or PLZF‐ZNF1,3‐GA expressing CRBN−/− HEK293T cells treated with DMSO or thalidomide (Tha) for 16 h.
Data information: All relative AlphaScreen (AS) signals are expressed as a relative luminescent signal with a luminescent signal of DMSO as one. Error bars denote ± standard deviation (n = 3). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (NS = not significant, ****P < 0.0001).
5‐Hydroxythalidomide induces degradation of PLZF and SALL4, but not IKZF1
5‐Hydroxythalidomide (Fig 4A) is produced as a primary metabolite of thalidomide by several types of human CYPs (Yamamoto et al, 2009; Chowdhury et al, 2010). However, it remains unclear whether 5‐hydroxythalidomide is involved in CRBN‐dependent effects, including thalidomide teratogenicity. We investigated whether 5‐hydroxythalidomide induces interaction between CRBN and protein. Surprisingly, 5‐hydroxythalidomide induced CRBN‐PLZF and CRBN‐SALL4 interactions (Fig 4B, middle and bottom panels, respectively), whereas it did not induce an interaction between CRBN and IKZF1 (Fig 4B, top panel). Furthermore, 5‐hydroxythalidomide induced the degradation of PLZF and SALL4 in cells, but not the degradation of IKZF1 (Fig 4C). Endogenous PLZF and SALL4 were also degraded in HuH7 and THP‐1 cells, respectively, after treatment with 5‐hydroxythalidomide (Fig 4D and E). Endogenous IKZF1 showed no change in the presence of 5‐hydroxythalidomide (Fig 4E), even though PLZF was degraded under the same conditions. Furthermore, 5‐hydroxythalidomide did not induce protein degradation of CK1α and ZFP91, which are thalidomide‐ and derivative‐dependent CRBN substrates (Fig 4F). Notably, a dose‐dependent biochemical experiment using thalidomide and 5‐hydroxythalidomide revealed that the CRBN‐SALL4 interaction with 5‐hydroxythalidomide was stronger than that of thalidomide at low concentrations (Fig 4B, middle panel). This high affinity of 5‐hydroxythalidomide was also observed in cell‐based analysis (Fig 4D and F). In contrast, the interaction between PLZF and CRBN with 5‐hydroxythalidomide was weaker than that of thalidomide (Fig 4B, bottom panel). Degradation of PLZF by 5‐hydroxythalidomide in cells occurred, albeit slightly weaker compared to thalidomide (Fig 4D–F). Taken together, these results suggested that 5‐hydroxythalidomide has the potential to induce protein degradation of PLZF and SALL4 in humans.
Figure 4. 5‐Hydroxythalidomide induces degradation of PLZF and SALL4 by CRBN.
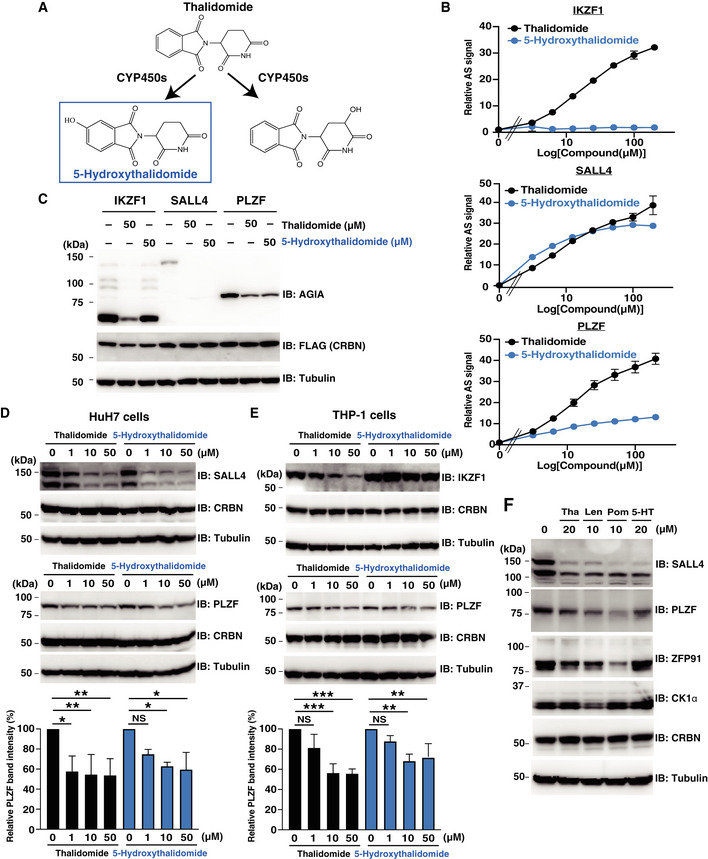
- Schematic diagram of thalidomide metabolites by CYPs.
- In vitro binding assay for thalidomide and 5‐hydroxythalidomide. Interaction between bls‐CRBN and FLAG‐GST‐IKZF1, FLAG‐GST‐SALL4 and FLAG‐GST‐PLZF in the presence of DMSO, thalidomide or 5‐hydroxythalidomide (3.125, 6.25, 12.5, 25, 50, 100 or 200 µM) was analysed using the AlphaScreen technology. All relative AlphaScreen (AS) signals are expressed as a relative luminescent signal with a luminescent signal of DMSO as one. Error bars denote ± standard deviation (n = 3).
- Immunoblot analysis of AGIA‐IKZF1, AGIA‐SALL4 or AGIA‐PLZF in FLAG‐CRBN expressing CRBN−/− HEK293T cells treated with DMSO, thalidomide or 5‐hydroxythalidomide for 16 h.
- Immunoblot analysis of endogenous SALL4 or PLZF protein levels in HuH7 cells treated with DMSO, thalidomide or 5‐hydroxythalidomide for 24 h.
- Immunoblot analysis of endogenous PLZF or IKZF1 protein levels in THP‐1 cells treated with DMSO, thalidomide or 5‐hydroxythalidomide for 24 h.
- Immunoblot analysis of endogenous SALL4, PLZF, ZFP91 or CK1α protein levels in HuH7 cells treated with DMSO, thalidomide (Tha), lenalidomide (Len), pomalidomide (Pom) or 5‐hydroxythalidomide (5‐HT) for 24 h.
Data information: Data in bar graphs in (D) and (E) were calculated as relative PLZF band intensity with PLZF band intensity of DMSO as 100. Error bars denote ± standard deviation (n = 3), and P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (NS = not significant, *P < 0.05, **P < 0.01 and ***P < 0.001).
PLZF is a substrate of CRBN with thalidomide in chicken embryos
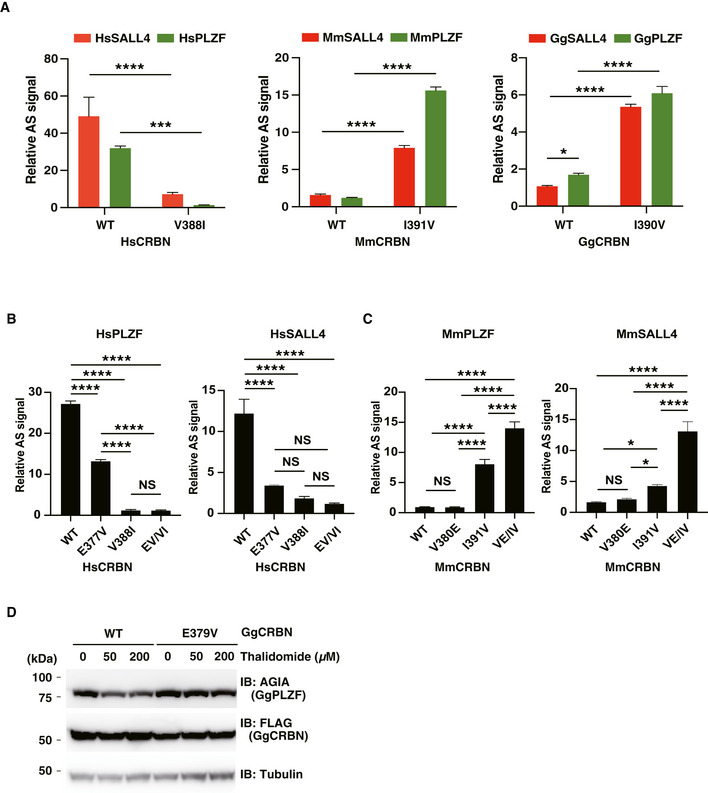
Thalidomide‐induced teratogenesis occurs in several species, including zebrafish, chicken and rabbit (Ito et al, 2010; Mahony et al, 2013; Matyskiela et al, 2018). The ZNF1/3 in PLZF (Fig 3) and ZNF2 in SALL4 (Donovan et al, 2018; Matyskiela et al, 2018) are important regions for the interaction with CRBN. We compared these amino acid sequences among vertebrate species. The sequences of ZNF1 and ZNF3 and other ZNFs in PLZF were significantly conserved among many species (Fig EV4A), although the SALL4‐ZNF2 sequence was not (Fig EV4B). To investigate the thalidomide‐dependent degradation of PLZF or SALL4 by CRBN from other animals, the recombinant proteins of CRBN, PLZF (ZBTB16 in chicken) and SALL4 from mouse (Mm) and chicken (Gg) were biochemically analysed. In addition, Val388 of human CRBN is a key residue for thalidomide‐dependent CRBN–protein interaction (Krönke et al, 2015; Donovan et al, 2018; Matyskiela et al, 2018). The biochemical analysis revealed that the binding ability of HsCRBN‐V388I was dramatically decreased compared with that of the WT HsCRBN (HsCRBN‐WT; Fig EV5A). Since the corresponding residue in MmCRBN and GgCRBN is isoleucine (Fig EV4C), we substituted Ile with Val in both to produce MmCRBN‐I391V and GgCRBN‐I390V. In the case of mouse proteins, although MmCRBN‐WT did not bind with either protein in the presence of thalidomide (Fig EV5A, middle panel), MmCRBN‐I391V displayed avid thalidomide‐dependent binding with both MmPLZF and MmSALL4. Interestingly, GgCRBN‐WT weakly bound to GgPLZF in the presence of thalidomide (Fig EV5A, right panel), whereas GgSALL4 did not bind to GgCRBN with thalidomide. Therefore, both MmCRBN‐I391V and GgCRBN‐I390V displayed thalidomide‐dependent binding with both SALL4 and PLZF.
Figure EV4. Sequence comparisons of thalidomide‐related regions in vertebrate PLZF, SALL4 and CRBN.

- Alignment of amino acid sequence of ZNF1 and ZNF3 in PLZF among human, rabbit, mouse, chicken and zebrafish. The red letters indicate different sequences of ZNF1,3 in PLZF among the species. The white letters indicate critical Gly residue for interaction between PLZF and CRBN with thalidomide or its derivatives.
- Alignment of amino acid sequence of ZNF2 in SALL4 among the species above. The red letters indicate different sequences of ZNF2 in SALL4 among the species. The white letters indicate critical Gly residues for interaction between SALL4 and CRBN with thalidomide or its derivatives.
- Alignment of amino acid sequence in CRBN among the species above. The blue and red letters indicate different sequences of thalidomide‐binding region in CRBN among the species.
Figure EV5. Interaction and protein degradation analyses between PLZF or SALL4 and CRBN with thalidomide among human, mouse and chicken.
-
AIn vitro binding assay using human, mouse and chicken proteins. Thalidomide‐dependent interaction between biotinylated HsCRBN, MmCRBN or GgCRBN, and FLAG‐GST‐SALL4 or FLAG‐GST‐PLZF (Hs, Mm or Gg) in the presence of DMSO or 200 µM thalidomide was analysed using the AlphaScreen technology.
-
B, CIn vitro binding assay using human or mouse proteins. Thalidomide‐dependent interaction in the presence of DMSO or 50 µM thalidomide was analysed using the same procedure used in Fig EV5A.
-
DImmunoblot analysis of AGIA‐GgPLZF in FLAG‐GgCRBN‐WT or FLAG‐GgCRBN‐E379V expressing CRBN−/− HEK293T cells treated with DMSO, 50 µM or 200 µM thalidomide for 16 h.
Data information: All relative AS (AlphaScreen) signals are expressed as a relative luminescent signal with a luminescent signal of DMSO as one. Error bars denote ± standard deviation (n = 3). P‐values were calculated by one‐way or two‐way ANOVA with Tukey's post hoc test (NS = not significant, *P < 0.05, ***P < 0.001 and ****P < 0.0001).
Source data are available online for this figure.
Thalidomide induced GgCRBN‐WT and GgPLZF interaction, although it did not facilitate an interaction between MmCRBN and MmPLZF (Fig EV5A). To investigate this, we compared the amino acid sequences of a thalidomide‐binding region among human, mouse and chicken proteins (Fig EV4C). Glu377 in HsCRBN was conserved in GgCRBN but Glu in MmCRBN was substituted to Val. It was reported that Glu377 in HsCRBN was an important amino acid for interaction between CRBN and GSPT1 (Matyskiela et al, 2016). Therefore, we investigated whether Glu377 is important for the interaction between CRBN and PLZF or SALL4. An in vitro binding assay showed that substitution of Glu to Val in HsCRBN significantly decreased binding ability to both SALL4 and PLZF (Fig EV5B). In MmCRBN, double substitution of Val380 to Glu and Ile391 to Val significantly increased the binding ability to SALL4 and PLZF, although single substitution of Val to Glu did not significantly increase (Fig EV5C).
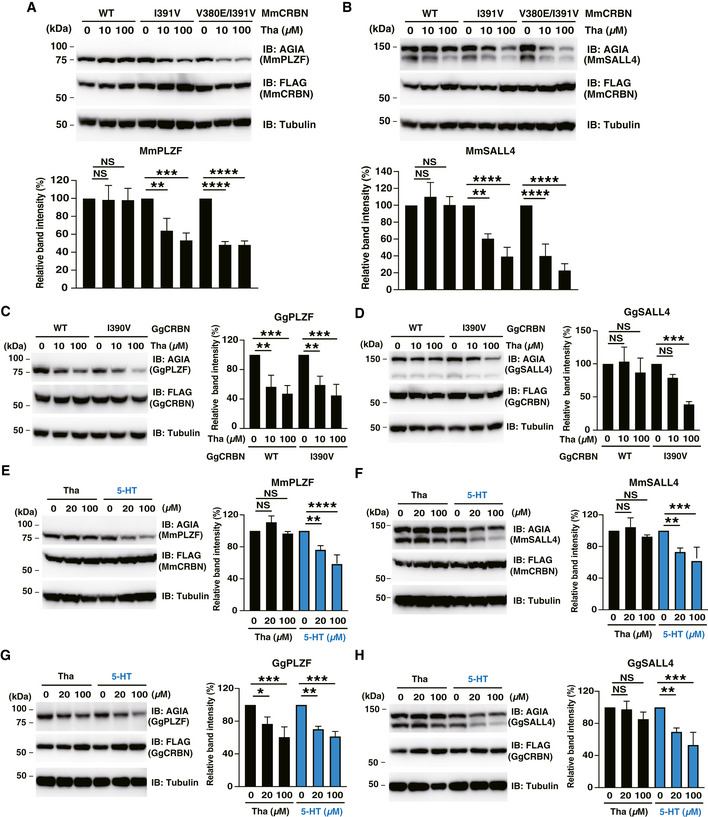
Next, each protein pair was transiently expressed in CRBN‐deficient HEK293T cells. In the mouse pair, MmCRBN‐WT did not degrade either MmPLZF or MmSALL4, whereas the MmCRBN‐I391V and MmCRBN‐V380E/I391V mutants degraded both proteins (Fig 5A and B, respectively). In the chicken pair, GgCRBN‐WT significantly induced the thalidomide‐dependent degradation of GgPLZF (Fig 5C), while GgSALL4 degradation was minimal in GgCRBN‐WT (Fig 5D). GgCRBN‐I390V also degraded GgPLZF and GgSALL4 with thalidomide (Fig 5C and D, respectively). In contrast, the GgCRBN‐E379V mutant did not induce GgPLZF degradation (Fig EV5D). The collective data supported the conclusion that Glu377 in the thalidomide‐binding region of CRBN is an important amino acid for thalidomide‐dependent interactions with PLZF and SALL4, and that this conserved amino acid sequence of the thalidomide‐binding region in CRBN was also required for GgPLZF degradation by GgCRBN. In addition, these results suggested that thalidomide‐dependent PLZF degradation, rather than SALL4 degradation, occurs in many species. However, further in vivo studies are required to demonstrate PLZF degradation in various species.
Figure 5. CRBN‐dependent degradation of PLZF and SALL4 from mouse and chicken by treatment with thalidomide and 5‐hydroxythalidomide.
-
A, BImmunoblot analysis of AGIA‐MmPLZF (A) or AGIA‐MmSALL4 (B) in FLAG‐MmCRBN‐WT, FLAG‐MmCRBN‐I391V or FLAG‐MmCRBN‐V380E/I391V expressing CRBN−/− HEK293T cells treated with DMSO or thalidomide (Tha) for 16 h.
-
C, DImmunoblot analysis of AGIA‐GgPLZF (C) or AGIA‐GgSALL4 (D) in FLAG‐GgCRBN‐WT or FLAG‐GgCRBN‐I390V expressing CRBN−/− HEK293T cells treated with DMSO or thalidomide (Tha) for 16 h.
-
E, FImmunoblot analysis of AGIA‐MmPLZF (E) or AGIA‐MmSALL4 (F) in FLAG‐MmCRBN‐WT expressing CRBN−/− HEK293T cells treated with indicated concentration of DMSO, thalidomide (Tha) or 5‐hydroxythalidomide (5‐HT) for 16 h.
-
G, HImmunoblot analysis of AGIA‐GgPLZF (G) or AGIA‐GgSALL4 (H) in FLAG‐GgCRBN‐WT expressing CRBN−/− HEK293T cells treated with indicated concentration of DMSO, thalidomide (Tha) or 5‐hydroxythalidomide (5‐HT) for 16 h.
Data information: Data in bar graphs in (A–H) were calculated as relative (Gg or Mm) PLZF or SALL4 band intensity with (Gg or Mm) PLZF or SALL4, respectively. The band intensity of DMSO was 100. Error bars denote ± standard deviation (n = 3). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Recently, humanised‐CYP3A mice, but not WT controls, were reported to show modest abnormal limb development in embryo culture, albeit not in vivo, after treatment with thalidomide (Kazuki et al, 2016), suggesting that CYP3A‐dependent thalidomide metabolites might induce teratogenesis. Therefore, we investigated the function of 5‐hydroxythalidomide in mouse and chicken CRBN‐dependent degradation. Surprisingly, 5‐hydroxythalidomide induced CRBN‐dependent degradation of both PLZF (Fig 5E and G) and SALL4 (Fig 5F and H), whereas thalidomide had no effect on MmCRBN and GgSALL4 degradation by GgCRBN. Furthermore, thalidomide and 5‐hydroxythalidomide did not induce the downregulation of Plzf and Sall4 mRNA expression in HuH7 cells (Appendix S2). These results suggested that 5‐hydroxythalidomide, rather than thalidomide, may potentially cause the degradation of both PLZF and SALL4 in many species. This needs to be validated in vivo to determine whether protein degradation of PLZF and SALL4 initiated by 5‐hydroxythalidomide occurs in mice and chickens.
PLZF is important in chicken limb development
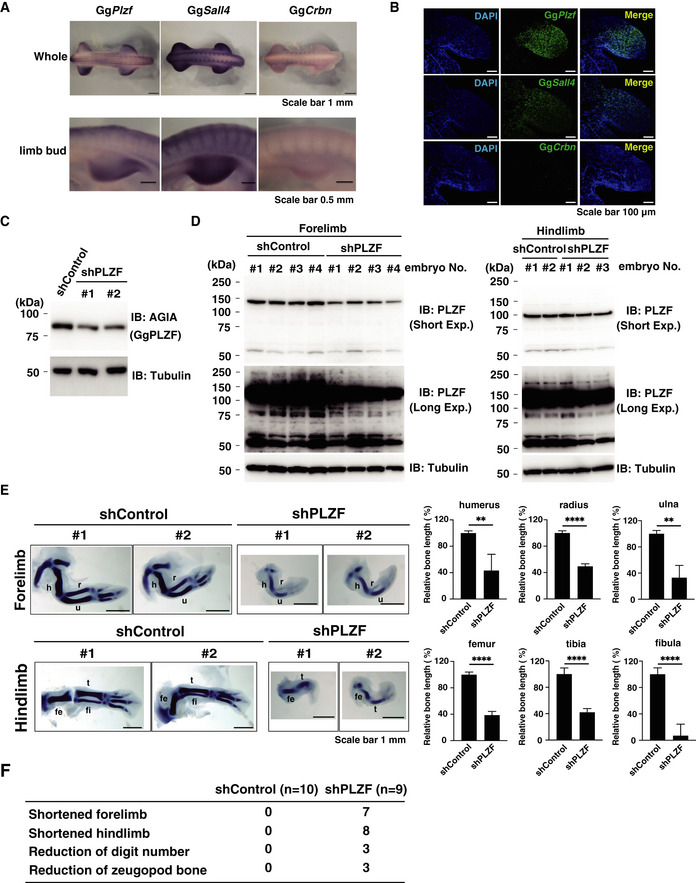
Both PLZF and Plzf mRNA are expressed in the limb buds of mouse and rat embryos (Barna et al, 2000; Liška et al, 2016), suggesting a direct function of CRBN‐thalidomide on PLZF in the developing limb bud. We investigated whether PLZF is important in the development of the chick limb bud. We first examined the expression of Plzf in the chick limb bud and confirmed Plzf mRNA expression by whole‐mount in situ hybridisation (Fig 6A). Expression of Sall4 and Crbn genes was also observed in the same region (Fig 6A). Expression of Plzf and Sall4 mRNA was confirmed in limb mesenchyme by section in situ hybridisation (Fig 6B). To investigate whether downregulation of Plzf mRNA induces limb teratogenicity in chicken embryos, we constructed an shRNA expression vector of GgPLZF. Immunoblot analysis showed that the constructed short hairpin RNA (shRNA) vector downregulated the protein expression of overexpressed GgPLZF in DF‐1 chicken embryonic fibroblasts (Fig 6C). Next, to elucidate the developmental role of PLZF in chick limb development, the RCAN retrovirus, which expresses shRNA (#2) against chick PLZF, was transfected into blastoderm cells containing prospective lateral plate mesoderm cells that give rise to the limb bud. Immunoblot analysis revealed that PLZF protein expression level was downregulated in the chicken embryos treated with PLZF (Fig 6D). PLZF shRNA transfected into limb buds caused several types of malformations (28%, n = 32; Fig 6E). Forelimb and hindlimb were shortened compared to limb buds transfected with control shRNA (green fluorescent protein [GFP] shRNA; 0%, n = 10). We also observed that only one bone was formed in the zeugopod, and the digit number was also reduced in limb buds transfected with PLZF shRNA‐infected limb bud (Fig 6F). These results suggested that chick PLZF plays a pivotal role in limb bud outgrowth and that downregulation of PLZF causes teratogenicity.
Figure 6. Downregulation of PLZF causes abnormal limb development in chicken embryo.
- Sall4, Plzf or Crbn mRNA expression in E4 chicken embryos was analysed by whole‐mount in situ hybridisation. Upper panel shows whole chicken embryo, and lower panel shows right forelimb bud.
- Sall4, Plzf or Crbn mRNA expression in E4 chicken limb bud was analysed by section in situ hybridisation.
- Immunoblot analysis of AGIA‐GgPLZF in AGIA‐GgPLZF expressing DF‐1 cells transfected with shControl (shGFP) or shPLZF expression vector.
- Immunoblot analysis of PLZF from tissue of chicken forelimb or hindlimb bud. Endogenous PLZF protein expression was detected by immunoblot using E4.5 chicken embryos infected with RCAN virus packaging shControl or shPlzf (forelimb shControl [n = 4], forelimb shPLZF [n = 4], hindlimb shControl [n = 2] or hindlimb shPlzf [n = 3]).
- Skeletal patterning of forelimb and hindlimb in E6 chicken embryos infected RCAN virus packaging shControl (n = 10) or shPLZF (n = 9) were analysed by Victoria blue staining. h; humerus, r; radius, u; ulna, fe; femur, fi; fibula, t; tibia. Data in bar graphs were calculated as relative bone length with bone length of shControl as 100. Error bars denote ± standard deviation (forelimb and hindlimb shControl [n = 3], forelimb shPLZF [n = 5] or hindlimb shPLZF [n = 6]). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (**P < 0.01 and ****P < 0.0001).
- Teratogenic phenotypes of chicken embryos in Fig 6E.
Source data are available online for this figure.
Thalidomide reduces PLZF protein levels in the abnormal limb buds of chickens
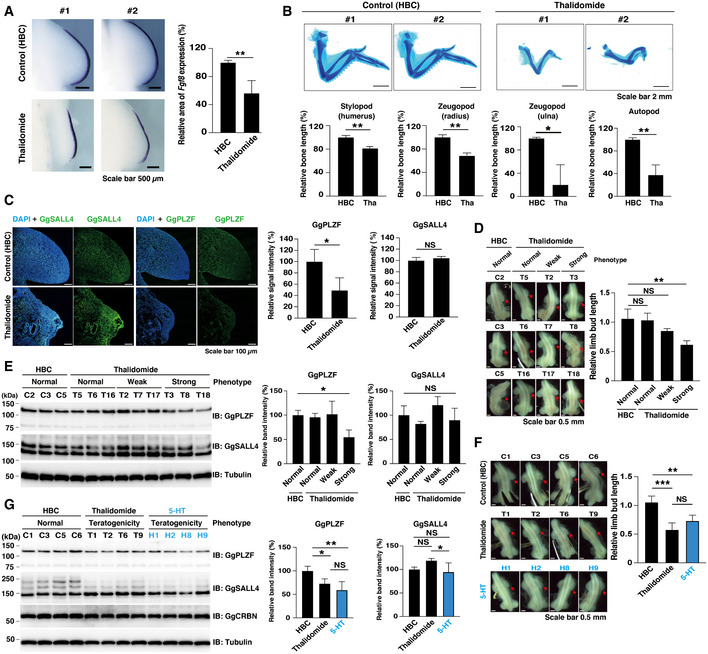
Next, we investigated whether thalidomide targets PLZF in the chick limb bud. In a previous report, decreased expression of the Fgf8 gene was identified as an indicator of abnormal limb bud development following thalidomide treatment (Ito et al, 2010). We confirmed a reduced/skewed pattern of Fgf8 expression in truncated limb buds after thalidomide treatment (Fig 7A). In addition, limb skeletal defects were observed under this condition (Fig 7B). Next, we investigated whether the amount of PLZF protein changes in these abnormal limb buds. Interestingly, reduced PLZF protein levels were observed in abnormal limb buds, although immunostaining of the SALL4 protein showed no change (Fig 7C). To confirm the protein levels of PLZF and SALL4, chick limb buds after thalidomide treatment were collected and characterised. Immunoblotting analysis also revealed the predominance of the reduction of PLZF protein in teratogenic limb buds (T3, T8 and T18 in Fig 7D and E). The observation that PLZF reduction by shRNA caused phenotypes of shortened forelimbs and hindlimbs (Fig 6E and F) suggests that PLZF is a CRBN substrate involved in thalidomide teratogenesis in chicken. In addition, we investigated whether 5‐hydroxythalidomide induces teratogenicity in chicken embryos. 5‐Hydroxythalidomide also induced teratogenicity and immunoblot analysis indicated that the degradation of PLZF and SALL4 induced by 5‐hydroxythalidomide was slightly stronger than that of thalidomide (Fig 7F and G). The morphological phenotypes of chicken embryos provided between 5‐hydroxythalidomide and thalidomide were very similar (Fig 7D and F), suggesting that a stronger degradation of SALL4 initiated by 5‐hydroxythalidomide is required to cause severe teratogenic phenotypes in chicken embryos.
Figure 7. Thalidomide induces degradation of PLZF in abnormal chick limb buds.
- Fgf8 mRNA expression in E4 chicken right limb bud embryo was analysed by whole‐mount in situ hybridisation. The two independent samples of control (2‐hydroxypropyl‐beta‐cyclodextrin [HBC], n = 4, upper panel) and thalidomide‐treated (1 µg/µl [3.9 mM], n = 7, lower panel) chicken embryos are shown. Data in bar graph were calculated as relative Fgf8 mRNA expressions with Fgf8 mRNA expressions of HBC as 100. Error bars denote ± standard deviation (HBC, n = 3 or thalidomide, n = 4). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (**P < 0.01).
- Limb skeletal stained with Alcian blue. Skeletal patterning of right forelimb in E10 chicken embryos treated with the vehicle for thalidomide, HBC (control, n = 6) or 1 µg/µl (3.9 mM) thalidomide (Tha; n = 6) were analysed by Alcian blue staining. Data in bar graphs were calculated as relative bone length with bone length of DMSO as 100. Error bars denote ± standard deviation (n = 3). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (*P < 0.05 and **P < 0.01).
- Immunohistochemical staining of PLZF or SALL4 in chicken forelimb bud. In left panels, endogenous PLZF or SALL4 protein expression were detected using forelimb bud section in chicken embryos treated with the vehicle for thalidomide, HBC (n = 4) or 1 µg/µl (3.9 mM) thalidomide (n = 4). In the data in the right bar graphs, relative intensities of IHC signals on forelimb bud sections in chick embryos were calculated as relative signal intensity with signal intensity of HBC as 100. Error bars denote ± standard deviation (PLZF [n = 3] or SALL4 [n = 4]), and P‐values were calculated by Student's t‐test (NS = not significant and *P < 0.05).
- Photographs show E4 chicken embryos treated with the vehicle for thalidomide, HBC (control, n = 6, C2, C3 and C5) or 1 µg/µl (3.9 mM) thalidomide (n = 18, [normal; T5, T6 and T16], [weak phenotype; T2, T7 and T17] or [strong phenotype; T3, T8 and T18]) corresponding to immunoblot analysis in Fig 7E. Red arrows show treated regions.
- Immunoblot analysis of PLZF or SALL4 from tissue of E4 chicken forelimb bud. Endogenous PLZF or SALL4 protein expression was detected by immunoblot using chicken embryos treated with the vehicle for thalidomide, HBC (n = 6) or 1 µg/µl (3.9 mM) thalidomide (n = 18).
- Photographs show E4 chicken embryos treated with HBC (control, n = 10, C1, C3, C5 and C6), 1 µg/µl (3.9 mM) thalidomide (n = 11, T1, T2, T6 and T9) or 1 µg/µl (3.6 mM) 5‐hydroxythalidomide (5‐HT; n = 10, H1, H2, H8 and H9) corresponding to immunoblot analysis in Fig 7G. Red arrows show treated regions.
- Immunoblot analysis of PLZF or SALL4 from tissue of E4 chicken forelimb bud. Endogenous PLZF or SALL4 protein expression was detected by immunoblot using chicken embryos treated with HBC (n = 10), 1 µg/µl (3.9 mM) thalidomide (n = 11) and 1 µg/µl (3.6 mM) 5‐hydroxythalidomide (5‐HT; n = 10).
Data information: Data in bar graphs in (D) and (F) were calculated as relative limb bud length with left limb bud length as one. Error bars denote ± standard deviation ((D), n = 3 or (F), n = 4). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (NS = not significant, **P < 0.01, and ***P < 0.001). Data in bar graphs in (E) and (G) were calculated as relative GgPLZF or GgSALL4 band intensity with GgPLZF or GgSALL4, respectively. The band intensity of HBC was 100. Error bars denote ± standard deviation ((E), n = 3 or (G), n = 4). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (NS = not significant, *P < 0.05, and **P < 0.01).
PLZF degradation is involved in thalidomide‐induced teratogenicity in chickens
Finally, to confirm that PLZF degradation is one of the reasons for thalidomide‐induced teratogenicity, we performed a rescue experiment by overexpression of GgPLZF in the chick forelimb bud (Fig 8B, F, J and N) and investigated the limb phenotypes and Fgf10/Fgf8 expression, which are affected by thalidomide treatment (Ito et al, 2010). In control embryos, expression of Fgf10 in the mesenchyme (Fig 8C and D, yellow arrowhead) and Fgf8 at the apical ectodermal ridge (AER; Fig 8C and D, pink arrowhead) was observed. Thalidomide treatment induced shortened limb bud (Fig 8E) concomitant with reduced expression of Fgf10 (Fig 8G and H, yellow arrowhead) and Fgf8 (Fig 8G and H, pink arrowhead). Overexpression of GgPLZF resulted in the normal development of the limb bud (Fig 8I) and expression of Fgf10 and Fgf8 (Fig 8K and L, yellow and pink arrowhead, respectively). When GgPLZF was overexpressed in the embryos treated with thalidomide, the reduction in limb elongation was rescued (Fig 8M) and normal expression levels of Fgf10 and Fgf8 were observed (Fig 8O and P, yellow and pink arrowhead, respectively). These results demonstrated that overexpression of GgPLZF partially rescued thalidomide‐induced inhibition of limb bud elongation (Fig 8Q and R). The findings implicated PLZF as an important target of thalidomide‐induced limb malformation.
Figure 8. PLZF overexpression rescues thalidomide‐induced limb phenotypes.
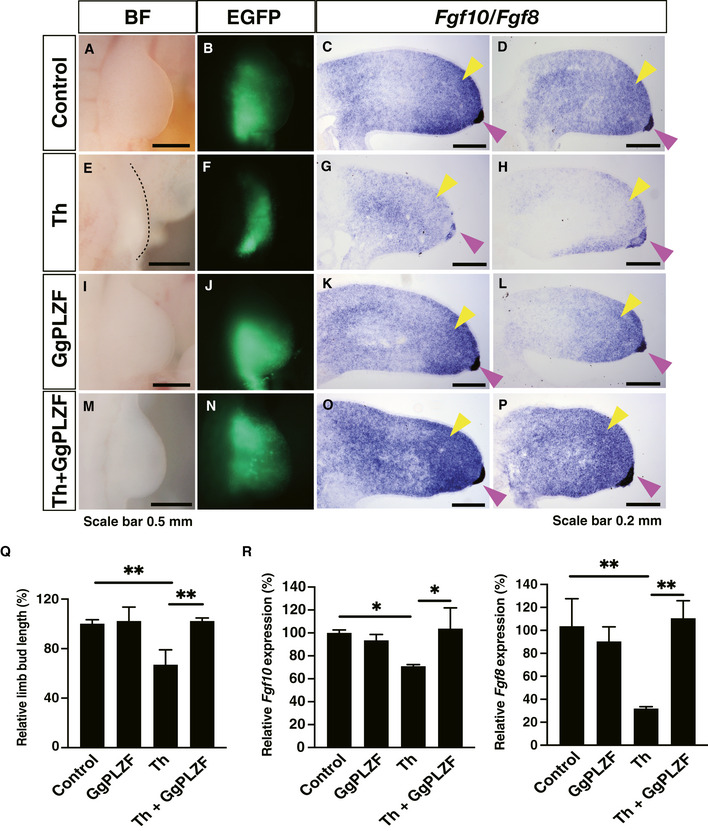
E3 chicken forelimb bud electroporated with empty pCAGGS or pCAGGS‐GgPLZF was treated with HBC or 1 µg/µl (3.9 mM) thalidomide, and the chicken forelimb bud phenotypes were observed in bright field (BF) or fluorescence of enhanced GFP (EGFP). Fgf10/Fgf8 expression was analysed by section by in situ hybridisation.
-
A–P(A–D) Empty vector and HBC (n = 8), (E–H) empty vector, and thalidomide (n = 4), (I–L) GgPLZF vector, and HBC (n = 5), (M–P) GgPLZF vector and thalidomide (n = 7). The dashed line in (E) indicates limb bud. The yellow and pink arrowheads show the expression of Fgf10 in the mesenchyme and expression of Fgf8 at the apical ectodermal ridge (AER), respectively.
-
QQuantitative analysis of phenotypes of chicken limb bud in Fig 8A, E, I and M. Data in bar graphs were calculated as relative limb bud length with limb bud length of control embryo as 100. Error bars denote ± standard deviation (n = 3). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (**P < 0.01).
-
RQuantitative analysis of Fgf10/Fgf8 expression in Fig 8C, D, G, H, K, L, O and P. Data in bar graphs were calculated as relative Fgf10/Fgf8 mRNA expressions with Fgf10/Fgf8 mRNA expressions of control embryo as 100. Error bars denote ± standard deviation (n = 3). P‐values were calculated by one‐way ANOVA with Tukey's post hoc test (*P < 0.05 and **P < 0.01).
Discussion
Many research groups have attempted to identify thalidomide‐dependent substrates of CRBN (Lu et al, 2014; Krönke et al, 2014, 2015; An et al, 2017; Donovan et al, 2018; Matyskiela et al, 2018; Sievers et al, 2018). The methodologies they have used have largely been based on cell‐based assays, such as stable isotope labelling by amino acids in cell culture, and transient expression systems. In this study, we performed a biochemical assay using cell‐free HuTFPA and AlphaScreen technologies. These methods easily detected thalidomide‐dependent interactions between CRBN and PLZF, SALL4 or IKZF1, without the need for protein purification (Fig 1) as well as the variations in the biochemical response of CRBN–protein depending on the presence of thalidomide, its derivatives or its 5‐hydroxythalidomide metabolite (Figs 3 and 4). The thalidomide‐dependent interaction between CRBN and PLZF was related to two ZNF domains (Fig 3) and the rate of IMiD‐dependent protein degradation, suggesting that the PLZF interaction is more complicated and may be different from known substrates, such as IKZF1, IKZF3 and SALL4. These results provide a reason why PLZF was not identified by traditional methods using cell‐based assays or in silico methods and suggests that our new biochemical approach using a protein array could be useful in identifying such substrates. However, our assay system cannot identify substrates that require interaction with CRBN in a protein complex. Therefore, our cell‐free approach and conventional cell‐based assays are complementary. We believe that these two approaches can be used together in future research for exploration and confirmation of substrates. Furthermore, as IKZF1 interacts with CRBN in the presence of thalidomide, but not 5‐hydroxythalidomide, this method would be useful for the analysis of compound‐dependent protein–protein interactions in the development of novel drugs, including thalidomide derivatives.
We identified PLZF as a new target of the thalidomide‐CRBN system. In chicken embryos, downregulation of PLZF produced hypoplasia of the limb bud (Fig 6E and F), indicating that PLZF is required for proper chicken limb development. Furthermore, thalidomide and 5‐hydroxythalidomide treatments decreased the protein level of PLZF, but SALL4 did not induce protein degradation in the abnormal limb buds of chicken embryos (Fig 7C, E and G). More importantly, PLZF overexpression in the chicken limb bud rescued the downregulation of thalidomide‐affected genes, such as Fgf10 and Fgf8 (Fig. 8). Based on these findings, we developed a model for the teratogenesis of chick limb buds (Fig 9A). Plzf −/−‐deficient mice display major musculoskeletal limb defects (Barna et al, 2000). PLZF deficiency alters Hoxds or Bmps expression in developing limbs (Barna et al, 2000). Bmp proteins function as regulators in programming cell death and cell death induced by Plzf/Gli3 deficiency (Barna et al, 2005). These findings suggest that degradation of PLZF by thalidomide treatment may affect cell proliferation in limb development. In contrast, in the case of Sall4 conditional knockout mice (Sall4‐CKO) driven by T‐Cre, which express early mesoderm, severe teratogenic phenotypes were observed in the hindlimb but did not show any phenotype in the forelimb (Akiyama et al, 2015). Furthermore, only 5% of Plzf −/− mice showed a forelimb phenotype featuring autopod abnormalities (Barna et al, 2000). This genetic evidence indicates that thalidomide‐induced teratogenicity in species that are sensitive to thalidomide, such as humans and rabbits, cannot be completely explained by the results of from a single knockout mouse for either Sall4 or Plzf. In a recent report, Sall4 and Plzf double knockout mice (Chen et al, 2020) showed phenotypes featuring remarkable reductions of the stylopod and zeugopod. These phenotypes are similar to those of phocomelia, which is a typical phenotype of thalidomide embryopathy. Therefore, we expect that protein degradation of both SALL4 and PLZF by thalidomide causes severe teratogenicity observed in the limbs of species that are sensitive to thalidomide (Fig 9B). Taken together, the results indicate that the variation in the protein sensitivity to thalidomide and/or differences in the genes that are necessary for normal limb development between species would result in differences in the phenotype between species in thalidomide teratology.
Figure 9. Schematic diagram of thalidomide teratogenicity based upon limb bud development in embryo.
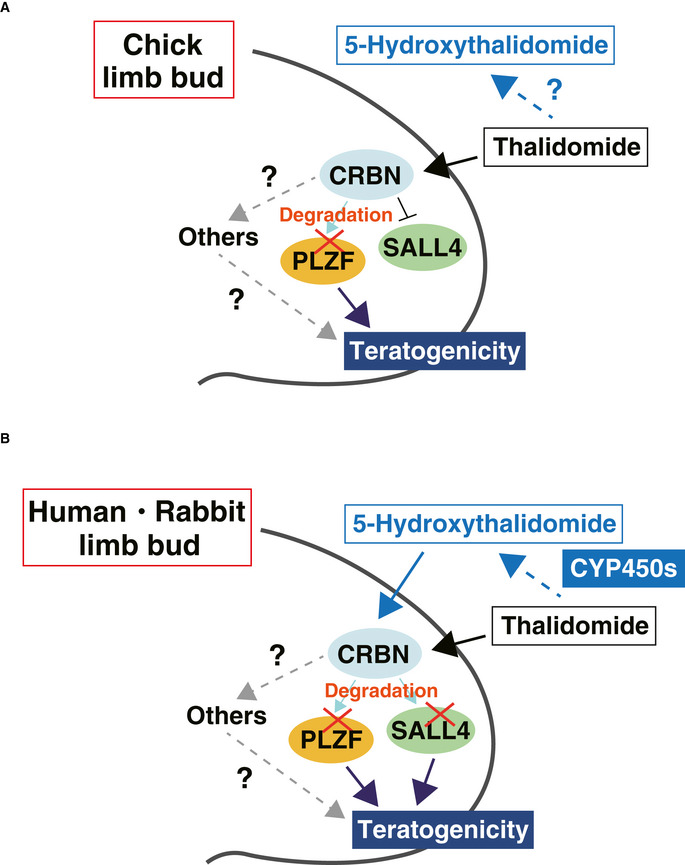
- Model of thalidomide teratogenicity based upon limb bud development in chicken embryo.
- Model of thalidomide teratogenicity based upon limb bud development in human or rabbit embryo.
PLZF is expressed in many cell types (Uhlén et al, 2005). PLZF is a multifunctional TF that modulates many developmental biological processes (Suliman et al, 2012), including cellular proliferation, cell cycle control, myeloid and lymphoid cell development and differentiation, programming of natural and killer T cells and invariant natural killer T cells, spermatogenesis and spermatogonial stem cell renewal, haematopoiesis, musculoskeletal limb development, megakaryocytic development and cytokine production. Notably, PLZF functions as a regulator of many immune responses (Savage et al, 2008; Xu et al, 2009; Serafini et al, 2015). Thalidomide is an immunomodulatory imide drug, and IKZF1 and IKZF3 are thought to be key targets of immunomodulation by thalidomide and its derivatives (Fink et al, 2018). Therefore, dysfunction of PLZF initiated by thalidomide may also result from its function as an IMiD. Further analyses will be required to confirm this.
Many spontaneous hydrolysis products and enzymatically formed hydroxylated metabolites of thalidomide are generated in vivo (Lu et al, 2003). In addition to thalidomide itself, several thalidomide metabolites also play important roles in thalidomide teratogenicity. For example, the hydrolysis metabolites 2‐phthalimidoglutaramic acid and 2‐phthalimidoglutaric acid induce oxidative stress, resulting in teratogenic phenotypes in rabbit embryos (Parman et al, 1999; Lee et al, 2011). In the case of enzymatic metabolites, thalidomide is metabolised to 5‐hydroxythalidomide or 5′‐hydroxythalidomide by CYP450 isoforms, such as CYP2C and 3A (Yamamoto et al, 2009; Chowdhury et al, 2010). An unstable arene oxide (epoxide) produced from 5‐hydroxythalidomide may cause developmental disorders (Gordon et al, 1981; Fort et al, 2000). Currently, CRBN is the only protein known to directly bind to thalidomide. CRBN interacts with thalidomide mainly via the glutarimide ring (Fischer et al, 2014). However, CRBN‐dependent thalidomide effects by 5‐hydroxythalidomide have not been thoroughly investigated, although 5‐hydroxythalidomide has a glutarimide ring and has a chemical structure similar to that of thalidomide derivatives. Recently, we have reported that 5‐hydroxythalidomide induced selective protein degradation of SALL4 (Yamanaka et al, 2020) and have showed structural insight on selectivity and strong degradation ability of 5‐hydroxythalidomide (Furihata et al, 2020). Therefore, we focussed on CRBN‐dependent thalidomide teratogenicity by 5‐hydroxythalidomide and revealed that 5‐hydroxythalidomide induced PLZF degradation at almost the same level as thalidomide and SALL4 degradation at a higher level than that induced by thalidomide (Fig 4). The previous and present results suggest that 5‐hydroxythalidomide has the potential for teratogenesis. Research on the generation and action of 5‐hydroxythalidomide will be important to understand the function of thalidomide. As well, research will be required to demonstrate how metabolites, including 5‐hydroxythalidomide, are involved in the range of thalidomide teratogenicity.
Thalidomide is a typical drug that shows species specificity (Ito et al, 2010). The species specificity of thalidomide has been thought to be provided by the difference of a thalidomide‐binding sequence in CRBN (Krönke et al, 2015; Donovan et al, 2018; Matyskiela et al, 2018). Consistent with these reports, in this study, humanised MmCRBN and GgCRBN induced thalidomide‐dependent interactions and degradation of PLZF and SALL4 proteins (Figs EV5 and 5A–D). In addition to the sequence difference between CRBN and neosubstrate, the metabolic cascade of thalidomide metabolites is also important in determining species specificity. In this study, 5‐hydroxythalidomide showed higher potential than thalidomide to degrade both PLZF and SALL4 proteins in the mouse and chicken (Figs 5E–H and 7G). Furthermore, one report described that humanised‐CYP3A mice undergo abnormal limb development in embryo culture at a high rate (> 40%) after thalidomide treatment (Kazuki et al, 2016), although thalidomide generally does not produce teratogenesis in mice (Krönke et al, 2015; Donovan et al, 2018; Matyskiela et al, 2018). Humans, in contrast, are highly sensitive to thalidomide (Beckman & Kampf, 1961; Ito et al, 2010). These reports and our results suggest that the thalidomide metabolic cascade, including metabolic rate and the types of metabolites, may play a role in the species specificity of thalidomide.
In conclusion, our findings support a hypothesis that the degradations of both PLZF and SALL4 by thalidomide and 5‐hydroxythalidomide produce CRBN‐dependent severe teratogenic phenotypes in species sensitive to thalidomide (Fig 9B).
Materials and Methods
Reagents
Thalidomide (Sigma‐Aldrich and Tokyo Chemical Industry Co., Ltd), pomalidomide (Sigma‐Aldrich), lenalidomide (FUJIFILM Wako Pure Chemical), 5‐hydroxythalidomide (5‐hydroxythalidomide was prepared according to a previously published method (Yamamoto et al, 2009)), MG132 (Peptide Institute) and MLN4924 (Chemscene) were dissolved in DMSO (FUJIFILM Wako Pure Chemical) at 2–100 mM and stored at −20°C as stock solutions. All drugs were diluted 1,000‐ or 500‐fold for in vivo experiments, or diluted 200‐fold for in vitro experiments.
Production of recombinant proteins using the cell‐free system
In vitro transcription and wheat cell‐free protein synthesis were performed using a WEPRO1240 expression kit (Cell‐Free Sciences). Transcripts were conducted using SP6 RNA polymerase with plasmids or DNA fragments as templates. The translation reaction was performed in bilayer mode using a WEPRO1240 expression kit (Cell‐Free Sciences), according to the manufacturer's instructions. For biotin labelling, 1 µl of cell‐free synthesised crude biotin ligase (BirA), produced by the wheat cell‐free expression system, was added to the lower layer, and 0.5 µM (final concentration) of d‐biotin (Nacalai Tesque) was added to both the upper and lower layers, as described previously (Sawasaki et al, 2008).
Interaction analysis of CRBN‐IMiD‐substrate using AlphaScreen technology
IMiD at the concentrations indicated in each figure and 0.5 µl of biotinylated HsCRBN, MmCRBN or GgCRBN were mixed in a 15 µl of AlphaScreen buffer containing 100 mM Tris (pH 8.0), 0.01% Tween‐20, 100 mM NaCl and 1 mg/ml BSA. Then, 5 µl of substrate mixture containing 0.8 µl of FLAG‐GST‐substrate in AlphaScreen buffer was added, and 20 µl of the reaction mixture was incubated at 26°C for 1 h in a 384‐well AlphaPlate (PerkinElmer). Subsequently, 5 µl of detection mixture containing 0.2 µg/ml anti‐DYKDDDDK mouse mAb (Wako), 0.08 µl of streptavidin‐coated donor beads, and 0.08 µl of protein A‐coated acceptor beads (PerkinElmer) in AlphaScreen buffer were added to each well. After incubation at 26°C for 1 h, luminescent signals were detected using an EnVision plate reader (PerkinElmer).
Production of the human transcription factor protein array (HuTFPA)
For the construction of human TF protein array, we prepared pEU‐E01‐FLAG‐GST‐K1‐02 vector containing FLAG tag, GST tag, SG linker and AsiSI restriction enzyme site at 5′ upstream of multiple cloning site. cDNA clones coding proteins with DNA‐binding domains were selected from cDNA resources collected by Kazusa DNA research institute (Nagase et al, 2008) (Table EV1). The plasmid of each clone was digested by combination of AsiSI and an appropriate restriction enzyme such as XhoI, SalI or NotI. The DNA fragment was inserted into pEU‐E01‐FLAG‐GST‐K1‐02 vector digested by same restriction enzymes. After subcloning, pEU expression plasmids were arranged in 96‐well format and stored as glycerol stock. Transcription template DNA fragments were amplified directly by PCR using PrimeStar Max PCR polymerase (Takara Bio), SPu‐2 (5′‐CAGTAAGCCAGATGCTACAC) and AODA2306 (5′‐AGCGTCAGACCCCGTAGAAA) primers and diluted glycerol stocks as template. Transcription and translation reactions were conducted using WEPRO7240 expression kit (Cell‐Free Sciences) in micro‐titre plate format. Transcription reaction mixture was prepared by mixing 1.4 µl of transcription buffer LM, 0.7 µl of NTP mixture (25 mM each), 0.07 µl RNase Inhibitor (Promega), 0.26 µl SP6 polymerase (Promega) and 1.4 µl PCR product in 96‐well plate. The transcription reaction was incubated at 37°C for 18 h. Translation reaction mixture containing 2.5 µl of mRNA, 1.67 µl of WEPRO7240 wheat germ extract, 0.14 µl of creatine kinase (20 mg/ml; Roche diagnostics) and 0.11 µl RNase Inhibitor was prepared and overlaid with 44 µl of SUB‐AMIX SGC solution (Cell‐Free Sciences) in V‐bottom 384‐well plate. The translation reaction was incubated at 26°C for 18 h. Expression of each protein product was confirmed by Western blotting using anti‐DYKDDDDK tag antibody (FUJIFILM Wako Pure Chemical).
High‐throughput screening using AlphaScreen technology
We added 20 µl of bait mixture, containing 50 µM thalidomide and 0.5 µl of biotinylated HsCRBN in AlphaScreen buffer, to 384‐well AlphaPlates using a FlexDrop Precision Reagent Dispenser (PerkinElmer). We next added 0.8 µl of FLAG‐GST‐transcription factor proteins to 384‐well AlphaPlates using a NanoHead (PerkinElmer) and a Janus Workstation (PerkinElmer). After the 384‐well AlphaPlates were incubated at 26°C for 1 h, 5 µl of detection mixture containing 0.2 µg/ml anti‐DYKDDDDK mouse mAb (FUJIFILM Wako Pure Chemical), 0.08 µl of streptavidin‐coated donor beads and 0.08 µl of protein A‐coated acceptor beads (PerkinElmer) in AlphaScreen buffer were added to each well using a FlexDrop precision reagent dispenser. After incubation at 26°C for 1 h, luminescent signals were detected using an EnVision plate reader (PerkinElmer).
Plasmids
Plasmids pDONR221 and pcDNA3.1(+), based on Gateway technology, or pCAGGS were purchased from Invitrogen or RIKEN, respectively, and the pEU vector for the wheat cell‐free system was constructed in our laboratory, as previously described (Sawasaki et al, 2002). Plasmids pcDNA3.1(+)‐FLAG‐GW, pcDNA3.1(+)‐FLAG‐MCS, pcDNA3.1(+)‐AGIA‐MCS, pCAGGS‐MCS, pEU‐bls‐GW and pEU‐bls‐MCS were constructed based on each original vector by PCR and using the In‐Fusion system (Takara Bio), or PCR and restriction enzymes. pEU‐FLAG‐GST‐IKZF1, pEU‐FLAG‐GST‐SALL4, pEU‐FLAG‐GST‐PLZF, pEU‐FLAG‐GST‐SALL1, pEU‐FLAG‐GST‐SALL2, pEU‐FLAG‐GST‐ZBTB17, pEU‐FLAG‐GST‐ZBTB20, pEU‐FLAG‐GST‐ZBTB38, pEU‐FLAG‐GST‐ZBTB48, pEU‐FLAG‐GST‐HNRNPK, pEU‐FLAG‐GST‐HMGB2 and pEU‐FLAG‐GST‐ELF5 were purchased from the Kazusa DNA Research Institute. Plasmids HsSALL4, HsPLZF and HsIKZF1 were amplified, and restriction enzyme sites were added by PCR and cloned into pcDNA3.1(+)‐AGIA‐MCS. The open reading frame of HsCRBN was purchased from the Mammalian Gene Collection (MGC) and MmCRBN, MmSALL4 and MmPLZF were did from Functional Annotation of Mouse (FAMTOM), respectively (Takahashi et al, 2016; Yano et al, 2016). The open reading frame of GgCRBN was artificially synthesised by IDT, and pcDNA3.1(+)‐GgSALL4‐DYKDDDDK and pcDNA3.1(+)‐GgPLZF‐DYKDDDDK were purchased from GenScript. HsCRBN was amplified, and the BP reaction sequence (attB and attP) was added by PCR and cloned into pDONR221 using BP recombination (Invitrogen). Then, pDONR221‐HsCRBN was recombined to pEU‐bls‐GW or pcDNA3.1(+)‐FLAG‐GW using LR recombination (attL and attR). MmCRBN and GgCRBN were amplified, and restriction enzyme sites were added by PCR and cloned into pEU‐bls‐MCS or pcDNA3.1(+)‐FLAG‐MCS. MmSALL4, GgSALL4, MmPLZF and GgPLZF were amplified, and restriction enzyme sites were added by PCR and cloned into pEU‐FLAG‐GST‐MCS or pcDNA3.1(+)‐AGIA‐MCS. Domain swapped HsPLZF was constructed by inverse PCR and the In‐Fusion system (Takara Bio). Deletion mutation and amino acid mutation of each protein was performed by inverse PCR.
For in situ hybridisation, the cDNAs for chicken Fgf8, Crbn, Sall4 and Plzf were obtained by PCR using the following primers: Fgf8 (NM_001012767.1), 5′‐attacgcgtATGGACCCCTGCTCCTCGCTCTTCA‐3′ and 5′‐attgataTCATGGGCGCAGGGAGGCGCTGGAG‐3′; Crbn (XM_015293204.2), 5′‐ctataggctagaattcacgcgtATGGCCGCCGAGGAGGGAGGTGACGGA‐3′ and 5′‐cactaaagggaagcggccgcgatatcTTACAAGCAGAGTAACGGAGATC‐3′; Sall4 (NM_001080872.1), 5′‐ctataggctagaattcacgcgtATGTCGCGACGGAAGCAGGCGAAGCCC‐3′ and 5′‐cactaaagggaagcggccgcgatatcTTAACTAACGGCAATTTTGTTCT‐3′; Plzf (XM_015298212.1), 5′‐ctataggctagaattcacgcgtATGGATTTGACTAAGATGGGCATGATA‐3′ and 5′‐cactaaagggaagcggccgcgataTCAGACGTAGCAGAGGTAGAGATAG‐3′. The amplified fragments of Fgf8 were digested by MluI‐EcoRV and subcloned into pCMS‐EGFP vector (Clontech). The amplified fragments of Crbn, Sall4 and Plzf were inserted into MluI‐EcoRV site of pCMS‐EGFP vector by In‐fusion (Takara Bio).
For knockdown of chick Plzf, the shRNA sequences of chick Plzf (#1: 5′‐GGAAATCGAGGTACATCAAGG‐3′ or #2: 5′‐GATTACTCGGCCATGATCAAA‐3′) were used. The following DNA oligos: #1: 5′‐gatcccGGAAATCGAGGTACATCAAGGgcttcctgtcacCCTTGATGTACCTCGATTTCCtttttta‐3′ and 5′‐agcttaaaaaaGGAAATCGAGGTACATCAAGGgtgacaggaagc CCTTGATGTACCTCGATTTCCgg‐3′; #2: 5′‐gatcccGATTACTCGGCCATGATCAAAgcttcctgtcacTTTGATCATGGCCGAGTAATCtttttta‐3′ and 5′‐agcttaaaaaaGATTACTCGGCCATGATCAAAgtgacaggaagc TTTGATCATGGCCGAGTAATgg‐3′ were purchased from Invitrogen. The DNA oligo pairs were annealed and inserted into pEntryCla12‐chickU6 shuttle vector using BamHI/HindIII site.
For rescue experiment by GgPLZF overexpression, GgPLZF was amplified and restriction enzyme sites were added by PCR and cloned into pCAGGS‐MCS.
Cell culture and transfection
HEK293T cells were cultured in DMEM (low glucose; FUJIFILM Wako Pure Chemical) supplemented with 10% foetal bovine serum (FBS; FUJIFILM Wako Pure Chemical), 100 unit/ml penicillin and 100 µg/ml streptomycin (Gibco) at 37°C under 5% CO2. HEK293T cells were transfected using TransIT‐LT1 transfection reagent (Mirus Bio) or PEI Max: Polyethyleneimine “Max” (MW 40,000; PolyScience, Inc.).
HuH7 cells were cultured in DMEM (high glucose; FUJIFILM Wako Pure Chemical) supplemented with 10% FBS (Wako), 100 unit/ml penicillin, 100 µg/ml streptomycin (Gibco), 1 mM Sodium Pyruvate (Gibco), 10 mM HEPES (Gibco) and 1× MEM NEAA (Gibco) at 37°C under 5% CO2.
THP‐1 cells were cultured in RPMI160 GlutaMAX medium (Gibco) supplemented with 10% FBS (FUJIFILM Wako Pure Chemical), 100 unit/ml penicillin and 100 µg/ml streptomycin (Gibco) at 37°C under 5% CO2.
TK, HT, BJAB, SU‐DHL‐4, MT‐4 and Raji cells were cultured in RPMI1640 GlutaMAX medium supplemented with 10% FBS (FUJIFILM Wako Pure Chemical), 100 unit/ml penicillin, 100 µg/ml streptomycin (Gibco) and 55 µM 2‐mercaptoethanol (Gibco) at 37°C under 5% CO2.
DF‐1 cells were cultured in DMEM (low glucose; FUJIFILM Wako Pure Chemical) supplemented with 10% FBS (Wako), 100 unit/ml penicillin and 100 µg/ml streptomycin (Gibco) at 37°C under 5% CO2. DF‐1 cells were transfected using TransIT‐LT1 transfection reagent (Mirus Bio).
Immunoblot and antibodies
Protein lysates were separated by SDS–PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). After the membranes were blocked using 5% skimmed milk (Megmilk Snow Brand) in TBST (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% Tween‐20) at room temperature for 1 h, the following antibodies were used. Anti‐FLAG mouse mAb (HRP‐conjugated, Sigma‐Aldrich, A8592) and anti‐AGIA rabbit mAb (Yano et al, 2016) (HRP‐conjugated, produced in our laboratory) were used to detect epitope‐tagged proteins. Anti‐α‐tubulin rabbit pAb (HRP‐conjugated, MBL, PM054‐7) was used to detect α‐tubulin. Biotinylated proteins were detected by anti‐biotin (HRP‐conjugated, Cell Signaling Technology, #7075). Anti‐CRBN rabbit mAb (Cell Signaling Technology, #71810), anti‐PLZF rabbit mAb (Cell Signaling Technology, #39784), anti‐PLZF rabbit pAb (GeneTex, GTX111046), anti‐PLZF goat pAb (R&D System, AF2944), anti‐SALL4 rabbit pAb (Abcam, ab29112), anti‐SALL4 mouse mAb (Santa Cruz Biotechnology, sc‐101147), anti‐DDB1 mouse mAb (Santa Cruz Biotechnology, sc‐376860), anti‐CUL4 mouse mAb (Santa Cruz Biotechnology, sc‐377188), anti‐RBX1 mouse mAb (Santa Cruz Biotechnology, sc‐393640) and anti‐ubiquitin mouse mAb (P4D1, Cell Signaling Technology, #3936) were used as primary antibodies. Anti‐rabbit IgG (HRP‐conjugated, Cell Signaling Technology, # 7074), anti‐mouse IgG (HRP‐conjugated, Cell Signaling Technology, # 7076) and anti‐goat IgG (HRP‐conjugated, Invitrogen/Thermo Fisher Scientific, #81‐1620) were used as secondary antibodies. Immobilon (Millipore) or ImmunoStar LD (FUJIFILM Wako Pure Chemical) was used as substrate HRP, and luminescent signals were detected using an ImageQuant LAS 4000mini (GE Healthcare). To perform re‐probing, Stripping Solution (FUJIFILM Wako Pure Chemical) was used and re‐blocked using 5% slim milk in TBST.
For immunoblot analysis of extract from chicken embryo, a right forelimb bud was dissected from HH st. 22/23 (embryonic day 4/E4) embryos and boiled in 50 μl of buffer (50 mM Tris–HCl pH 7.5, 4% SDS) at 98°C for 10 min. Protein concentration of each lysate was quantified using BCA assay (Thermo Fisher Scientific).
Immunoblot data were obtained using Image Quant LAS 4000mini and analysed by ImageJ (https://imagej.net/). For quantification of immunoblot analysis, each band intensity was measured by ImageJ (https://imagej.net/).
In vitro pull‐down assay of CRBN and substrate
To confirm the thalidomide‐dependent interactions between IKZF1, PLZF or SALL4 and CRBN, we performed pull‐down assays using Dynabeads M‐280 Streptavidin (Invitrogen). Biotinylated CRBN‐WT and CRBN‐YW/AA were synthesised using the wheat cell‐free system as described above. We then mixed 5 µl of Dynabeads M‐280 Streptavidin with 5 µl of biotinylated CRBN‐WT or CRBN‐YW/AA and diluted this 10‐fold with PBS containing 0.05% Tween‐20, and incubated this at room temperature for 1 h. The beads were washed three times in 500 µl PBS containing 0.05% Tween‐20, and substrate–thalidomide mixture was added containing 10 µl of FLAG‐GST‐IKZF1, FLAG‐GST‐SALL4 or FLAG‐GST‐PLZF and 200 µM thalidomide (0.5% DMSO) in 300 µl of AlphaScreen buffer containing 100 mM NaCl, 0.01% Tween‐20 and 1 mg/ml BSA. After rotation at room temperature for 90 min, the beads were washed four times in 500 µl of 1× Lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X‐100) and proteins were eluted by boiling in 1× sample buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol) containing 5% 2‐mercaptoethanol. The proteins were then analysed by immunoblot.
Construction of CRBN‐KO HEK293T cells
For analysis using CRBN‐KO cells, we selected the guide nucleotide sequence 5′‐ACTCCGGGCGGTTACCAGGC‐3′ from the human CRBN gene. Then, CRBN‐KO HEK293T cells were generated by genome editing based on CRISPR/Cas9 (Yamanaka et al, 2020).
In vivo IMiD‐dependent degradation assay of substrates
To confirm IMiD‐dependent degradation of PLZF, HEK293T or HEK293T‐CRBN−/− cells were cultured in 48‐well plates and transfected with 200 ng pcDNA3.1(+)‐FLAG‐CRBN‐WT or 200 ng pcDNA3.1(+)‐FLAG‐CRBN‐YW/AA and 20 ng pcDNA3.1(+)‐AGIA‐PLZF or pcDNA3.1(+)‐AGIA‐PLZF variants or pcDNA3.1(+)‐AGIA‐SALL4. After the cells were transfected for 8 h, they were treated with IMiD or dimethyl sulphoxide (DMSO, 0.1%) in the culture medium at the times and concentrations indicated in each figure.
To show that the IMiD‐dependent PLZF degradation was caused by CRL4CRBN and the 26S proteasome, the cells were treated with 2 µM MLN4924 and 10 µM MG132 (0.2% DMSO) at the times indicated in Fig 2A.
To examine the degradation of endogenous PLZF or SALL4, HEK293T, HuH7 or THP‐1 cells were cultured in 24‐ or 48‐well plates and treated with lenalidomide or DMSO (0.1%) in the culture medium at the times and concentrations indicated in each figure.
To examine the degradation of endogenous PLZF in TK, HT, BJAB, SU‐DHL‐4, MT‐4 and Raji cells, we cultured the cells in 12‐well plates and treated them with lenalidomide, pomalidomide or DMSO (0.1%) in the culture medium at the times and concentrations indicated in Fig EV5. The cells were lysed with RIPA buffer containing a protease inhibitor cocktail (Sigma‐Aldrich). The protein concentration of each lysate was quantified using a BCA assay (Thermo Fisher Scientific).
To examine the 5‐hydroxythalidomide‐dependent degradation of overexpressed PLZF, SALL4 or IKZF1, HEK293T‐CRBN−/− cells were cultured in 48‐well plates and transfected with 200 ng pcDNA3.1(+)‐FLAG‐CRBN‐WT and 20 ng pcDNA3.1(+)‐AGIA‐SALL4, pcDNA3.1(+)‐AGIA‐PLZF or pcDNA3.1(+)‐AGIA‐IKZF1. After the cells were transfected for 8 h, they were treated with thalidomide, 5‐hydroxythalidomide or DMSO (0.1%) in the culture medium at the indicated times and concentrations (Fig 4C). For endogenous SALL4, PLZF, IKZF1, HuH7 or THP‐1 cells were cultured in 48‐well plates and treated with thalidomide, 5‐hydroxythalidomide or DMSO (0.1%) in the culture medium at the indicated times and concentrations (Fig 4D and E).
To examine the species specificity of IMiD‐dependent protein degradation, HEK293T‐CRBN−/− cells were cultured in 48‐well plates and transfected with 200 ng pcDNA3.1(+)‐FLAG‐(mouse or chicken) CRBN‐WT or CRBN‐IV and 20 ng pcDNA3.1(+)‐AGIA‐(mouse or chicken) PLZF or pcDNA3.1(+)‐AGIA‐(mouse or chicken) SALL4. After the cells were transfected for 8 h, they were treated with thalidomide or DMSO (0.1%) in the culture medium for the times and concentrations indicated in Fig 5A–D.
To examine whether 5‐hydroxythalidomide induced the degradation of mouse or chicken SALL4 or PLZF, HEK293T‐CRBN−/− cells were cultured in 48‐well plates and transfected with 200 ng pcDNA3.1(+)‐FLAG‐(mouse or chicken) CRBN‐WT and 20 ng pcDNA3.1(+)‐AGIA‐(mouse or chicken) SALL4 or pcDNA3.1(+)‐AGIA‐(mouse or chicken) PLZF. After the cells were transfected for 8 h, they were treated with thalidomide, 5‐hydroxythalidomide or DMSO (0.1%) in the culture medium for the times and concentrations indicated in Fig 5E–H.
In all experiments, cells were lysed by boiling in 1× sample buffer containing 5% 2‐mercaptoethanol and the lysates were analysed by immunoblotting.
Quantitative real‐time PCR (qRT–PCR)
To demonstrate decreased post‐translational level of PLZF protein, PLZF mRNA expression in HEK293T, HuH7 or THP‐1 cells treated with DMSO or lenalidomide for 24 h was assessed by qRT–PCR. To analyse SALL4 or PLZF mRNA expression, HuH7 cells treated with DMSO, thalidomide or 5‐hydroxythalidomide for 24 h were assessed by qRT–PCR. Total RNA was isolated from HEK293T, HuH7 or THP‐1 cells treated with DMSO or lenalidomide for 24 h using a SuperPrep cell lysis kit (Toyobo). cDNA was synthesised using a SuperPrep RT kit (Toyobo) according to the manufacturer's protocol. RT–PCR was performed using a KOD SYBR qPCR Mix (Toyobo), and data were normalised against glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) mRNA levels. PCR primers were as follows: PLZF FW 5′‐GCACAGTTTTCGAAGGAGGA‐3′, PLZF RV 5′‐GGCCATGTCAGTGCCAGT‐3′, SALL4 FW 5′‐GGTCCTCGAGCAGATCTTGT‐3′, SALL4 RV 5′‐GGCATCCAGAGACAGACCTT‐3′, GAPDH FW 5′‐AGCAACAGGGTGGTGGAC‐3′, GAPDH RV 5′‐GTGTGGTGGGGGACTGAG‐3′.
Co‐immunoprecipitation of CRL4CRBN‐IMiD‐PLZF
To examine whether PLZF interacts with CRL4CRBN‐thalidomide or CRL4CRBN‐lenalidomide, HEK293T cells were cultured in a 15‐cm dish and transfected with 12 µg pcDNA3.1(+)‐FLAG‐CRBN and 12 µg pcDNA3.1(+)‐AGIA‐PLZF. After HEK293T cells were treated with DMSO, 10 or 100 µM thalidomide or lenalidomide in the presence of 10 µM MG132 for 8 h, cells were lysed in 1.5 ml IP Lysis buffer (Pierce; 25 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP‐40, 5% glycerol) containing a protease inhibitor cocktail (Sigma‐Aldrich) and incubated on ice for 15 min. Lysates were clarified by centrifugation at 16,100 g for 15 min, and CRL4CRBN was immunoprecipitated using anti‐FLAG M2 magnetic beads (Sigma‐Aldrich) with DMSO or 10 or 100 thalidomide or lenalidomide in the presence of 10 µM MG132. After overnight rotation at 4°C, the beads were washed three times with 800 µl of IP Lysis buffer (Pierce) and the proteins were eluted with 1× sample buffer. After eluted proteins were transferred to another tube, 2‐mercaptoethanol (final concentration is 5%) was added and boiled at 98°C for 5 min. The proteins were then analysed by immunoblot.
In vivo ubiquitination assay
To detect polyubiquitination of PLZF in cells, HEK293T cells were cultured in a 10‐cm dish and transfected with 5 µg pcDNA3.1(+)‐FLAG‐CRBN and 5 µg pcDNA3.1(+)‐AGIA‐PLZF. After HEK293T cells were treated with DMSO, 10 or 100 µM thalidomide or lenalidomide in the presence of 10 µM MG132 for 10 h, cells were lysed in 600 µl of SDS Lysis buffer (50 mM Tris–HCl pH 7.5, 1% SDS) containing a protease inhibitor cocktail (Sigma‐Aldrich) and boiled at 90°C for 15 min. Denatured lysates were treated with Benzonase Nuclease (Sigma‐Aldrich) at 37°C for 1 h, and the lysates were clarified by centrifugation at 16,100 g for 15 min and then diluted 10‐fold with IP Lysis buffer (Pierce). The proteins were immunoprecipitated overnight with Dynabeads Protein G (Invitrogen) interacting anti‐AGIA antibody at 4°C, which were then washed three times with 800 µl of IP Lysis buffer (Pierce). Proteins were eluted by boiling in 1× sample buffer containing 5% 2‐mercaptoethanol. The proteins were then analysed by immunoblot.
In vitro binding and ubiquitination assay
To confirm that PLZF is a direct substrate, we performed an in vitro binding and ubiquitination assay as described previously (Lu et al, 2014). HEK293T cells were cultured in a 15‐cm dish and transfected with 28 µg pcDNA3.1(+)‐FLAG‐CRBN or empty vector. HEK293T cells were cultured in two 15‐cm dishes and transfected with 25 µg pcDNA3.1(+)‐AGIA‐PLZF per one dish. After 24 h, the cells were lysed in 1.6 ml/dish of IP Lysis buffer (Pierce) containing a protease inhibitor cocktail (Sigma‐Aldrich), and 400 µl of FLAG‐CRBN or empty vector lysates were mixed with 400 µl of AGIA‐PLZF lysates in the presence of DMSO or 100 µM lenalidomide. The FLAG‐CRBN and AGIA‐PLZF mixtures were immunoprecipitated using anti‐FLAG M2 magnetic beads by rotating overnight at 4°C. As a negative control, empty vector and AGIA‐PLZF mixtures were immunoprecipitated using anti‐AGIA‐conjugated magnetic beads (produced in our laboratory) by rotating overnight at 4°C. The beads were washed three times with 800 µl of IP Lysis buffer (Pierce) and washed twice with 600 µl of 1× ubiquitin reaction buffer (50 mM Tris–HCl pH 7.5, 5 mM KCl, 5 mM MgCl2, 0.5 mM DTT), then resuspended in 20 µl of 1× ubiquitin reaction buffer containing 200 nM UBE1 E1 (R&D systems, U‐110), 1 µM UbcH5a E2 (Enzo, BML‐UW9050‐100), 1 µM UbcH5b (Enzo, BML‐UW9060‐100), 5 mM ATP, 10 µM HA‐ubiquitin (Boston Biochem, U‐110), 10 µM MG132, protease inhibitor cocktail and DMSO or 200 µM lenalidomide. Then, in vitro ubiquitination was performed at 30°C for 3 h; the proteins were denatured in 2% SDS by boiling at 95°C for 15 min. The proteins were diluted 20‐fold with IP Lysis buffer (Pierce) and immunoprecipitated anti‐AGIA‐conjugated magnetic beads (produced in our laboratory) at 4°C for 4 h. The beads were washed four times with 800 µl of IP Lysis buffer (Pierce), and the proteins were eluted with 750 µl (lane 1–4) or 20 µl (lane 5–6) of 1× sample buffer. After elution, proteins were transferred to another tube, 2‐mercaptoethanol (final concentration is 5%) was added, and were boiled at 98°C for 5 min. The proteins were then analysed by immunoblot.
Plzf knockdown in DF‐1 cells
To confirm the efficiency of shRNA, DF‐1 cells were cultured in a 48‐well plate and transfected with 50 ng pcDNA3.1(+)‐AGIA‐GgPLZF and 400 ng shRNA vector. After 6 h, the culture medium was exchanged with fresh culture medium and the DF‐1 cells were harvested after 48 h of transfection. The lysates were denatured by boiling at 98°C for 5 min and analysed by immunoblotting.
Animals
Fertilised eggs of white leghorn chicken (Gallus gallus domesticus) purchased from a domestic poultry farm (Kakeien, Sendai, Japan) or the Avian Bioscience Research Center (ABRC), Nagoya University, Japan, were incubated at 38°C until the appropriate developmental stage was reached. Embryos were staged according to the HH criteria of Hamburger and Hamilton (1951). All animal experiments were conducted in accordance with the guidelines of Tohoku University and Nagoya University.
Plzf knockdown in chicken embryos
Five micrograms of RCAN(A) retrovirus vector containing chick U6 promoter expressing shRNA for gfp or Plzf was transfected into M/O chicken strain‐derived embryonic fibroblast cells (CEF) cultured in 60‐mm dishes using Fugene HD transfection reagent (Promega). CEF was cultured in DMEM‐high glucose containing 10% FBS, 2% chicken serum and 1% penicillin–streptomycin, and spread three times. After the growth in ten 10‐cm dishes reached confluence, the medium was changed to DMEM‐high glucose containing 2% FBS and maintained for 24 h. The supernatant was then harvested and centrifuged at 100,000 g for 3 h at 4°C to concentrate the retrovirus. Isolated virus virions were stored at −80°C. To infect the retrovirus into the chick embryo, virions were sprinkled on the blastderm cells at HH st. 8/embryonic day 1 (E1) M/O chick embryo and incubated for 5 days. Fertilised M/O chicken eggs were provided by the National BioResource Project (NBRP) “Chicken/Quail” at Nagoya University.
Thalidomide treatment of chicken embryos
A solid crystal of thalidomide (Tokyo Chemical Industry Co., Ltd) was resolved in 45% 2‐hydroxypropyl‐beta‐cyclodextrin (HBC, FUJIFILM Wako Pure Chemical) for 1–2 h at 60°C to make thalidomide (2 μg/μl, 7.8 mM) or 5‐hydroxythalidomide (2 μg/μl, 7.3 mM) stock solution. This stock was mixed with the same volume of 2× Hanks buffer as a working solution (1 μg/μl, 3.9 mM thalidomide or 3.6 mM 5‐hydroxythalidomide). To apply thalidomide to an embryo, a small hole was opened at the amnion above the right forelimb bud at HH st. 18 (E3), and the working solution was injected in a space between the amnion and right forelimb bud. In the case of samples for in situ hybridisation, to induce a strong effect on target proteins of thalidomide or 5‐hydroxythalidomide for short exposure time, immunofluorescence and immunoblot analyses, embryos were treated with 30 μl of working solution and incubated until HH st. 22/23 (E4). In the case of samples of skeletal pattern analysis, to reduce lethality and increase the collection rate of chicken embryos showing teratogenic phenotypes in limb bud at HH st. 36 (E10), embryos were treated three times with 10 μl of working solution every 12 h and incubated until approximately HH st. 36 (E10).
In situ hybridisation
Embryos for whole‐mount in situ hybridisation and fluorescent in situ hybridisation in sections (FISH) were fixed by 4% paraformaldehyde (PFA)/phosphate‐buffered saline (PBS) at 4°C for 12 h. Digoxigenin (DIG)‐labelled RNA probes were prepared according to the manufacturer's instructions (Roche). Whole‐mount in situ hybridisation was performed as previously described (Tonegawa et al, 1997). In FISH, 10‐μm‐thick frozen sections were prepared with cryostat (LEICA CM3050S). FISH protocol was described previously (Sato et al, 2008), with additional processes in order to amplify fluorescent signal. Additional processes are as follows. After the reaction between DIG‐labelled RNA and anti‐DIG antibody‐conjugated horseradish peroxidase (HRP; Roche), sections were washed 3 times by TNT buffer (100 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.05% Tween‐20) and were treated with DIG amplification working solution in TSA plus DIG Kit (PerkinElmer) for 5 min at room temperature (RT) in accordance with manufacturer's instructions (PerkinElmer). After three times wash by TNT, samples were treated by anti‐DIG antibody‐conjugated HRP at 4°C for 12 h again. Images were processed by FIJI (https://fiji.sc), and Fgf8 expression area on limb bud regions was measured by the “Measure” command.
Immunofluorescence staining and quantification
Chicken embryos for immunofluorescence staining were fixed by 4% paraformaldehyde in PBS at 4°C for 12 h. Ten‐micrometre‐thick frozen sections were prepared using a cryostat. PLZF immunofluorescence staining was performed as previously described (Saito et al, 2012) using anti‐PLZF rabbit polyclonal antibody (GTX111046, 1:250 dilution; GeneTex) as the primary antibody and anti‐rabbit horseradish‐conjugated (1:500 dilution; GE Healthcare) as secondary antibody. To detect fluorescent signals, the TSA Plus Fluorescent System (PerkinElmer) was used for 5 min at room temperature. The sections were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI; Wako Pure Chemical Corporation) and sealed with FluorSave (Calbiochem).
SALL4 was detected as follows. After pre‐blocking with blocking solution consisting of 1% blocking reagent in TNT buffer (0.1 M Tris–HCl, pH 7.6, 0.15 M NaCl, 0.05% Tween 20) for 1 h at room temperature, the sections were incubated at 4°C overnight using an anti‐SALL4 rabbit polyclonal antibody (1:250 dilution, ab29112; Abcam). After three washes in TNT buffer, the specimens were incubated with anti‐rabbit IgG‐Alexa Fluor 488‐conjugated donkey antibody (Invitrogen) diluted 1:500 with blocking solution for 1 h at room temperature, followed by three washes using TNT buffer. The sections were stained with DAPI and sealed with FluorSave reagent. Images were processed by FIJI (https://fiji.sc), and intensities of fluorescent signals on limb bud regions were measured by the “Measure” command.
Limb skeletal staining of chicken embryo treated with thalidomide
Embryos for skeletal staining (Alcian blue or Victoria blue) were fixed with 10% formalin/tyrode. The Alcian blue staining protocol was described previously (Seki et al, 2015). Embryos were harvested in PBS and fixed in 10% formalin solution overnight. Fixed embryos were washed with 3% HCl in 70% ethanol three times and stained in 1% Victoria blue B (Sigma‐Aldrich) dissolved in 3% HCl in 70% ethanol overnight. Embryos were washed with 3% HCl in 70% ethanol overnight and then treated with methylsalicylate to render them transparent.
Imaging
Fluorescence in situ hybridisation and immunofluorescence staining of sections were obtained using a TCS SP5 confocal microscope (LEICA). Images of whole‐mount in situ hybridisation were obtained using a model M165FC fluorescent stereo microscope (LEICA) equipped with a model DP74 camera (Olympus). Images of skeletal staining were obtained using a SZX16 stereo microscope (Olympus) equipped with a model DP21 camera (Olympus).
Rescue experiment with GgPLZF overexpression in chicken limb bud
pCAGGS‐enhanced GFP (EGFP; 0.8 μg/ml), which is used for confirmation of expression area, with 2 μg/ml of pCAGGS‐GgPLZF or pCAGGS control plasmids were injected into the prospective forelimb field at HH st. 14 (E2). Electric pulses (60 ms pulse‐on and 999 ms pulse‐off at 8 V with three repetitions) were applied using the GEB14 electroporator (BEX). After 18‐h electroporation, 30 μl of 1 μg/μl (3.9 mM) thalidomide diluted in 45% HBC compound was dropped over the forelimb bud. After 12‐h incubation, embryos were harvested in PBS and EGFP expression was observed. Embryos in which EGFP expression was detected in the forelimb bud were treated with 10, 20 or 30% sucrose solution for 1 h. Embryos were embedded in the cryosection compound and frozen at −80°C. Frozen sections (20 μm) were cut using a cryostat, and in situ hybridisation was performed using digoxigenin‐labelled Fgf10 with Fgf8 probes. Images were processed by FIJI (https://fiji.sc), and intensities of Fgf10/Fgf8 on limb bud sections were measured by the “Measure” command.
Statistical analysis and reproducibility
The data are shown as the means ± standard deviation (SD) from the more than three technical replicates. Significant changes were analysed by one‐way analysis of variants (ANOVA) followed by Tukey's tests using GraphPad Prism (version 8) software (GraphPad, Inc.)
Author contributions
SY performed the biochemical, molecular and cellular biology experiments. TI and HTakeda constructed the human protein array. HTakahashi supported the screening. ET and NS synthesised and analysed the 5‐hydroxythalidomide. HM, DS, GA, TSu and KT performed the chicken teratogenesis studies. TSu performed knockdown and rescue experiments using chicken embryos. SY and TSa analysed the data, designed the study and wrote the paper, and all authors contributed to the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
TableEV1
Source Data for Expanded View/Appendix
Review Process File
Source Data for Figure 6
Acknowledgements
We thank H. Yamakawa (Kazusa DNA Research Institute) for the vector construction for the human protein array, C. Takahashi and C. Furukawa for technical assistance, and the Applied Protein Research Laboratory of Ehime University. We also thank Prof. F. Tokunaga and Dr. D. Oikawa (Osaka City University) for providing the DLBCL cell lines. This work was mainly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP19am0101077 (H. Takeda, T.S.), a Grant‐in‐Aid for Scientific Research on Innovative Areas (JP16H06579 for T.S.) from the Japan Society for the Promotion of Science (JSPS). This work was also partially supported by JSPS KAKENHI (JP17J08477 for S.Y., JP16H04729, JP19H03218 for T.S., JP17H06112 for N.S. and JP18H02446, JP18H04811, JP18H04756, JP20H05024 for K.T), a Grant‐in‐Aid for JSPS Research Fellow (JP17J08477 for S.Y) from JSPS., and Takeda Science Foundation. Establishment and screening of the human protein array used in this study was conducted in Proteo‐Science Center at Ehime University with the support of MEXT.
The EMBO Journal (2021) 40: e105375.
Data availability
There are no primary datasets or computer codes associated with this study. The data in this study are available from the corresponding author on reasonable request.
References
- Akiyama R, Kawakami H, Wong J, Oishi I, Nishinakamura R, Kawakami Y (2015) Sall4‐Gli3 system in early limb progenitors is essential for the development of limb skeletal elements. Proc Natl Acad Sci USA 112: 5075–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Ponthier CM, Sack R, Seebacher J, Stadler MB, Donovan KA, Fischer ES (2017) pSILAC mass spectrometry reveals ZFP91 as IMiD‐dependent substrate of the CRL4CRBN ubiquitin ligase. Nat Commun 8: 15398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Hawe N, Niswander L, Pandolfi PP (2000) Plzf regulates limb and axial skeletal patterning. Nat Genet 25: 166–172 [DOI] [PubMed] [Google Scholar]
- Barna M, Pandolfi PP, Niswander L (2005) Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature 436: 277–281 [DOI] [PubMed] [Google Scholar]
- Beckman R, Kampf HH (1961) Zur quantitativen Bestimmung und zum qualitativen Nachweis von N‐phthalyl‐glutaminsauremid (thalidomide). Arzneimittelforschung 11: 45–47 13688557 [Google Scholar]
- Chen KQ, Tahara N, Anderson A, Kawakami H, Kawakami S, Nishinakamura R, Pandolfi PP, Kawakami Y (2020) Development of the proximal‐anterior skeletal elements in the mouse hindlimb is regulated by a transcriptional and signaling network controlled by Sall4. Genetics 215: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G, Murayama N, Okada Y, Uno Y, Shimizu M, Shibata N, Guengerich FP, Yamazaki H (2010) Human liver microsomal cytochrome P450 3A enzymes involved in thalidomide 5‐hydroxylation and formation of a glutathione conjugate. Chem Res Toxicol 23: 1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KA, An J, Nowak RP, Yuan JC, Fink EC, Berry BC, Ebert BL, Fischer ES (2018) Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial syndrome. eLife 7: e38430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink EC, McConkey M, Adams DN, Haldar SD, Kennedy JA, Guirguis AA, Udeshi ND, Mani DR, Chen M, Liddicoat B et al (2018) CrbnI391V is sufficient to confer in vivo sensitivity to thalidomide and its derivatives in mice. Blood 132: 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Kohlhase J, Böhm D, Schweiger B, Hoffmann D, Heitmann M, Horsthemke B, Wieczorek D (2008) Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet 45: 731–737 [DOI] [PubMed] [Google Scholar]
- Fischer ES, Böhm K, Lydeard JR, Yang H, Stadler MB, Cavadini S, Nagel J, Serluca F, Acker V, Lingaraju GM et al (2014) Structure of the DDB1‐CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512: 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort DJ, Stover EL, Bantle JA, Finch RA (2000) Evaluation of the developmental toxicity of thalidomide using frog embryo teratogenesis assay–Xenopus (FETAX): biotransformation and detoxification. Teratog Carcinog Mutagen 20: 35–47 [DOI] [PubMed] [Google Scholar]
- Furihata H, Yamanaka S, Honda T, Miyauchi Y, Asano A, Shibata N, Tanokura M, Sawasaki T, Miyakawa T (2020) Structural bases of IMiD selectivity that emerges by 5‐hydroxythalidomide. Nat Commun 11: 4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GB, Spielberg SP, Blake DA, Balasubramanian V (1981) Thalidomide teratogenesis: evidence for a toxic arene oxide metabolite. Proc Natl Acad Sci USA 78: 2545–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL (1951) A series of normal stages in the development of the chick embryo. J Morphol 88: 49–92 [PubMed] [Google Scholar]
- Hideshima T, Chauhan D, Shima Y, Raje N, Davies FE, Tai YT, Treon SP, Lin B, Schlossman RL, Richardson P et al (2000) Thalidomide and its analogs overcome drug resistance of human multiple myeloma cells to conventional therapy. Blood 96: 2943–2950 [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H (2010) Identification of a primary target of thalidomide teratogenicity. Science 327: 1345–1350 [DOI] [PubMed] [Google Scholar]
- Kazuki Y, Akita M, Kobayashi K, Osaki M, Satoh D, Ohta R, Abe S, Takehara S, Kazuki K, Yamazaki H et al (2016) Thalidomide‐induced limb abnormalities in a humanized CYP3A mouse model. Sci Rep 6: 21419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhase J, Chitayat D, Kotzot D, Ceylaner S, Froster UG, Fuchs S, Montgomery T, Rösler B (2005) SALL4 mutations in Okihiro syndrome (Duane‐radial ray syndrome), acro‐renal‐ocular syndrome, and related disorders. Hum Mutat 26: 176–183 [DOI] [PubMed] [Google Scholar]
- Krönke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X et al (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343: 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, Chamberlain PP, Mani DR, Man HW, Gandhi AK et al (2015) Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523: 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJJ, Gonçalves LL, Wells PG (2011) Embryopathic effects of thalidomide and its hydrolysis products in rabbit embryo culture: evidence for a prostaglandin H synthase (PHS)‐dependent, reactive oxygen species (ROS)‐mediated mechanism. FASEB J 25: 2468–2483 [DOI] [PubMed] [Google Scholar]
- Liška F, Peterková R, Peterka M, Landa V, Zídek V, Mlejnek P, Šilhavý J, Šimáková M, Křen V, Starker CG et al (2016) Targeting of the Plzf gene in the rat by transcription activator‐like effector nuclease results in caudal regression syndrome in spontaneously hypertensive rats. PLoS One 11: e0164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Palmer BD, Kestell P, Browett P, Baguley BC, Muller G, Ching L‐M (2003) Thalidomide metabolite in mice and patients with multiple myeloma. Clin Cancer Res 9: 1680–1688 [PubMed] [Google Scholar]
- Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong K‐K, Bradner JE, Kaelin WG (2014) The myeloma drug lenalidomide promotes the cereblon‐dependent destruction of Ikaros proteins. Science 343: 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony C, Erskine L, Niven J, Greig NH, Figg WD, Vargesson N (2013) Pomalidomide is nonteratogenic in chicken and zebrafish embryos and nonneurotoxic in vitro . Proc Natl Acad Sci USA 110: 12703–12708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Lu G, Ito T, Pagarigan B, Lu C‐C, Miller K, Fang W, Wang N‐Y, Nguyen D, Houston J et al (2016) A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535: 252–257 [DOI] [PubMed] [Google Scholar]
- Matyskiela ME, Couto S, Zheng X, Lu G, Hui J, Stamp K, Drew C, Ren Y, Wang M, Carpenter A et al (2018) SALL4 mediates teratogenicity as a thalidomide‐dependent cereblon substrate. Nat Chem Biol 14: 981–987 [DOI] [PubMed] [Google Scholar]
- Miller MT, Strömland K (1999) Teratogen update: thalidomide: a review, with a focus on ocular findings and new potential uses. Teratology 60: 306–321 [DOI] [PubMed] [Google Scholar]
- Nagase T, Yamakawa H, Tadokoro S, Nakajima D, Inoue S, Yamaguchi K, Itokawa Y, Kikuno RF, Koga H, Ohara O (2008) Exploration of human ORFeome: high‐throughput preparation of ORF clones and efficient characterization of their protein products. Nat Med 15: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parman T, Wiley MJ, Wells P (1999) Free radical‐mediated oxidative DNA damage in the mechanism of thalidomide teratogenicity. Nat Med 5: 582–585 [DOI] [PubMed] [Google Scholar]
- Petzold G, Fischer ES, Thomä NH (2016) Structural basis of lenalidomide‐induced CK1α degradation by the CRL4CRBN ubiquitin ligase. Nature 532: 127–130 [DOI] [PubMed] [Google Scholar]
- Saito D, Takase Y, Murai H, Takahashi Y (2012) The dorsal aorta initiates a molecular cascade that instructs sympatho‐adrenal specification. Science 336: 1578–1581 [DOI] [PubMed] [Google Scholar]
- Sato Y, Watanabe T, Saito D, Takahashi T, Yoshida S, Kohyama J, Ohata E, Okano H, Takahashi Y (2008) Notch signaling mediates the segmental specification of angioblasts in somites and their directed migration toward the dorsal aorta in avian embryos. Dev Cell 14: 890–901 [DOI] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A (2008) The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasaki T, Ogasawara T, Morishita R, Endo Y (2002) A cell‐free protein synthesis system for high‐throughput proteomics. Proc Natl Acad Sci USA 99: 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasaki T, Kamura N, Matsunaga S, Saeki M, Tsuchimochi M, Morishita R, Endo Y (2008) Arabidopsis HY5 protein functions as a DNA‐binding tag for purification and functional immobilization of proteins on agarose/DNA microplate. FEBS Lett 582: 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki R, Kitajima K, Matsubara H, Suzuki T, Saito D, Yokoyama H, Tamura K (2015) AP‐2β is a transcriptional regulator for determination of digit length in tetrapods. Dev Biol 407: 75–89 [DOI] [PubMed] [Google Scholar]
- Serafini N, Vosshenrich CAJ, Di Santo JP (2015) Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol 15: 415–428 [DOI] [PubMed] [Google Scholar]
- Sievers QL, Petzold G, Bunker RD, Renneville A, Słabicki M, Liddicoat BJ, Abdulrahman W, Mikkelsen T, Ebert BL, Thomä NH (2018) Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362: eaat0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithells RW, Newman CG (1992) Recognition of thalidomide defects. J Med Genet 29: 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suliman BA, Xu D, Williams BRG (2012) The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Front Oncol 2: 74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro D, Takahama S, Shimizu K, Hayashi S, Endo Y, Sawasaki T (2010) Characterization of a caspase‐3‐substrate kinome using an N‐ and C‐terminally tagged protein kinase library produced by a cell‐free system. Cell Death Dis 1: e89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Uematsu A, Yamanaka S, Imamura M, Nakajima T, Doi K, Yasuoka S, Takahashi C, Takeda H, Sawasaki T (2016) Establishment of a wheat cell‐free synthesized protein array containing 250 human and mouse E3 ubiquitin ligases to identify novel interaction between E3 ligases and substrate proteins. PLoS One 11: e0156718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therapontos C, Erskine L, Gardner ER, Figg WD, Vargesson N (2009) Thalidomide induces limb defects by preventing angiogenic outgrowth during early limb formation. Proc Natl Acad Sci USA 106: 8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa A, Funayama N, Ueno N, Takahashi Y (1997) Mesodermal subdivision along the mediolateral axis in chicken controlled by different concentrations of BMP‐4. Development 124: 1975–1984 [DOI] [PubMed] [Google Scholar]
- Uhlén M, Björling E, Agaton C, Szigyarto CA‐K, Amini B, Andersen E, Andersson A‐C, Angelidou P, Asplund A, Asplund C et al (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: 1920–1932 [DOI] [PubMed] [Google Scholar]
- Xu D, Holko M, Sadler AJ, Scott B, Higashiyama S, Berkofsky‐Fessler W, McConnell MJ, Pandolfi PP, Licht JD, Williams BRG (2009) Promyelocytic leukemia zinc finger protein regulates interferon‐mediated innate immunity. Immunity 30: 802–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Shoya Y, Matsuoka S, Nishida‐Fukuda H, Shibata N, Sawasaki T (2020) An IMiD‐induced SALL4 degron system for selective degradation of target proteins. Commun Biol 3: 515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Shibata N, Sukeguchi D, Takashima M, Nakamura S, Toru T, Matsunaga N, Hara H, Tanaka M, Obata T et al (2009) Synthesis, configurational stability and stereochemical biological evaluations of (S)‐ and (R)‐5‐hydroxythalidomides. Bioorg. Med Chem Lett 19: 3973–3976 [DOI] [PubMed] [Google Scholar]
- Yano T, Takeda H, Uematsu A, Yamanaka S, Nomura S, Nemoto K, Iwasaki T, Takahashi H, Sawasaki T (2016) AGIA tag system based on a high affinity rabbit monoclonal antibody against human dopamine receptor D1 for protein analysis. PLoS One 11: e0156716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
TableEV1
Source Data for Expanded View/Appendix
Review Process File
Source Data for Figure 6
Data Availability Statement
There are no primary datasets or computer codes associated with this study. The data in this study are available from the corresponding author on reasonable request.


