Abstract
Pain is a serious clinical challenge, and is associated with a significant reduction in quality of life and high financial costs for affected patients. Research efforts have been made to explore the etiological basis of pain to guide the future treatment of patients suffering from pain conditions. Findings from studies using KA (kainate) receptor agonist, antagonists and receptor knockout mice suggested that KA receptor dysregulation and dysfunction may govern both peripheral and central sensitization in the context of pain. Additional evidence showed that KA receptor dysfunction may disrupt the finely-tuned process of glutamic acid transmission, thereby contributing to the onset of a range of pathological contexts. In the present review, we summarized major findings in recent studies which examined the roles of KA receptor dysregulation in nociceptive transmission and in pain. This timely overview of current knowledge will help to provide a framework for future developing novel therapeutic strategies to manage pain.
Keywords: Pain, kainate receptors, neuron, plasticity, modulation
Introduction
Pain is a normal sensory function that is necessary for survival. It is intended to protect the individual from continued or current injury; however, when the sensation becomes aberrant and develops into a more chronic nature, it transitions into a dysfunctional sensation that handicaps the sufferer, severely affecting quality of life.1 The mechanisms underlying pain have been extensively studied in both humans and animal models. It is well established that glutamatergic transmission is essential to nociceptive transmission.2,3 Glutamate receptors are composed of two major families: ionotropic (iGluRs) and metabotropic glutamate receptors (mGluRs).4 While iGluRs form an ion channel pore that becomes activated upon ligand binding, mGluRs do not conduct ion flux, instead they regulate G-proteins to control biochemical processes within cells.
There are three main classes of iGluRs which are structurally and pharmacologically different: NMDA (N-methyl-D-aspartate), AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid) and kainate (KA) receptors.5,6 NMDA receptors are important for initiating long-term neuronal plasticity such as long-term potentiation,7,8 whereas AMPA receptors control fast excitatory synaptic transmission and plasticity.9,10 Both of these receptors were found to be associated with pain development, and both NMDA and AMPA receptor antagonists were aid in the treatment of pain. Unfortunately, the systemic administration of these drugs in patients has been associated with significant adverse effects including ataxia, loss of motor coordination, memory impairment, and psychotomimetic effects, ataxia.11–13
Unlike NMDA and APMA receptors, KA receptors primarily exert modulatory roles in the peripheral and central nervous system, serving as an important regulator of nociceptive transmission and integration, and thus may represent a more promising therapeutic target than NMDA and AMPA receptors in the context of pain.14–16 Indeed, KA receptor antagonists exhibited analgesic activity in a variety of pain modalities in both preclinical and clinical studies.17,18 In contrast, KA receptor dysfunction and its associated abnormalities in glutamic acid transmission were shown to disrupt the finely-tuned process of neurotransmission, and contribute to the onset of pain.19–21
In line with the findings that dysregulated KA receptor function or abundance can contribute to pain sensitization, recent studies suggested that targeting KA receptor dysfunctional may also play a central role in attenuating pain.22 Here, we reviewed recent studies pertaining to the regulation of KA receptors and associated proteins at the cellular level, and explored the correlations between such regulation with pain transmission and sensitization. Through this work, we summarized what is known, what remains to be determined, and provided a framework for future research of KA receptors and their roles in pain.
KA receptor structure and function – A brief overview
KA receptors are homo- or heteromeric tetramers which can be composed of five different subunit proteins including GluK1, GluK2, GluK3, GluK4, and GluK5, previously also known as GluR5, GluR6, GluR7, KA1, and KA2.23 Each subunit protein is composed of an extracellular N-terminal domain (NTD), an intracellular C-terminal domain (CTD), and four hydrophobic domains (M1-M4), with M1, M3, and M4 being transmembrane domains and M2 forming a hairpin loop in the pore-forming domain.23,24 The ligand-binding domain (LBD) is formed by the terminal 150 amino acids of the NTD together with the extracellular loop between M3 and M4. These subunits have been groups into low-affinity (GluK1-3) and high-affinity categories (GluK4-5) based upon ligand affinity. In recombinant model systems, GluK1-3 subunits can form functional homomeric or heteromeric ion channels, whereas GluK4 and GluK5 must partner with GluK1-3 subunits to form functional receptors.25 KA receptor crystal structures have recently been unraveled, providing a wealth of detailed structural information which has been summarized previously.26 Briefly, the transmembrane domains of these subunits form the ion channel pore, while the CTD is important to downstream interactions with auxiliary/scaffold proteins and for synaptic incorporation. The extracellular domain of the receptor is commonly utilized for developing antibodies for immunohistochemistry staining.26,27
Physiological studies have shown that KA receptors serve as key regulators of synaptic transmission and plasticity.28,29 These receptors mediate postsynaptic depolarization and neuronal excitation. In specific excitatory synapse subsets, they can also carry a portion of the synaptic current.30 KA receptors can also function as modulators of the presynaptic release of neurotransmitters including both glutamate and γ-aminobutyric acid (GABA).31,32 In addition, they can facilitate macromolecule and molecular aggregate anchoring, thereby influencing long-term synaptic plasticity in the hippocampus, cortex, and amygdalas.33,34
Owing to a range of regulatory actions, KA receptors can profoundly influence the homeostatic balance between inhibition and excitation in neuronal networks.35 Accordingly, KA receptor dysfunction and dysregulation may drive the development of pathological conditions, such as pain. The expression of functional KA receptors has been detected via immunohistochemistry and electrophysiology along pain neuraxis including the DRG (Dorsal Root Ganglion), spinal cord dorsal horn, thalamus, and cortex wherein they control nociceptive transmission and pain modulation.36 The development of research tools including KA receptor agonist, antagonist, and transgenic mice in which these receptors were knocked out helped to unravel important roles of KA receptor dysregulation and dysfunction in peripheral and central sensitization in the context of pain.37,38 Intriguingly, besides functioning as ion channels, KA receptors were also found to activate certain G-proteins, thereby may influence long-term changes in synaptic transmission and plasticity, underscoring a potential dual signaling mechanisms for KA receptors to regulate pain.39,40 The details of mechanistic understanding for such noncanonical metabotropic signaling, and the factors that determine the roles of KA receptors in the context of pain remain unclear.
The post-transcriptional regulation of KA receptors
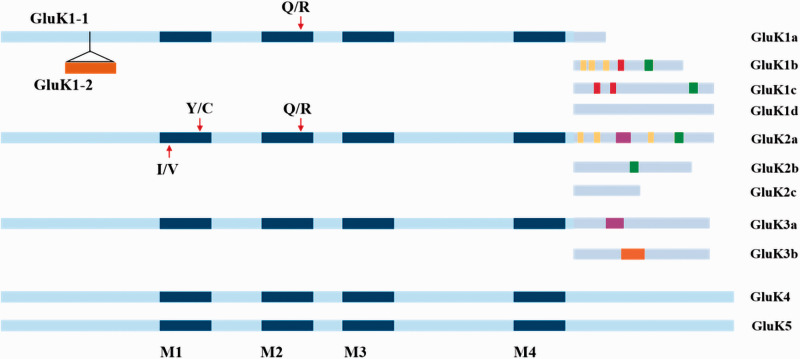
The alternative splicing of the GluK1-3 subunits gives rise to additional KA receptor isoforms, whereas GluK4 and GluK5 were not thought to undergo alternative splicing, Figure 1.41 NTD and CTD regions of GluK1-3 subunits are the primary sites of alternative RNA splicing. For example, the GluK1 extracellular NTD can be alternatively spliced to give rise to the GluK1-1 and GluK1-2 variants, while the CTD exhibits four such variants (GluK1a, GluK1b, GluK1c, and GluK1d).42,43 GluK2 and GluK3 similarly exhibit CTD splice variants (GluK2a/GluK2b/GluK2c and GluK3a/GluK3b, respectively).41,44 These splice variants are associated with significant changes in KA receptor exit from the ER (Endoplasmic Reticulum) and surface accumulation, thereby receptor function and neuronal excitability. Because these variants also enable altered interactions between proteins, they may tune KA receptor function in a site-specific manner.45–47
Figure 1.
Splice variants and RNA editing of KA receptors. GluK1 extracellular NTD can be alternatively spliced to give rise to the GluK1-1 and GluK1-2 variants. The CTD exhibits four such variants (GluK1a, GluK1b, GluK1c, and GluK1d). GluK2 and GluK3 similarly exhibit CTD splice variants (GluK2a/GluK2b/GluK2c and GluK3a/GluK3b, respectively). GluK4 and GluK5 may not undergo alternative splicing. Splice variants are depicted including reported interaction sites with other proteins, post-translationally modified residues, and trafficking motifs. Color index: red: retention motif; purple: forward trafficking motif; orange: endocytosis; yellow: post-translational modification; green: residues or regions reported to interact with other proteins. Blue (dark or light) bars within schematic drawings of receptors represent either transmembrane domains or the re-entrant loop. Editing sites for GluK1 and GluK2 can undergo editing at the “Q/R” site, while GluK2 can also be edited at “I/V” and “Y/C” sites in the M1 transmembrane domain, thereby altering amino acid substitutions at critical sites within these receptor subunits.
A significant amount of research has been conducted to evaluate how splice variants of growth factors and ion channels affect pain.48 For example, three TRPV1 (Transient Receptor Potential Vanilloid-1) splice variants were found to in the DRG or trigeminal ganglia, and may play a role in nociceptive processing. Splicing affects the N-terminal domain in these splice variants, resulting in a loss of activation by capsaicin and other activators such as protons, resiniferatoxin, or temperature, thus yield a dominant negative channel.49 The expression of different splice variants of voltage-gated calcium channel, particularly Cav2.2, is also enriched in nociceptors. The presence of variants also increases sensitivity to neuronal inhibition through opioid and GABA receptors.50 NMDA receptor splice variants (including NR1-1b, NR1-3b, and NR1-4b) colocalize with NK1 (Neurokinin-1) receptors in projection neurons of the spinal cord dorsal horn, indicating that they may play a role in spinal nociceptive processing.51,52 Yet, formalin-induced nociception was not affected by alternative NR1 splicing.53 In addition, no change in pain behavior or anxiety was observed in mice after knocking in a mutant mGluR7 splice variant (mGluR7a) which lacks the PDZ ((PSD-95, Drosophila disc large tumor suppressor (Dlg1), and zonula occludens-1 protein (zo-1))) domain, indicating that function of this isoform is not required for normal nociceptive processing.54 However, relatively little is known regarding the roles of KA receptor splice variants in pain. Research is needed to establish whether KA receptor splice variants may play a role in the development of pain.
The RNA editing of GluK1 and GluK2 subunit further broadens the KA receptor functional repertoire. GluK1 can undergo editing at the channel pore-forming P-loop (the ‘Q/R’ site), and GluK2 can be edited at the I/V and Y/C sites in the M1 transmembrane domain wherein an isoleucine (ATT) is replaced with a valine (ITT) and a tyrosine (TAC) is replaced with a cysteine (TIC), Figure 1.55,56 These modifications result in amino acid substitutions at critical sites within the subunits, thereby altering ion channel properties and influencing the degree to which KA receptors allow Ca2+ ions to pass through.57 Since Q/R editing can impair the oligomerization, ER export, cell surface expression, and stability of homomeric GluK2 KA receptors, KA receptor editing is also linked to the trafficking of these receptors to the cell surface.58 The susceptibility of these KA receptors to cytoplasmic polyamine- and cis-unsaturated fatty acid-mediated inhibition is regulated by Q/R site editing.59 RNA editing thus controls KA receptor synaptic activity. Importantly, increasing amount of evidence suggests that this form of post-transcriptional regulation is disrupted in the context of pain.60,61 For example, Guo et al. showed that increased levels of GluK2 (Q) species can facilitate inflammatory hyperalgesia. Because GluK2 subunit Q/R editing suppresses the ability of calcium to pass through KA receptor channels, an increase of GluK2 (Q) variants will increase calcium influx in spinal cord neurons and enhance channel conductance after inflammation. The resultant rises in intracellular calcium level in turn trigger kinase and receptor phosphorylation, thereby bolstering neuronal excitability.62 RNA editing has also been shown to influence neuropathic pain in an L5 spinal nerve transection (SNT) model. SNT markedly decreased the Q/R editing of GluK2 mediated by adenosine deaminase acting on RNA (ADAR, Adenosine Deaminase Acting on RNA) enzyme in the injured DRG neurons. Furthermore, targeting of these ADAR enzymes was sufficient to achieve pain inhibition in SNT model.22 Overall, these studies provided important evidence that post-transcriptional modifications of KA receptors significantly altered receptor functionality, highlighting a warrant of future studies of these modifications as targets for pain treatment.
The post-translational regulation of KA receptors
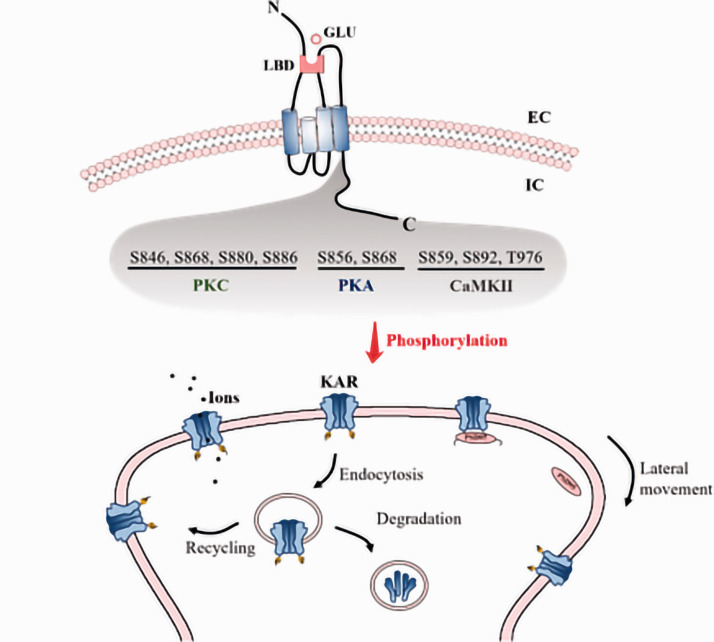
KA receptors also undergo post-translational modifications such as phosphorylation, which is the most common form of post-translational modification that can alter protein activity, localization, and interactions with other proteins.63,64 A number of residues within KA receptor subunits were shown to be phosphorylated, including S846, S856, S859, S868 S880, S886, S892, and T976.32 Of these, protein kinase C (PKC)-mediated S846, S868, S880, and S886 phosphorylation has been shown to directly impact receptor function. For example, Dildy-Mayfield and Harris first demonstrated that PKC phosphorylated recombinant GluK2, thereby reducing kainite-evoked currents.65 In contrast, Cho et al. showed that activation of PKC by mGluR5 enhanced GluK1-containing KA receptor-mediated excitatory postsynaptic potential (EPSP) in perirhinal cortical neurons.66 KA receptor activation further stimulated PKC-induced GluK1-2b S880 and/or S886 phosphorylation, leading to the internalization of these subunits.67 Evidence also showed that PKC-mediated S868 phosphorylation was associated with endocytosed GluK2 recycling back to the plasma membrane, suggesting that the phosphorylation may regulate the membrane localization of this subunit in a context-dependent manner.68 Such a bidirectional signaling may serve as an important feedback mechanism to prevent KA receptor-mediated neuronal overactivation.67
In addition to PKC, cAMP-dependent protein kinase (PKA) can directly modulate KA receptor functionality. For example, PKA-mediated GluK2 S856 and S868 phosphorylation was shown to potentiate kainite-evoked currents through recombinant KA receptors.69–72 In parallel with the actions of PKA and PKC, CaMKII-induced GluK5 S859, S892, and T976 phosphorylation uncouples these KA receptors from postsynaptic density 95 (PSD-95), improving the overall lateral mobility of these receptors by freeing them from synaptic incorporation, Figure 2.71
Figure 2.
Post-translational modifications (phosphorylation) of KA receptors. A number of residues within KA receptor subunits were shown to be phosphorylated, including S846/S868/S880/S886 (PKC), S856/S868 (PKA), S859/S892/T976 (CaMKII). All of these phosphorylation is shown to directly impact the kainite-evoked currents, and is also illustrated to be related to the endocytosis of KA receptors, leading to recycling of KA receptors to the membrane or degradation. Moreover, phosphorylation uncouples KA receptors from postsynaptic density 95 (PSD-95), improving the overall lateral mobility of these receptors by freeing them from synaptic incorporation. LBD ligand binding domain, GLU glutamate, N N-terminal domain, C C-terminal domain, P phosphorylation, EC extracellular, IC intracellular.
Overall, KA receptor phosphorylation is a key regulator of the trafficking of these receptors to synapses, and thus affect synaptic plasticity with respect to both integration and transmission. Consequently, KA receptor phosphorylation plays an important role in long term synaptic plasticity. For example, postsynaptic KA receptors at thalamocortical synapses were rapidly downregulated during the induction of long term potentiation via a mechanism that requires PKC. In perirhinal cortex layer 2/3 pyramidal neurons, a form of long term depression was associated with decreased KA receptor activation, characterized by a rapid reduction in KA receptor-mediated synaptic transmission. Interestingly, this long term depression also requires PKC.73 Recent work suggested a role of GluK2 phosphorylation in regulating KA receptor channel opening and subsequent pro-apoptotic signaling in the context of brain ischemia, suggesting it may play a role in the context of ischemic stroke.74 AMPA/KA receptor-mediated PKA and PKC activation is also required for pain sensation.72 Given that KA receptors can further drive PKC-mediated phosphorylation of KA receptors, this post-translational regulatory mechanism may contribute to thermal stimulus-evoked allodynia. Future research needs to establish whether this mechanism may be targeted to regulate neuronal hyperexcitability in the context of pain.
The regulation of KA receptor activity by interacting proteins
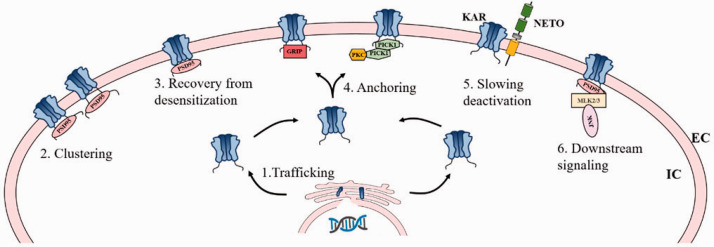
KA receptors do not function in a vacuum. Instead, they participate in a large macromolecular complex at the plasma membrane surface that contains trafficking chaperones, molecular scaffolds, and signaling enzymes capable of shaping the nature of the downstream responses.75 KA receptor-interacting proteins include BTB-Kelch or PDZ/CUB domain-containing proteins.76–78 The major interactions of auxiliary proteins with KA receptors and their relevance in the context of pain are summarized below, Figure 3.
Figure 3.
Schematic of domain organization of PSD95, GRIP, PICK1, and Netos with KA receptors. PDZ domain proteins (PSD95, GRIP and PICK1) are shown to interact with KA receptors through the PDZ domain-mediated interactions, which are necessary for the appropriate regulation of KA receptor-mediated synaptic functionality. Interactions with PSD95 enable KA receptors to recover more rapidly following receptor desensitization. Such interactions also drive KA receptor clustering, and KA receptors-PSD95 complex can additionally interact with mixed-lineage kinases 2 and 3 (MLK2 and MLK3), which drives JNK kinase activation. GRIP regulates KA receptor anchoring at synapses, and that the PICK1-targeted phosphorylation of KA receptors by PKC stabilizes GRIP binding. NETOs (NETO1/2) interact with KA receptors via their PDZ-ligand domains and CUB domains, thereby forming stable complexes with KA receptors. Netos interaction with KA receptors typically slow KA receptor deactivation kinetics, and such interactions also represents a regulator of KA receptor trafficking. EC extracellular, IC intracellular.
PDZ domain proteins
PSD-95 interacts with KA receptors through the PDZ domain-mediated interactions,79 interacting with PDZ domains within the CTD of GluK1, GluK2, and GluK5.80 Interactions with PSD-95 enable GluK2 homomeric receptors and GluK2/GluK5 heteromeric receptors to recover more rapidly following receptor desensitization.81 Such PSD-95 interactions drive KA receptor clustering, as evidenced by the reduction in KA receptors at MF-CA3 synapses in mice lacking PSD-95 expression.82 GluK5 and PSD-95 interaction are also necessary for long-term depression at MF-CA3 synapses.71
Owing to these regulatory functions of KA receptors, the interactions between KA receptors and PSD-95 were also suggested to be involved in a range of pathological contexts. For example, a PDZ inhibitor peptide was shown to protect against the neuron apoptotic death due to ischemia/reperfusion, suggesting that disrupting GluK2-PSD-95 interactions may represent an effective approach for neuroprotection.83 The GluK2-PSD-95 complex can additionally interact with mixed-lineage kinases 2 and 3 (MLK2 and MLK3), which bind to the PSD-95 Src homology 3(SH3) domain.84 The resultant interaction then drives JNK kinase activation,85,86 which is known to be important to chronic inflammatory pain and neuropathic pain.87,88 Interactions between KA receptors and PSD-95 may thus regulate pain at multiple steps by both directly impacting the functionality of these receptors and by influencing downstream signaling pathways.
A number of other PDZ domain-containing proteins may also interact with KA receptors to influence their functionality. For example, Hirbec et al. showed that GluK1-2b, GluK1-2c, and GluK2 bind to the PDZ domain-containing proteins PICK1 (protein interacting with PRKCA 1), GRIP (glutamate receptor-interacting protein 1), and syntenin via their CTDs.80 GRIP and PICK1 are necessary for the appropriate regulation of KA receptor-mediated synaptic functionality at mossy fiber-CA3 synapses.80 Disrupting these interactions interferes with synaptic transmission facilitated by these KA receptors. GRIP can also directly bind to kinesin motor proteins, indicating it may control the transportation and trafficking of KA receptors.89 Hiberc et al. also suggested that GRIP regulates KA receptor anchoring at synapses, and that the PICK1-targeted phosphorylation of GluK1-2b S880 and/or S886 by PKC stabilizes GRIP binding, as evidenced by the disruption of KA receptor-mediated currents from inhibiting PICK1 interaction or PKC activity.80 A lack of appropriate interactions between PDZ domain-containing protein and KA receptors thus interferes KA receptor plasma membrane stability and functionality, which may lead to network instability, neuron hyperexcitability, and pathological changes including pain.
CUB domain proteins
Neto1 and Neto2 are neuropilin- and tolloid-like (Neto) proteins that contain CUB (complement subcomponent C1r, C1s/sea urchin embryonic growth factor Uegf/bone morphogenetic protein 1)-domains,90 and interact with KA receptors. Both Neto1 and Neto2 are auxiliary proteins that interact with scaffolding proteins via their PDZ-ligand domains and CUB domains, thereby forming stable complexes with KA receptors.91 How these Neto proteins impact the properties of KA receptor channels has been shown previously.92–95 In brief, Neto proteins typically slow KA receptor deactivation kinetics, explaining why these receptors exhibited distinct properties in vivo from that in cell lines which are lack of Neto protein expression. These Neto proteins additionally control neuronal network inhibition via regulating somatodendritic and presynaptic KA receptors in somatostatin, cholecystokinin, cannabinoid receptor 1, and parvalbumin-containing interneurons.96 Yet, the specific roles of Neto proteins in the context of KA receptor trafficking and synaptic incorporation remain to be defined. Early studies suggested that Neto1/2 had minimal impact on GluK2 surface expression in a heterologous system, nor were GluK2/GluK5 abundance impacted in PSD fractions obtained from mice lacking Neto1 expression.97,98 In addition, Neto1 and Neto2 co-expression failed to bolster exogenous CA1 pyramidal neuron KA receptor responses, even though these cells typically lack postsynaptic KA receptor EPSCs. These findings suggest that Neto proteins have no impact on GluK2-containing KA receptor synaptic incorporation.99 However, hippocampal synaptic GluK2 was found to be reduced in Neto1 knockout mice, and similar findings were found in cerebellar PSD fractions from mice lacking Neto2, indicating that Neto proteins may be important regulators of GluK2 synaptic targeting.100,101 Neto1 and Neto2 have also been shown to increase the cell surface expression of GluK2 in HEK293 cells, and injecting GluK2 and Neto2 into oocytes also enhanced the surface expression of GluK2.102 These data suggest that Neto proteins may play a role in controlling KA receptor cell surface localization in at least certain contexts.
Given these conflicting findings, exactly how Neto proteins influence the trafficking of GluK2-containing KA receptors remains to be established. The variable model systems used in previous studies may partially cause the discrepancy. In addition, this may also reflect the complex nature of interactions between KA receptors and Neto proteins, which can be impacted by differential subunit expression and cell type-specific interacting protein expression. GluK2 undergoes a range of post-translational modifications such as phosphorylation, ubiquitination, SUMOylation, and palmitoylation, all of which can have a direct or indirect impact on Neto protein activity.103,104 For instance, phosphor-deficient mutant Neto2 S409A impeded GluK1 trafficking to synapses, suggesting that Neto2 Ser-409 phosphorylation inhibited synaptic targeting of GluK1.105 Disrupting Neto protein activity may thus adversely impact KA receptor functionality and the synaptic networks regulated by these receptors.
Several studies have highlighted the impact of such disruptions in pathological contexts. For example, Neto2-knockout mice exhibit decreased pentylenetetrazole (PTZ)-induced seizure latency and increased severity of seizures.106 Furthermore, Sargin D suggested that Neto2-mediated KA receptor modulation is a key driver of fear memory in mice. They found that a lack of Neto2 expression was associated with decreased synaptic KA receptor subunit accumulation at synapses in the brain fear center, driving the development of behavior phenotypes consistent with post-traumatic stress disorder in animals.107 Vernon and Swanson also found that mice lacking Neto1 and Neto2 expression exhibited normal thresholds for acute thermal and mechanical pain, indicating that KA receptors are not important to acute pain signaling. In contrast, a delayed upregulation of Neto2 following sciatic nerve crush indicated that KA receptors may contribute to the development of chronic neuropathic pain which is Neto-dependent.108
In addition to Neto1/2, SEZ6 (Seizure protein 6) is another CUB domain-containing protein that is capable of interacting with KA receptors.109 Membrane proteome analyses conducted in neurons lacking SEZ6 expression revealed that cell surface GluK2 and GluK3 levels were selectively reduced, and that GluK2 post-ER transport in the secretory pathway had been altered in these neurons. SEZ6 knockout also decreased kainite-evoked currents in CA1 pyramidal neurons in acute hippocampal sections.110 From a mechanistic perspective, SEZ6 may represent a regulator of KA receptor trafficking. When interactions between these proteins and KA receptors are dysregulated, neuropathies caused by KA receptor-dysfunction may develop. Notably, SEZ6 proteins are widely expressed throughout the brain, and have been implicated in neurodevelopmental and psychiatric disorders.111 It was shown that a lack of SEZ6 family proteins also impaired motor functions, short-term memory, and cognitive flexibility.112 Most importantly, recent evidence suggested that SEZ6 is a driver of the onset of inflammatory hyperalgesia.113 Although there has been no direct evidence supporting the role of SEZ6 interactions with KA receptors in pain, the aforementioned findings strongly suggest the notion that SEZ6 may control KA receptor functionality in this context.
Conclusions
Herein, we provide an updated overview about roles of KA receptors in synaptic transmission and plasticity, through regulating of the receptor trafficking toward and from the synaptic membrane, ion channel gating, and downstream signaling. KA receptor function can be regulated via a host of mechanisms including post-transcriptional RNA editing and alternative splicing, post-translational phosphorylation, and interactions with accessory proteins including PSD-95, NETO, and SEZ6. These regulatory mechanisms have been suggested to play a role in nociceptive signaling, pain transmission and sensitization, highlighting KA receptors as a promising target of pharmacological interventions for pain treatment.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Natural Science Foundation of China (81771181, 81571065); the Beijing Natural Science Foundation (7202053); Beijing Hospitals Authority Youth Program (QML20180105); and Scientific Research Common Program of Beijing Municipal Commission of Education (KM201910025018).
ORCID iDs: Huili Li https://orcid.org/0000-0001-9316-4087
References
- 1.Lee GI, Neumeister MW. Pain: pathways and physiology. Clin Plast Surg 2020; 47: 173–180. [DOI] [PubMed] [Google Scholar]
- 2.Nijs J, Leysen L, Vanlauwe J, Logghe T, Ickmans K, Polli A, Malfliet A, Coppieters I, Huysmans E. Treatment of central sensitization in patients with chronic pain: time for change? Expert Opin Pharmacother 2019; 20: 1961–1970. [DOI] [PubMed] [Google Scholar]
- 3.Kung LH, Gong K, Adedoyin M, Ng J, Bhargava A, Ohara PT, Jasmin L. Evidence for glutamate as a neuroglial transmitter within sensory ganglia. PLoS One 2013; 8: e68312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen TJ, Kukley M. Glutamate receptors and glutamatergic signalling in the peripheral nerve. Neural Regen Res 2020; 15: 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu W, Jiang X, Zu Y, Yang Y, Liu Y, Sun X, Xu Z, Ding H, Zhao Q. A comprehensive description of GluN2B-selective N-methyl-D-aspartate (NMDA) receptor antagonists. Eur J Med Chem 2020; 200: 112447. [DOI] [PubMed] [Google Scholar]
- 6.Griego E, Galván EJ. Metabotropic glutamate receptors at the aged mossy fiber – CA3 synapse of the hippocampus. Neuroscience 2020; S0306–4522(19)30864-4. [DOI] [PubMed] [Google Scholar]
- 7.Lutzu S, Castillo PE. Modulation of NMDA receptors by G-protein-coupled receptors: role in synaptic transmission, plasticity and beyond. Neuroscience 2020; S0306–4522(20)30105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warnet XL, Bakke Krog H, Sevillano-Quispe OG, Poulsen H, Kjaergaard M. The C-terminal domains of the NMDA receptor: how intrinsically disordered tails affect signalling, plasticity and disease. Eur J Neurosci Epub ahead of print 2020. DOI:10.1111/ejn.14842 [DOI] [PubMed]
- 9.Lüscher C, Frerking M. Restless AMPA receptors: implications for synaptic transmission and plasticity. Trends Neurosci 2001; 24: 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Bramham CR. Bidirectional dysregulation of AMPA receptor-mediated synaptic transmission and plasticity in brain disorders. Front Synaptic Neurosci 2020; 12: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Childers WE, Jr, Baudy RB. N-methyl-D-aspartate antagonists and neuropathic pain: the search for relief. J Med Chem 2007; 50: 2557–2562. [DOI] [PubMed] [Google Scholar]
- 12.Kurian R, Raza K, Shanthanna H. A systematic review and meta-analysis of memantine for the prevention or treatment of chronic pain. Eur J Pain 2019; 23: 1234–1250. [DOI] [PubMed] [Google Scholar]
- 13.Hara K, Haranishi Y, Terada T. Intrathecally administered perampanel alleviates neuropathic and inflammatory pain in rats. Eur J Pharmacol 2020; 872: 172949. [DOI] [PubMed] [Google Scholar]
- 14.Falcón-Moya R, Sihra TS, Rodríguez-Moreno A. Kainate receptors: role in epilepsy. Front Mol Neurosci 2018; 11: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuo M. Cortical. Cortical kainite receptors and behavioral anxiety. Mol Brain 2017; 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhangoo SK, Swanson GT. Kainate receptor signaling in pain pathways. Mol Pharmacol 2013; 83: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen AM, Bunch L. Medicinal chemistry of competitive kainite receptor antagonists. ACS Chem Neurosci 2011; 2: 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matute C. Therapeutic potential of kainite receptors. CNS Neurosci Ther 2011; 17: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takenouchi T, Hashida N, Torii C, Kosaki R, Takahashi T, Kosaki K. 1p34.3 deletion involving GRIK3: further clinical implication of GRIK family glutamate receptors in the pathogenesis of developmental delay. Am J Med Genet A 2014; 164A: 456–460. [DOI] [PubMed] [Google Scholar]
- 20.Bliss TV, Collingridge GL, Kaang BK, Zhuo M. Synaptic plasticity in the anterior cingulate cortex in acute and chronic pain. Nat Rev Neurosci 2016; 17: 485–496. [DOI] [PubMed] [Google Scholar]
- 21.Zhuo M. Ionotropic glutamate receptors contribute to pain transmission and chronic pain. Neuropharmacology 2017; 112: 228–234. [DOI] [PubMed] [Google Scholar]
- 22.Uchida H, Matsumura S, Okada S, Suzuki T, Minami T, Ito S. RNA editing enzyme ADAR2 is a mediator of neuropathic pain after peripheral nerve injury. FASEB J 2017; 31: 1847–1855. [DOI] [PubMed] [Google Scholar]
- 23.Valbuena S, Lerma J. Kainate receptors, homeostatic gatekeepers of synaptic plasticity. Neuroscience 2019; S0306–4522(19)30840-1. [DOI] [PubMed] [Google Scholar]
- 24.Fu H, Chen Z, Josephson L, Li Z, Liang SH. Positron emission tomography (PET) ligand development for ionotropic glutamate receptors: Challenges and opportunities for radiotracer targeting N-Methyl-d-aspartate (NMDA), α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), and kainate receptors. J Med Chem 2019; 62: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrais D, Veran J, Mulle C. Gating and permeation of kainate receptors: differences unveiled. Trends Pharmacol Sci 2010; 31: 516–522. [DOI] [PubMed] [Google Scholar]
- 26.Møllerud S, Frydenvang K, Pickering DS, Kastrup JS. Lessons from crystal structures of kainate receptors. Neuropharmacology 2017; 112: 16–28. [DOI] [PubMed] [Google Scholar]
- 27.Karakas E, Regan MC, Furukawa H. Emerging structural insights into the function of ionotropic glutamate receptors. Trends Biochem Sci 2015; 40: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sihra TS, Flores G, Rodríguez-Moreno A. Kainate receptors: multiple roles in neuronal plasticity. Neuroscientist 2014; 20: 29–43. [DOI] [PubMed] [Google Scholar]
- 29.Huettner JE. Kainate receptors and synaptic transmission. Prog Neurobiol 2003; 70: 387–407. [DOI] [PubMed] [Google Scholar]
- 30.Chamberlain SE, Jane DE, Jones RS. Pre- and post-synaptic functions of ainite receptors at glutamate and GABA synapses in the rat entorhinal cortex. Hippocampus 2012; 22: 555–576. [DOI] [PubMed] [Google Scholar]
- 31.Sihra TS, Rodríguez-Moreno A. Presynaptic kainate receptor-mediated bidirectional modulatory actions: mechanisms. Neurochem Int 2013; 62: 982–987. [DOI] [PubMed] [Google Scholar]
- 32.Pahl S, Tapken D, Haering SC, Hollmann M. Trafficking of kainate receptors. Membranes (Basel) 2014; 4: 565–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihály A. The reactive plasticity of hippocampal ionotropic glutamate receptors in animal epilepsies. Int J Mol Sci 2019; 20: 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Contractor A, Mulle C, Swanson GT. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci 2011; 34: 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerma J, Marques JM. Kainate receptors in health and disease. Neuron 2013; 80: 292–311. [DOI] [PubMed] [Google Scholar]
- 36.Wu LJ, Ko SW, Zhuo M. Kainate receptors and pain: from dorsal root ganglion to the anterior cingulate cortex. Curr Pharm Des 2007; 13: 1597–1605. [DOI] [PubMed] [Google Scholar]
- 37.Chappell AS, Iyengar S, Lobo ED, Prucka WR. Results from clinical trials of a selective ionotropic glutamate receptor 5 (iGluR5) antagonist, LY5454694 tosylate, in 2 chronic pain conditions. Pain 2014; 155: 1140–1149. [DOI] [PubMed] [Google Scholar]
- 38.Qiu CS, Lash-Van Wyhe L, Sasaki M, Sakai R, Swanson GT, Gereau IV RW. Antinociceptive effects of MSVIII-19, a functional antagonist of the GluK1 kainate receptor. Pain 2011; 152: 1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negrete-Díaz JV, Sihra TS, Flores G, Rodríguez-Moreno A. Non-canonical mechanisms of presynaptic kainate receptors controlling glutamate release. Front Mol Neurosci 2018; 11: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rozas JL. Metabotropic actions of kainate receptors in dorsal root ganglion cells. Adv Exp Med Biol 2011; 717: 69–80. [DOI] [PubMed] [Google Scholar]
- 41.Jaskolski F, Coussen F, Nagarajan N, Normand E, Rosenmund C, Mulle C. Subunit composition and alternative splicing regulate membrane delivery of kainate receptors. J Neurosci 2004; 24: 2506–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bettler B, Boulter J, Hermans-Borgmeyer I, O'Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 1990; 5: 583–595. [DOI] [PubMed] [Google Scholar]
- 43.Pinheiro P, Mulle C. Kainate receptors. Cell Tissue Res 2006; 326: 457–482. [DOI] [PubMed] [Google Scholar]
- 44.Schiffer HH, Swanson GT, Heinemann SF. Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron 1997; 19: 1141–1146. [DOI] [PubMed] [Google Scholar]
- 45.Ren Z, Riley NJ, Needleman LA, Sanders JM, Swanson GT, Marshall J. Cell surface expression of GluR5 kainate receptors is regulated by an endoplasmic reticulum retention signal. J Biol Chem 2003; 278: 52700–52709. [DOI] [PubMed] [Google Scholar]
- 46.Coussen F, Perrais D, Jaskolski F, Sachidhanandam S, Normand E, Bockaert J, Marin P, Mulle C. Co-assembly of two GluR6 kainate receptor splice variants within a functional protein complex. Neuron 2005; 47: 555–566. [DOI] [PubMed] [Google Scholar]
- 47.Jaskolski F, Normand E, Mulle C, Coussen F. Differential trafficking of GluR7 kainate receptor subunit splice variants. J Biol Chem 2005; 280: 22968–22976. [DOI] [PubMed] [Google Scholar]
- 48.Hulse RP, Drake RA, Bates DO, Donaldson LF. The control of alternative splicing by SRSF1 in myelinated afferents contributes to the development of neuropathic pain. Neurobiol Dis 2016; 96: 186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu G, Henderson D, Liu L, Reinhart PH, Simon SA. TRPV1b, a functional human vanilloid receptor splice variant. Mol Pharmacol 2005; 67: 1119–1127. [DOI] [PubMed] [Google Scholar]
- 50.Bates DO, Morris JC, Oltean S, Donaldson LF. Pharmacology of modulators of alternative splicing. Pharmacol Rev 2017; 69: 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luque JM, Bleuel Z, Malherbe P, Richards JG: Alternatively spliced isoforms of the N-methyl-D-aspartate receptor subunit 1 are differentially distributed within the rat spinal cord. Neuroscience 1994; 63: 629–635. [DOI] [PubMed] [Google Scholar]
- 52.Benoliel R, Tanaka M, Caudle RM, Iadarola MJ. Co-localization of N-methyl-D-aspartate receptors and substance P (neurokinin-1) receptors in rat spinal cord. Neurosci Lett 2000; 291: 61–64. [DOI] [PubMed] [Google Scholar]
- 53.Gaunitz C, Schüttler A, Gillen C, Allgaier C. Formalin-induced changes of NMDA receptor subunit expression in the spinal cord of the rat. Amino Acids 2002; 23: 177–182. [DOI] [PubMed] [Google Scholar]
- 54.Donaldson LF, Beazley-Long N. Alternative RNA splicing: contribution to pain and potential therapeutic strategy. Drug Discov Today 2016; 21: 1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Köhler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron 1993; 10: 491–500. [DOI] [PubMed] [Google Scholar]
- 56.Barbon A, Barlati S. Glutamate receptor RNA editing in health and disease. Biochemistry (Mosc) 2011; 76: 882–889. [DOI] [PubMed] [Google Scholar]
- 57.Egebjerg J, Heinemann SF. Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc Natl Acad Sci USA 1993; 90: 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ball SM, Atlason PT, Shittu-Balogun OO, Molnár E. Assembly and intracellular distribution of kainate receptors is determined by RNA editing and subunit composition. J Neurochem 2010; 114: 1805–1818. [DOI] [PubMed] [Google Scholar]
- 59.Wilding TJ, Zhou Y, Huettner JE. Q/R site editing controls kainate receptor inhibition by membrane fatty acids. J Neurosci 2005; 25: 9470–9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Filippini A, Bonini D, La Via L, Barbon A. The good and the bad of glutamate receptor RNA Editing. Mol Neurobiol 2017; 54: 6795–6805. [DOI] [PubMed] [Google Scholar]
- 61.Gurung S, Evans AJ, Wilkinson KA, Henley JM. ADAR2-mediated Q/R editing of GluK2 regulates kainate receptor upscaling in response to suppression of synaptic activity. J Cell Sci 2018; 131: jcs222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo W, Zou S, Tal M, Ren K. Activation of spinal kainate receptors after inflammation: behavioral hyperalgesia and subunit gene expression. Eur J Pharmacol 2002; 452: 309–318. [DOI] [PubMed] [Google Scholar]
- 63.Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int J Mol Med 2017; 40: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W, Zhang W, Wang X. Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol J 2017; 15: 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dildy-Mayfield JE, Harris RA. Activation of protein kinase C inhibits kainate-induced currents in oocytes expressing glutamate receptor subunits. J Neurochem 1994; 62: 1639–1642. [DOI] [PubMed] [Google Scholar]
- 66.Cho K, Francis JC, Hirbec H, Dev K, Brown MW, Henley JM, Bashir ZI. Regulation of kainate receptors by protein kinase C and metabotropic glutamate receptors. J. Physiol 2003; 548: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rivera R, Rozas JL, Lerma J. PKC-dependent autoregulation of membrane kainate receptors. EMBO J 2007; 26: 4359–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chamberlain SE, González-González IM, Wilkinson KA, Konopacki FA, Kantamneni S, Henley JM, Mellor JR. SUMOylation and phosphorylation of GluK2 regulate kainate receptor trafficking and synaptic plasticity. Nat Neurosci 2012; 15: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Traynelis SF, Wahl P. Control of rat GluR6 glutamate receptor open probability by protein kinase A and calcineurin. J Physiol 1997; 503: 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falcón-Moya R, Losada-Ruiz P, Rodríguez-Moreno A. Kainate receptor-mediated depression of glutamate release involves protein kinase A in the cerebellum. Int J Mol Sci 2019; 20: 4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carta M, Opazo P, Veran J, Athané A, Choquet D, Coussen F, Mulle C. CaMKII-dependent phosphorylation of GluK5 mediates plasticity of kainate receptors. EMBO J 2013; 32: 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones TL, Sorkin LS. Activated PKA and PKC, but not CaMKIIalpha, are required for AMPA/Kainate-mediated pain behavior in the thermal stimulus model. Pain 2005; 117: 259–270. [DOI] [PubMed] [Google Scholar]
- 73.Nasu-Nishimura Y, Jaffe H, Isaac JT, Roche KW. Differential regulation of kainate receptor trafficking by phosphorylation of distinct sites on GluR6. J Biol Chem 2010; 285: 2847–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu QJ, Kong FS, Xu H, Wang Y, Du CP, Sun CC, Liu Y, Li T, Hou XY. Tyrosine phosphorylation of GluK2 up-regulates kainate receptor-mediated responses and downstream signaling after brain ischemia. Proc Natl Acad Sci U S A 2014; 111: 13990–13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheng N, Shi YS, Nicoll RA. Amino-terminal domains of kainate receptors determine the differential dependence on Neto auxiliary subunits for trafficking. Proc Natl Acad Sci USA 2017; 114: 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheng N, Bemben MA, Díaz-Alonso J, Tao W, Shi YS, Nicoll RA. LTP requires postsynaptic PDZ-domain interactions with glutamate receptor/auxiliary protein complexes. Proc Natl Acad Sci USA 2018; 115: 3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li YJ, Duan GF, Sun JH, Wu D, Ye C, Zang YY, Chen GQ, Shi YY, Wang J, Zhang W, Shi YS. Neto proteins regulate gating of the kainate-type glutamate receptor GluK2 through two binding sites. J Biol Chem 2019; 294: 17889–17902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marshall J, Blair LA, Singer JD. BTB-Kelch proteins and ubiquitination of kainate receptors. Adv Exp Med Biol 2011; 717: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron 1998; 21: 727–739. [DOI] [PubMed] [Google Scholar]
- 80.Hirbec H, Francis JC, Lauri SE, Braithwaite SP, Coussen F, Mulle C, Dev KK, Couthino V, Meyer G, Isaac JT, Collingridge GL, Henley JM. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron 2003; 37: 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bowie D, Garcia EP, Marshall J, Traynelis SF, Lange GD. Allosteric regulation and spatial distribution of kainate receptors bound to ancillary proteins. J Physiol 2003; 547: 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki E, Kamiya H. PSD-95 regulates synaptic kainate receptors at mouse hippocampal mossy fiber-CA3 synapses. Neurosci Res 2016; 107: 14–19. [DOI] [PubMed] [Google Scholar]
- 83.Yin XH, Yan JZ, Yang G, Chen L, Xu XF, Hong XP, Wu SL, Hou XY, Zhang G. PDZ1 inhibitor peptide protects neurons against ischemia via inhibiting GluK2-PSD-95-module-mediated Fas signaling pathway. Brain Res 2016; 1637: 64–70. [DOI] [PubMed] [Google Scholar]
- 84.Savinainen A, Garcia EP, Dorow D, Marshall J, Liu YF. Kainate receptor activation induces mixed lineage kinase-mediated cellular signaling cascades via post-synaptic density protein 95. J Biol Chem 2001; 276: 11382–11386. [DOI] [PubMed] [Google Scholar]
- 85.Hirai Si, Katoh M, Terada M, Kyriakis JM, Zon LI, Rana A, Avruch J, Ohno S. MST/MLK2, a member of the mixed lineage kinase family, directly phosphorylates and activates SEK1, an activator of c-Jun N-terminal kinase/stress-activated protein kinase. J Biol Chem 1997; 272: 15167–15173. [DOI] [PubMed] [Google Scholar]
- 86.Nagata Ki, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J 1998; 17: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Q, Liu S, Li L, Ji X, Wang M, Zhou J. Spinal IL-36γ/IL-36R participates in the maintenance of chronic inflammatory pain through astroglial JNK pathway. Glia 2019; 67: 438–451. [DOI] [PubMed] [Google Scholar]
- 88.Zhao Y, Xin Y, Chu H. MC4R is involved in neuropathic pain by regulating JNK signaling pathway after chronic constriction injury. Front Neurosci 2019; 13: 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 2002; 417: 83–87. [DOI] [PubMed] [Google Scholar]
- 90.Koromina M, Flitton M, Blockley A, Mellor IR, Knight HM. Damaging coding variants within kainate receptor channel genes are enriched in individuals with schizophrenia, autism and intellectual disabilities. Sci Rep 2019; 9: 19215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Polenghi A, Nieus T, Guazzi S, Gorostiza P, Petrini EM, Barberis A. Kainate receptor activation shapes short-term synaptic plasticity by controlling receptor lateral mobility at glutamatergic synapses. Cell Rep 2020; 31: 107735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Copits BA, Swanson GT. Dancing partners at the synapse: auxiliary subunits that shape kainate receptor function. Nat Rev Neurosci 2012; 13: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tomita S, Castillo PE. Neto1 and Neto2: auxiliary subunits that determine key properties of native kainate receptors. J Physiol 2012; 590: 2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Howe JR. Modulation of non-NMDA receptor gating by auxiliary subunits. J Physiol 2015; 593: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han L, Howe JR, Pickering DS. Neto2 influences on kainate receptor pharmacology and function. Basic Clin Pharmacol Toxicol 2016; 119: 141–148. [DOI] [PubMed] [Google Scholar]
- 96.Wyeth MS, Pelkey KA, Yuan X, Vargish G, Johnston AD, Hunt S, Fang C, Abebe D, Mahadevan V, Fisahn A, Salter MW, McInnes RR, Chittajallu R, McBain CJ. Neto auxiliary subunits regulate interneuron somatodendritic and presynaptic kainate receptors to control network inhibition. Cell Rep 2017; 20: 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, Tomita S. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron 2009; 61: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Straub C, Hunt DL, Yamasaki M, Kim KS, Watanabe M, Castillo PE, Tomita S. Distinct functions of kainate receptors in the brain are determined by the auxiliary subunit Neto1. Nat Neurosci 2011; 14: 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheng N, Shi YS, Lomash RM, Roche KW, Nicoll RA. Neto auxiliary proteins control both the trafficking and biophysical properties of the kainate receptor GluK1. Elife 2015; 4: e11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wyeth MS, Pelkey KA, Petralia RS, Salter MW, McInnes RR, McBain CJ. Neto auxiliary protein interactions regulate kainate and NMDA receptor subunit localization at mossy fiber-CA3 pyramidal cell synapses. J Neurosci 2014; 34: 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang M, Ivakine E, Mahadevan V, Salter MW, McInnes RR. Neto2 interacts with the scaffolding protein GRIP and regulates synaptic abundance of kainate receptors. PLoS One 2012; 7: e51433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palacios-Filardo J, Aller MI, Lerma J. Synaptic targeting of kainate receptors. Cereb Cortex 2016; 26: 1464–1472. [DOI] [PubMed] [Google Scholar]
- 103.Maraschi A, Ciammola A, Folci A, Sassone F, Ronzitti G, Cappelletti G, Silani V, Sato S, Hattori N, Mazzanti M, Chieregatti E, Mulle C, Passafaro M, Sassone J: Parkin regulates kainate receptors by interacting with the GluK2 subunit. Nat Commun 2014; 5: 5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pickering DS, Taverna FA, Salter MW, Hampson DR. Palmitoylation of the GluR6 kainate receptor. Proc Natl Acad Sci USA 1995; 92: 12090–12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lomash RM, Sheng N, Li Y, Nicoll RA, Roche KW. Phosphorylation of the kainate receptor (KAR) auxiliary subunit Neto2 at serine 409 regulates synaptic targeting of the KAR subunit GluK1. J Biol Chem 2017; 292: 15369–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mahadevan V, Dargaei Z, Ivakine EA, Hartmann AM, Ng D, Chevrier J, Ormond J, Nothwang HG, McInnes RR, Woodin MA: Neto2-null mice have impaired GABAergic inhibition and are susceptible to seizures. Front Cell Neurosci 2015; 9: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sargin D. Heightened fear in the absence of the kainate receptor auxiliary subunit NETO2: implications for PTSD. Neuropsychopharmacology 2019; 44: 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vernon CG, Swanson GT. Neto2 assembles with kainate receptors in DRG neurons during development and modulates neurite outgrowth in adult sensory neurons. J Neurosci 2017; 37: 3352–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pigoni M, Wanngren J, Kuhn PH, Munro KM, Gunnersen JM, Takeshima H, Feederle R, Voytyuk I, De Strooper B, Levasseur MD, Hrupka BJ, Müller SA, Lichtenthaler SF. Seizure protein 6 and its homolog seizure 6-like protein are physiological substrates of BACE1 in neurons. Mol Neurodegener 2016; 11: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pigoni M, Hsia HE, Hartmann J, Rudan Njavro J, Shmueli MD, Müller SA, Güner G, Tüshaus J, Kuhn PH, Kumar R, Gao P, Tran ML, Ramazanov B, Blank B, Hipgrave Ederveen AL, Von Blume J, Mulle C, Gunnersen JM, Wuhrer M, Rammes G, Busche MA, Koeglsperger T, Lichtenthaler SF. Seizure protein 6 controls glycosylation and trafficking of kainate receptor subunits GluK2 and GluK3. EMBO J 2020; 39: e103457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paracchini L, Beltrame L, Boeri L, Fusco F, Caffarra P, Marchini S, Albani D, Forloni G. Exome sequencing in an Italian family with Alzheimer's disease points to a role for seizure-related gene 6 (SEZ6) rare variant R615H. Alzheimers Res Ther 2018. 12; 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nash A, Aumann TD, Pigoni M, Lichtenthaler SF, Takeshima H, Munro KM, Gunnersen JM. Lack of Sez6 family proteins impairs motor functions, short-term memory, and cognitive flexibility and alters dendritic spine properties. Cereb Cortex 2020; 30: 2167–2184. [DOI] [PubMed] [Google Scholar]
- 113.Roitman M, Edgington-Mitchell LE, Mangum J, Ziogas J, Adamides AA, Myles P, Choo-Bunnett H, Bunnett NW, Gunnersen JM. Sez6 levels are elevated in cerebrospinal fluid of patients with inflammatory pain-associated conditions. Pain Rep 2019; 4: e719. [DOI] [PMC free article] [PubMed] [Google Scholar]