Abstract
SARS‐CoV and SARS‐CoV‐2 encode four structural and accessory proteins (spike, envelope, membrane and nucleocapsid proteins) and two polyproteins (pp1a and pp1ab). The polyproteins are further cleaved by 3C‐like cysteine protease (3CLpro) and papain‐like protease (PLpro) into 16 nonstructural proteins (nsps). PLpro is released from nsp3 through autocleavage, and then it cleaves the sites between nsp1/2, between nsp2/3 and between nsp3/4 with recognition motif of LXGG, and the sites in the C‐terminus of ubiquitin and of protein interferon‐stimulated gene 15 (ISG15) with recognition motif of RLRGG. Alone or together with SARS unique domain (SUD), PLpro can stabilize an E3 ubiquitin ligase, the ring‐finger, and CHY zinc‐finger domain‐containing 1 (RCHY1), through domain interaction, and thus, promote RCHY1 to ubiquitinate its target proteins including p53. However, a dilemma appears in terms of PLpro roles. On the one hand, the ubiquitination of p53 is good for SARS‐CoV because the ubiquitinated p53 cannot inhibit SARS‐CoV replication. On the other hand, the ubiquitination of NF‐κB inhibitor (IκBα), TNF receptor‐associated factors (TRAFs), and stimulator of interferon gene (STING), and the ISGylation of targeted proteins are bad for SARS‐CoV because these ubiquitination and ISGylation initiate the innate immune response and antiviral state. This mini‐review analyzes the dilemma and provides a snapshot on how the viral PLpro smartly manages its roles to avoid its simultaneously contradictory actions, which could shed lights on possible strategies to deal with SARS‐CoV‐2 infections.
Keywords: ISG15, PLpro, p53, SARS‐CoV, SARS‐CoV‐2, ubiquitin
1. INTRODUCTION
Being the largest positive‐sense RNA virus, 1 severe acute respiratory syndrome coronavirus (SARS‐CoV) encodes four structural and accessory proteins: spike, envelope, membrane, and nucleocapsid proteins, and two polyproteins: pp1a and pp1ab, which are further cleaved by two proteases into 16 nonstructural proteins (nsps). 2 , 3 These two proteases, 3C‐like cysteine protease (3CLpro) and papain‐like protease (PLpro), come from nsp5 and nsp3, respectively. 2 Of 16 nsps, nsp3 is the longest one with 1922 amino acid residues, 2 and can be further cleaved into functional components 4 with identified domains 5 (top of Figure 1).
FIGURE 1.
Roles of SARS‐CoV PLpro. The green dotted frames labeled with bad effects on SARS‐CoV are the scenarios, which are unlikely to happen when SARS‐CoV prevails
Initially, the domains in nsp3 of SARS‐CoV were predicted and identified as follows: N‐terminal glutamic acid rich acidic‐domain (Ac), 6 X, 6 , 7 PLpro, and Y. 2 , 6 Thereafter, severe acute respiratory syndrome (SARS) unique domain (SUD) was added into the list of domains in nsp3. 8 Eventually, the arrangement of domains from N‐terminus include (a) the first ubiquitin‐like domain (Ubl‐1), (b) a Ac, (c) ADP‐ribose‐1”‐phosphatase (ADRP) domain (X domain), (d) SUD, (e) the second ubiquitin‐like domain at the N‐terminus (Ubl‐2), (f) nucleic acid binding domain (NAB), (g) the marker domain (G2M), (h) a metal‐binding region (ZF) domain formed by four predicted transmembrane domains (TM1‐TM4), and (i) Y domain. 5 , 9
The release of 3CLpro and PLpro from pp1a/pp1ab is done through autocleavage at flanking sites on each protease, 4 , 6 , 10 , 11 , 12 and hereafter, they perform the role of transcleavage 13 (top of Figure 1). As ORF1b can encode polyprotein only if a ribosomal frameshift from ORF1a into ORF1b occurs, 14 , 15 presumably there will be a single copy of PLpro and 3CLpro when the frameshift does not occur but two copies of PLpro and 3CLpro when the frameshift occurs within a replication cycle of SARS‐CoV because the range from 18% to 30% was found for the frameshift frequency, of which less than 1% can synthesize pp1ab. 16
It is generally considered that PLpro has two roles: (a) the role of protease, by which it cleaves three sites between nsp1/2 (glycine‐180/alanine‐181), between nsp2/3 (glycine‐818/alanine‐819) and between nsp3/4 (glycine‐2740/lysine‐2741) 13 (top of Figure 1), and (b) the role of deubiquitinating (DUB) and deISGylating enzyme, by which it deconjugates ubiquitin (the downward arrow in left part of Figure 1) and ubiquitin‐like (UBL) protein interferon‐stimulated gene 15 (ISG15) from their substrates 17 (the downward arrow in right‐middle part of Figure 1).
Here, the functionality of PLpro is still not very clear: does a PLpro do several tasks throughout its lifetime or does a PLpro do only a single task during its lifetime? This question comes from the fact that PLpro domain alone is enough to cleave nsp1/2 and nsp2/3 sites with different efficiencies, but PLpro needs the downstream hydrophobic domain (amino acid residues 2207 to 2365) inserted into membranes in order to cleave nsp3/4 site in SARS‐CoV 13 , 18 (right top part of Figure 1). As PLpro is left from nsp3 through autocleavage, it must have a certain mechanism that PLpro associates with the downstream hydrophobic domain in order to cleave nsp3/4 site in SARS‐CoV. Since it is not clear whether or not PLpro is disassociated from the downstream hydrophobic domain, the open question is that two cleavage mechanisms may exist because the association of PLpro with the hydrophobic domain may change PLpro 3D structure.
Although the job of cleavaging nsp1/2, nsp2/3, and nsp3/4 by PLpro is needed for coronavirus RNA synthesis, 19 another important role played by PLpro is directly related to its deubiquitinating and deISGylating functions because the deubiquitinating function is closely related to inhibiting the innate immune response to SARS‐CoV infection 17 , 20 , 21 (the blue frame in left bottom part of Figure 1), and the deISGylating function is closely related to inhibiting the antiviral state 22 (the violet dotted frame in middle bottom part of Figure 1). In fact, PLpro is structurally similar to the cellular deubiquitinase, herpesvirus‐associated ubiquitin‐specific protease (HAUSP)/USP7/USP14, 23 , 24 which is one of more than hundred human deubiquitinating enzymes (DUBs) 25 encoded by human genome. 26
As a matter of fact, the deubiquitinating and deISGylating roles are essentially related to the proteolytic function of PLpro because both ubiquitin and ISG15 have the LXGG recognition motif at their C‐terminus, which are exactly as the same as the cleavage sites between nsp1/2, between nsp2/3 and between nsp3/4 13 (the blue dashed and violet dotted frames in lower part vs top part of Figure 1). These are very suggestive since the above mentioned literature indicates that the PLpro has multiple tasks to do in host cells.
It is generally considered that an enzyme usually works on a single type of substrate rather than several types of substrates simultaneously, thus, PLpro may have to make a choice as seen in most microorganisms. 27 , 28 In this mini‐review, we analyze the literature along this line of thought.
2. SWITCH BETWEEN TWO ROLES OF PLpro
X‐ray crystallography reveals that PLpro has both peptidase and isopeptidase activities, 29 which correspond to the two roles of PLpro (top of Figure 1). It is likely that the PLpro peptidase function is exclusively directed to the recognition motif of LXGG between nsp1/2, between nsp2/3, and between nsp3/4. By contrast, an isopeptide bond is often associated with the ε‐amino group of lysine because an ubiquitin has seven lysines. 30 It turns out RLRGG as recognition subsites at C‐terminus of ubiquitin, 29 which suggests a different mechanism for substrate recognition, 29 , 31 , 32 , 33 because the cleavage of RLRGG‐7‐amido‐4‐methylcoumarin (AMC) (k cat/Km = 5.5 × 103 M−1 s−1) is slower than ubiquitin‐AMC (k cat/Km = 7.5 × 104 M−1 s−1). 29 , 34 Surely, these are kinetic evidence that PLpro does not work equally on its substrates, and its catalytic order is ISG15, ubiquitin, and then, nsp1/2, nsp2/3, and nsp3/4 in terms of efficiency and rapidity. 29 , 33 , 34 When including chained ubiquitins, PLpro catalytic order is K48‐linked ubiquitin, ISG15, K63‐linked ubiquitin and ubiquitin. 29
Here, the recognition subsites with RLRGG may serve as the switch between peptidase and isopeptidase activities because RLRGG are located from R72 to G76 in ubiquitin and ISG15, while G76 would form an isopeptide bond with ε‐amino group of lysine side chains of another ubiquitin. Sometimes, C77 in ubiquitin can exist if a cysteine is attached, whose proteolysis could be inhibited by N‐ethylmaleimide (NEM) if we consider a similar case in 3CLpro. 10
Both ubiquitin and ISG15 play their roles in conjugating their target proteins through their C‐terminus. For example, K48‐linked ubiquitin conjugates NF‐κB inhibitor (IκBα) blocking the activation of the NF‐κB signaling pathway, 35 , 36 which initiates the immune response to viral infection 37 , 38 , 39 , 40 (the green dotted frame in left part of Figure 1). As another example, ISG15 conjugates its target proteins to establish the antiviral state 22 (the green dotted frame in bottom middle part of Figure 1).
Overall, the cleavage of these conjugations by PLpro through an isopeptide bond with the recognition subsites of RLRGG would inhibit a series of activities against SARS‐CoV. Indeed, the current treatment coronavirus disease 2019 (COVID‐19) heavily depends on the patient immune system. 41 For this reason, SARS‐CoV PLpro is considered as a strong antagonist against the ubiquitin‐dependent cellular responses to viral infection. 17
An interesting question is what form PLpro adopts when it cleaves the isopeptide bond with the recognition subsites RLRGG? This is not only because PLpro has two forms, one for the cleavage of nsp1/2 and nsp2/3, another for the cleavage of nsp3/4, which requires the attachment of PLpro to membrane, 13 , 18 but also because ubiquitin and ISG15 are in cytosol.
It showed that pp1a and pp1ab were difficult to be detected in vivo, and this difficulty is most probably due to the fact that pp1a and pp1ab were cotranslationally and autocatalytically processed into intermediates and nonstructural proteins, 6 although the number and the origin of intermediates need to be determined. 42 These findings implicate that the accumulation of pp1a and pp1ab is unlikely because various domains in nsps (nsp3 in our case) release immediately after the replication of SARS‐CoV.
3. THE THIRD ROLE OF PLpro
Theoretically, the conjugation of ubiquitin to its substrates requires the sequential three enzymes, E1 (ubiquitin‐activating enzymes) with consumption of ATP, E2 (ubiquitin‐conjugating enzymes) and E3 (ubiquitin ligases). 43 Very likely, the chained ubiquitins, such as K48‐linked ubiquitin and K63‐linked ubiquitin, also undergo E1‐E2‐E3 system (the green leftward arrow in Figure 1) as ISGs including ISG15 do so (the green downward arrow in Figure 1). 5 Consequently, the second role of PLpro as isopeptidase 29 works on the products of E1‐E2‐E3 system (the green dotted frames in middle left and lower parts of Figure 1).
The ring‐finger and CHY zinc‐finger domain‐containing 1 (RCHY1) 44 , 45 , 46 and mouse double minute 2 homolog (MDM2) are two E3 ubiquitin ligases. 47 , 48 , 49 RCHY1 acts on the central region of p53, and then, ubiquitinates p53 (the green rightward arrow in Figure 1), and thus, promotes p53 degradation independently of MDM2 49 (the violet dotted frame in right part of Figure 1). Similarly, MDM2 not only can induce polyubiquitination and degradation of p53 at high levels, but can also cause the monoubiquitination and nuclear export of p53 at low levels, 50 resulting in inhibition of apoptosis and innate immune signaling. 51 At this point, it is not clear whether the chained ubiquitins and ISG15 interact with RCHY1 or MDM2, or both, or other E3 ligases such as ECS E3 ligase complex (Elongin B/C‐Cul2/5‐SOCS‐box protein ubiquitin ligase complex). 52
It seems counter‐intuitive for the common concept on the ubiquitin system because the ubiquitin system in principle should play positive roles to conjugate useless or harmful proteins, 53 , 54 but here ubiquitin system conjugates active and helpful proteins. Since p53 inhibits the SARS‐CoV replication 55 and other virus infections, 52 , 56 p53 plays a positive role in antiviral infection (the green dotted frame in lower right part of Figure 1), for which E3 ubiquitin ligases should not ubiquitinate p53 leading to its degradation.
PLpro can stabilize RCHY1, 55 which has a half‐life of 3.5 hours, 57 and even more strongly stabilizes RCHY1 with SUD together through the interaction between residues 95‐144 of RCHY1 and 389‐652 of SUD (SUD‐NM) 55 (upper middle part of Figure 1). As PLpro is autoproteolytic cleaved from nsp3, a question raised here is which protease cleaves SUD from nsp3 owing to the fact that SUD does not have the recognition subsites when looking at the terminus of their sequences (accession number: 2W2G_A and 2W2G_B)? Although the autocleavage mechanism is not fully understood, 12 , 58 , 59 SUD is unlikely to be released from nsp3 through the autocleavage mechanism. Another mechanism, which needs to explore, is how PLpro is fused with SUD in vivo since PLpro should be released by autocleavage without SUD?
Evidently, a dilemma appears in consideration of PLpro role on E3 stabilization (central part of Figure 1). On the one side, the stabilization of E3 promotes the ubiquitination of p53 (the violet frame in middle right part of Figure 1), and thus, minimizes the role of p53 in inhibiting SARS‐CoV replication in host cells. 55 On the other side, the stabilization of E3 promotes the ubiquitination of IκBα by K48‐linked ubiquitin, the ubiquitination of TNF receptor‐associated factors (TRAFs) 60 and stimulator of interferon genes (STING) 61 by K63‐linked ubiquitin, and the ISGylation of targeted proteins by ISG15, and thus, initiates the innate immune response to SARS‐CoV infection and antiviral state (the green frames in left and middle‐bottom parts of Figure 1). This dilemma is that the ubiquitination of p53 is useful and helpful against SARS‐CoV infection, whereas the ubiquitination of IκBα, TRAFs, and STING, and the ISGylation of targeted proteins are bad against SARS‐CoV infection. In short, the role of PLpro/PLpro+SUD on stabilizing E3 is to promote ubiquitination and ISGylation, which consequently plays dual effects: promoting intercellular reaction to SARS‐CoV (innate immune system response and antiviral state), but promoting SARS‐CoV replication within host cells (virus replication).
Furthermore, the role of PLpro is to cleave the ubiquitinated IκBα, TRAFs, and STING, and the ISGylated targeted proteins through the recognition subsites with RLRGG. The deubiquitination of IκBα, TRAFs, and STING, and the deISGylation of targeted proteins are good for SARS‐CoV because these actions inhibit the innate immune response and antiviral state (the blue frame in left part and violet dotted frame in middle‐bottom part of Figure 1). However, PLpro does not deubiquitinate the ubiquitinated p53. 4 If such deubiquitination occurs, it will be bad for SARS‐CoV (the violet frame in right‐bottom part of Figure 1) because p53 inhibits SARS‐CoV replication. Although PLpro has the role of deubiquitination, it is not the case in this experiment. In short, the final consequence of second dilemma in effect is inhibiting intercellular reaction to SARS‐CoV, but inhibiting SARS‐CoV replication within host cells.
Collectively, PLpro has two important recognition functions, one for the recognition of LXGG in pp1a/pp1ab and of RLRGG in C‐terminus of ubiquitin and of ISG15, and the other for the recognition of a ring‐finger domain in E3. The latter enhances the functions of E3, which bring about counter effects on viral infection and on host‐cell reactions.
4. STRATEGY OF PLpro
The above analyzed dilemma reveals that PLpro may play contradictory roles in SARS‐CoV infection. It is very unlikely that the viruses evolve and conserve the codes whose products are harmful to themselves. Therefore, we need to figure out what strategy PLpro adapted to bypass this dilemma. If we arrange the roles of PLpro along the time course, we can find that these roles are well managed by SARS‐CoV (Figure 2).
FIGURE 2.
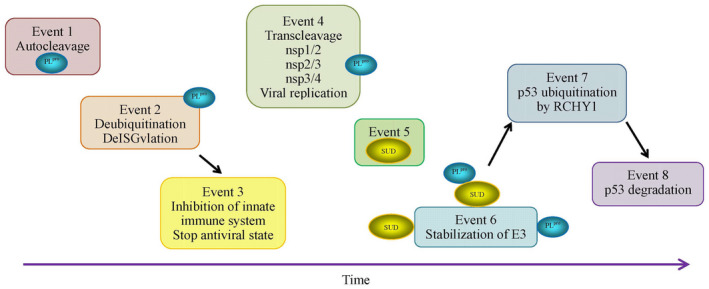
Proposed strategy of PLpro to bypass its contradictory roles in infected host cells according to its kinetic preference
SARS‐CoV PLpro has its choice to cleave the chained ubiquitins, ISG15, ubiquitins (if any of them exists), and the sites between nps1/2 and between nps2/3 because the PLpro should kinetically prefer to recognition subsites RLRGG than LXGG. Without knowing the kinetics of association of PLpro with its downstream hydrophobic domain for cleaving nsp3/4, this kinetic preference dictates viral PLpro to cleave chained ubiquitins, ISG15, ubiquitins before cleaving nps1/2, nps2/3, and nsp3/4 (Event 2 vs Event 4 in Figure 2). This strategy suggests that SARS‐CoV at first inhibits the innate immune response and the initiation of viral state in infected host cells, and then, begins its replication (Event 3 vs Event 4 in Figure 2). The release of SUD is likely to be simultaneously with the accomplishment of cleaving nsp1/2, nsp2/3, and nsp3/4 by PLpro (Event 5 in Figure 2). Thereafter, PLpro and SUD work alone or together to stabilize E3 (Event 6 in Figure 2), and finally, proceed to p53 ubiquitination and degradation (Events 7 and 8 in Figure 2). Hence, PLpro accomplishes its series of tasks for viral infection.
Accordingly, there may theoretically be several interesting points to explore what happens after the release of PLpro from nsp3. Could it be possible to apply the artificial peptides of RLRGG as an aptamer to the recognition subsites in the host cells in order to derail the cleavage of nps1/2, nps2/3, and nsp3/4 to prevent the replication of SARS‐CoV? Also, it is not clear whether PLpro fusion with SUD is reversible or not. If it would not be so, then mimic of SUD interactive domain with PLpro could be a way to move out PLpro from its active duty.
Although the above discussed mechanisms were obtained from the studies on SARS‐CoV, mainly in vitro studies, it is highly likely that the knowledge on SARS‐CoV is equally applicable to severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) because the domains in nsp3 were predicted to be conserved across all CoV. 62 Therefore, the development of antiviral drugs targeting PLpro is an important strategy. 5 , 63 , 64 , 65
Nevertheless, this mini‐review is exclusively directed to the roles of PLpro according to the current knowledge although many other players are also involved in the deubiquitinase activity, for example, the endoribonuclease from nsp15. 66
In conclusion, this review demonstrates that PLpro evolves a very efficient and clever strategy to suppress the host's response to SARS‐CoV infection, which sheds lights on understanding the roles of PLpro on SARS‐CoV‐2 infection as well as on possible strategies to deal with SARS‐CoV2 infections.
CONSENT FOR PUBLICATION
Both authors read and approved the manuscript.
5. COMPETING INTEREST
None of the authors has any competing interests in the manuscript.
AUTHOR CONTRIBUTIONS
G. Wu designed this review and wrote the first draft; S.M. Yan visualized the data, and both edited and finalized this review.
Yan S, Wu G. Spatial and temporal roles of SARS‐CoV PLpro—A snapshot. The FASEB Journal.2021;35:e21197 10.1096/fj.202002271
This article was fast‐tracked under a recently instituted interim policy in which editors may, at their discretion, accept coronavirus‐related manuscripts submitted for the Review, Perspectives, and Hypotheses categories without additional review.
REFERENCES
- 1. Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thiel V, Ivanov KA, Putics Á, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84(Pt 9):2305‐2315. [DOI] [PubMed] [Google Scholar]
- 4. Muramatsu T, Takemoto C, Kim Y‐T, et al. SARS‐CoV 3CL protease cleaves its C‐terminal autoprocessing site by novel subsite cooperativity. Proc Natl Acad Sci USA. 2016;113:12997‐13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Báez‐Santos YM, St John SE, Mesecar AD. The SARS‐coronavirus papain‐like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziebuhr J, Thiel V, Gorbalenya AE. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain‐like proteases that cleave the same peptide bond. J Biol Chem. 2001;276:33220‐33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorbalenya AE, Koonin EV. Comparative analysis of amino acid sequences of key enzymes of replication and expression of positive‐strand RNA viruses. Validity of approach and functional and evolutionary implications. Sov Sci Rev D Physicochem Biol. 1993;11:1‐84. [Google Scholar]
- 8. Tan J, Kusov Y, Mutschall D, et al. The “SARS‐unique domain” (SUD) of SARS coronavirus is an oligo(G)‐binding protein. Biochem Biophys Res Commun. 2007;364:877‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barretto N, Jukneliene D, Ratia K, et al. Deubiquitinating activity of the SARS‐CoV papain‐like protease. Adv Exp Med Biol. 2006;581:37‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piñón JD, Mayreddy RR, Turner JD, et al. Efficient autoproteolytic processing of the MHV‐A59 3C‐like proteinase from the flanking hydrophobic domains requires membranes. Virology. 1997;230:309‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen S, Jonas F, Shen C, et al. Liberation of SARS‐CoV main protease from the viral polyprotein: n‐terminal autocleavage does not depend on the mature dimerization mode. Protein Cell. 2010;1:59‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shan YF, Li SF, Xu GJ. A novel auto‐cleavage assay for studying mutational effects on the active site of severe acute respiratory syndrome coronavirus 3C‐like protease. Biochem Biophys Res Commun. 2004;324:579‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harcourt BH, Jukneliene D, Kanjanahaluethai A, et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain‐like protease activity. J Virol. 2004;78:13600‐13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brierley I, Boursnell ME, Binns MM, et al. An efficient ribosomal frame‐shifting signal in the polymerase‐encoding region of the coronavirus IBV. EMBO J. 1987;6:3779‐3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bredenbeek PJ, Pachuk CJ, Noten AF, et al. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV‐A59: a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825‐1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herold J, Raabe T, Schelle‐Prinz B, et al. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mielech AM, Chen Y, Mesecar AD, et al. Nidovirus papain‐like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han YS, Chang GG, Juo CG, et al. Papain‐like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS‐CoV): expression, purification, characterization, and inhibition. Biochemistry. 2005;44:10349‐10359. [DOI] [PubMed] [Google Scholar]
- 19. Kim JC, Spence RA, Currier PF, et al. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology. 1995;208:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calistri A, Munegato D, Carli I, et al. The ubiquitin‐conjugating system: multiple roles in viral replication and infection. Cells. 2014;3:386‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun LI, Xing Y, Chen X, et al. Coronavirus papain‐like proteases negatively regulate antiviral innate immune response through disruption of STING‐mediated signaling. PLoS One. 2012;7:e30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12:4‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sulea T, Lindner HA, Purisima EO, et al. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain‐like protease? J Virol. 2005;79(7):4550‐4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu M, Li P, Li M, et al. Crystal structure of a UBP‐family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041‐1054. [DOI] [PubMed] [Google Scholar]
- 25. Ratia K, Saikatendu KS, Santarsiero BD, et al. Severe acute respiratory syndrome coronavirus papain‐like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717‐5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550‐563. [DOI] [PubMed] [Google Scholar]
- 27. Deutscher J, Francke C, Postma PW. How phosphotransferase system‐related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613‐624. [DOI] [PubMed] [Google Scholar]
- 29. Ratia K, Kilianski A, Baez‐Santos YM, Baker SC, Mesecar A. Structural basis for the ubiquitin‐linkage specificity and deISGylating activity of SARS‐CoV papain‐like protease. PLoS Pathog. 2014;10:e1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203‐229. [DOI] [PubMed] [Google Scholar]
- 31. Baez‐Santos YM, Mielech AM, Deng X, et al. Catalytic function and substrate specificity of the PLpro domain of nsp3 from the Middle East Respiratory Syndrome Coronavirus (MERS‐CoV). J Virol. 2014;88:12511‐12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindner HA, Fotouhi‐Ardakani N, Lytvyn V, et al. The papain‐like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79:15199‐15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindner HA, Lytvyn V, Qi H. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain‐like protease. Arch Biochem Biophys. 2007;466:8‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barretto N, Jukneliene D, Ratia K, et al. The papain‐like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189‐15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frieman M, Ratia K, Johnston RE, et al. Severe acute respiratory syndrome coronavirus papain‐like protease ubiquitin‐like domain and catalytic domain regulate antagonism of IRF3 and NF‐kappaB signaling. J Virol. 2009;83:6689‐6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rahighi S, Ikeda F, Kawasaki M, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF‐kappaB activation. Cell. 2009;136:1098‐1109. [DOI] [PubMed] [Google Scholar]
- 37. Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Kasteren PB, Bailey‐Elkin BA, James TW, et al. Deubiquitinase function of arterivirus papain‐like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci USA. 2013;110:E838‐E847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng SC, Joosten LA, Netea MG. The interplay between central metabolism and innate immune responses. Cytokine Growth Factor Rev. 2014;25:707‐713. [DOI] [PubMed] [Google Scholar]
- 40. Davis ME, Gack MU. Ubiquitination in the antiviral immune response. Virology. 2015;479‐480:52‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leng Z, Zhu R, Hou W, et al. Transplantation of ACE2‐ mesenchymal stem cells improves the outcome of patients with COVID‐19 pneumonia. Aging Dis. 2020;11:216‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus‐encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853‐879. [DOI] [PubMed] [Google Scholar]
- 43. Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1‐E2‐E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81‐83. [DOI] [PubMed] [Google Scholar]
- 44. Jung YS, Qian Y, Chen X. The p73 tumor suppressor is targeted by Pirh2 RING finger E3 ubiquitin ligase for the proteasome‐dependent degradation. J Biol Chem. 2011;286:35388‐35395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jung YS, Qian Y, Yan W, et al. Pirh2 E3 ubiquitin ligase modulates keratinocyte differentiation through p63. J Invest Dermatol. 2013;133:1178‐1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Halaby MJ, Hakem R, Hakem A. Pirh2: an E3 ligase with central roles in the regulation of cell cycle, DNA damage response, and differentiation. Cell Cycle. 2013;12:2733‐2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296‐299. [DOI] [PubMed] [Google Scholar]
- 48. Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25‐27. [DOI] [PubMed] [Google Scholar]
- 49. Leng RP, Lin Y, Ma W, et al. Pirh2, a p53‐induced ubiquitin‐protein ligase, promotes p53 degradation. Cell. 2003;112:779‐791. [DOI] [PubMed] [Google Scholar]
- 50. Li M, Brooks CL, Wu‐Baer F, et al. Mono‐ versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972‐1975. [DOI] [PubMed] [Google Scholar]
- 51. Yuan L, Chen Z, Song S, et al. p53 degradation by a coronavirus papain‐like protease suppresses type I interferon signaling. J Biol Chem. 2015;290:3172‐3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sato Y, Kamura T, Shirata N, et al. Degradation of phosphorylated p53 by viral protein‐ECS E3 ligase complex. PLoS Pathog. 2009;5:e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503‐533. [DOI] [PubMed] [Google Scholar]
- 54. Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol. 2004;22:81‐127. [DOI] [PubMed] [Google Scholar]
- 55. Ma‐Lauer Y, Carbajo‐Lozoya J, Hein MY, et al. p53 down‐regulates SARS coronavirus replication and is targeted by the SARS‐unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc Natl Acad Sci USA. 2016;113:E5192‐E5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sato Y, Tsurumi T. Genome guardian p53 and viral infections. Rev Med Virol. 2013;23:213‐220. [DOI] [PubMed] [Google Scholar]
- 57. Duan S, Yao Z, Hou D, et al. Phosphorylation of Pirh2 by calmodulin‐dependent kinase II impairs its ability to ubiquitinate p53. EMBO J. 2007;26:3062‐3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin CW, Tsai CH, Tsai FJ, et al. Characterization of trans‐ and ciscleavage activity of the SARS coronavirus 3CLpro protease: basis for the in vitro screening of anti‐SARS drugs. FEBS Lett. 2004;574:131‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hsu MF, Kuo CJ, Chang KT, et al. Mechanism of the maturation process of SARS‐CoV 3CL protease. J Biol Chem. 2005;280:31257‐31266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li SW, Wang CY, Jou YJ, et al. SARS coronavirus papain‐like protease inhibits the TLR7 signaling pathway through removing Lys63‐linked polyubiquitination of TRAF3 and TRAF6. Int J Mol Sci. 2016;17:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain‐like protease inhibits the type I interferon signaling pathway through interaction with the STING‐TRAF3‐TBK1 complex. Protein Cell. 2014;5:369‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neuman BW, Joseph JS, Saikatendu KS, et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol. 2008;82:5279‐5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085‐4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Clasman JR, Báez‐Santos YM, Mettelman RC, et al. X‐ray structure and enzymatic activity profile of a core papain‐like protease of MERS coronavirus with utility for structure‐based drug design. Sci Rep. 2017;7:40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin MH, Moses DC, Hsieh CH, et al. Disulfiram can inhibit MERS and SARS coronavirus papain‐like proteases via different modes. Antiviral Res. 2018;150:155‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Volk A, Hackbart M, Deng X, et al. Coronavirus endoribonuclease and deubiquitinating interferon antagonists differentially modulate the host response during replication in macrophages. J Virol. 2020;94:e00178‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]


