Abstract
The regulatory circuitry underlying embryonic stem (ES) cell self‐renewal is well defined, but how this circuitry is disintegrated to enable lineage specification is unclear. RNA‐binding proteins (RBPs) have essential roles in RNA‐mediated gene regulation, and preliminary data suggest that they might regulate ES cell fate. By combining bioinformatic analyses with functional screening, we identified seven RBPs played important roles for the exit from pluripotency of ES cells. We characterized hnRNPLL, which mainly functions as a global regulator of alternative splicing in ES cells. Specifically, hnRNPLL promotes multiple ES cell‐preferred exon skipping events during the onset of ES cell differentiation. hnRNPLL depletion thus leads to sustained expression of ES cell‐preferred isoforms, resulting in a differentiation deficiency that causes developmental defects and growth impairment in hnRNPLL‐KO mice. In particular, hnRNPLL‐mediated alternative splicing of two transcription factors, Bptf and Tbx3, is important for pluripotency exit. These data uncover the critical role of RBPs in pluripotency exit and suggest the application of targeting RBPs in controlling ES cell fate.
Keywords: alternative splicing, embryonic stem cells, exit from pluripotency, hnRNPLL, RNA‐binding proteins
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Regenerative Medicine; Protein Biosynthesis & Quality Control
RNA‐binding protein hnRNPLL acts as a licensing factor for successful fate transition of embryonic stem cells.
Introduction
Mammalian embryonic stem (ES) cells are derived from the inner cell mass of the pre‐implantation embryo (Evans & Kaufman, 1981; Martin, 1981). These cells possess the remarkable property of retaining an unlimited self‐renewal capacity in culture while having the potential to generate cells of all somatic lineages under defined culture conditions (Jaenisch & Young, 2008; Graf & Enver, 2009). The transition away from self‐renewal and toward competency for lineage specification—a process known as exit from pluripotency—likely requires the disintegration of the naive pluripotency program (Respuela et al, 2016). It is not a simple switch from pluripotency to differentiation via the presence or absence of the pluripotency maintenance factors. A novel class of genes is required for exit‐from‐pluripotency to destruct the expression of the pluripotency maintenance genes. Several studies have used genetic screening methods to explore the roles of candidate genes responsible for exit from pluripotency (Yang et al, 2012; Betschinger et al, 2013; Leeb et al, 2014; Liu et al, 2017; Li et al, 2019). Some of the important regulators, such as Tcf3, Tfe3 (Betschinger et al, 2013), Pum1 (Leeb et al, 2014), and Whsc1(Tian et al, 2019) have been uncovered. Despite the growing body of data on the mechanisms underlying exit from pluripotency, and in contrast to the relatively extensive characterization of the genetic network for pluripotency maintenance (Martello & Smith, 2014; Weinberger et al, 2016; Li & Izpisua Belmonte, 2018), the key factors and the underlying genetic circuitry are still elusive.
Post‐transcriptional gene regulation is essential for cellular metabolism and for coordinating the maturation, transport, stability, and degradation of all classes of RNAs (Licatalosi & Darnell, 2010; Manning & Cooper, 2017). Each of these events is regulated by RNA‐binding proteins (RBPs), which can transiently or stably interact with RNA to form ribonucleoprotein complexes (Glisovic et al, 2008). Numerous RBPs and the associated post‐transcriptional regulation can help determine stem cell fate (Ye & Blelloch, 2014; Chen & Hu, 2017; Li & Izpisua Belmonte, 2018). RBPs that regulate splicing (e.g., FOX2, SON, and SFRS2) (Yeo et al, 2009; Lu et al, 2013; Lu et al, 2014), alternative polyadenylation (e.g., FIP1) (Lackford et al, 2014), RNA export (e.g., THOC2 and THOC5) (Wang et al, 2013), RNA modification (e.g., ADAR, METTL3 and METTL14) (Osenberg et al, 2010; Liu et al, 2014), and RNA stability (e.g., LIN28A and TRIM71) (Rybak et al, 2009; Chang et al, 2012; Loedige et al, 2013) have all been implicated in pluripotency maintenance. During exit from pluripotency, RBPs mediated repression of splicing, processing, export or translation of already transcribed self‐renewal gene transcripts may be the first step to repress pluripotency genes, earlier than the transcriptional repression. Which RBPs and the mediated post‐transcriptional regulations drive exit from pluripotency, however, remains unclear.
Here, we aimed to determine the precise role of RBPs in exit‐from‐pluripotency. To do so, we analyzed the dynamic expression of RBPs during transition from pluripotency toward differentiation of mouse ES cells and investigated their interaction with the pluripotency molecular network. We integrated known genome‐wide screening data (Yang et al, 2012; Betschinger et al, 2013; Leeb et al, 2014; Liu et al, 2017) with functional analyses performed in individual RBP knockout (KO) ES cells to identify whether any RBPs are essential for exit from pluripotency. We revealed several RBPs required for ES cell exit from pluripotency and characterized the function of hnRNPLL in promoting exon skipping of multiple critical genes to facilitate pluripotency exit. These data are the first to systematically demonstrate roles of RBPs in regulating exit from pluripotency.
Results
RBPs are differentially regulated during exit from pluripotency
To comprehensively investigate the role of RBPs in exit‐from‐pluripotency, we combined canonical RBPs included in the mouse RBP database (Cook et al, 2011) with the RBP repertoire of mouse ES cells revealed by the mRNA interactome (Kwon et al, 2013; Fig 1A). Of the 789 murine RBPs, 181 are included simultaneously in RBP database and RBP repertoire of mouse ES cells. Because RBPs are ubiquitously expressed across different tissues and cell types, we analyzed the expression of total 789 RBPs during ES cell exit‐from‐pluripotency and during various stages of the differentiation process, including EB (embryoid body) formation, neural (GSE44067), and cardiac differentiation (GSE47948) (Dataset EV1, Data ref: Alexander & Wamstad, 2013; Data ref: Zhang et al, 2013). Surprisingly, the variation analysis identified a total of 424 RBPs that were differentially expressed during these processes and they can be categorized into different subclasses according to their changing trend (Fig 1B).
Figure 1. RBPs are differentially regulated during exit from pluripotency.
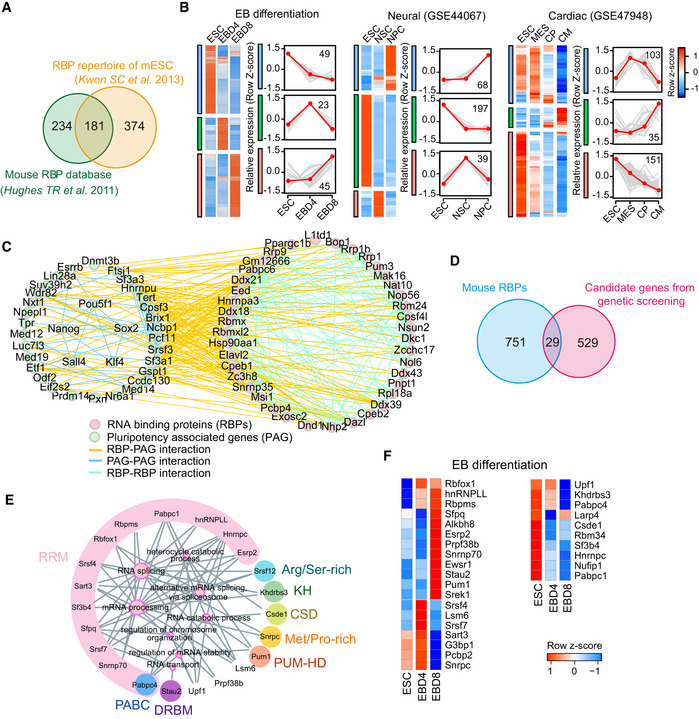
- Overlap of the mouse RBP database and the RBP repertoire revealed by the mRNA interactome in mouse ES cells. All the 789 RBPs were subsequently analyzed.
- The differentially expressed RBPs during embryoid body (EB) differentiation, neural differentiation, and cardiac differentiation, which are classified as different clusters. The right line plots show the dynamic expression pattern of each cluster, gray lines represent the expression of all genes in individual cluster, while the red line represents the average expression level. The number on the graph indicates the number of RBPs in the cluster. The expression value was firstly z‐score scaled before plotting. EBD4: embryoid body at day 4; EBD8: embryoid body at day 8; NSC: Neural Stem Cells; NPC: Neural Progenitor Cell; MES: mesodermal cells; CP: Cardiac Precursors; CM: Cardiomyocytes.
- The protein–protein interaction (PPI) network between differentially expressed RBPs (pink dot) during EB formation and pluripotency‐associated genes (PAG) (green dot). The interaction relationships were downloaded from STRING database.
- Overlap of the 789 RBPs with the published screen data on mouse ES cell exit from pluripotency. The overlapped 29 RBPs may function in exit from pluripotency.
- Gene ontology (GO) enrichment of the above 29 RBPs. The node size of enriched GO terms indicates the number of enriched RBPs, and RBPs with the same domains (e.g., RPM) are clustered and filled with the same background color. The domain names are labeled around enriched RBPs.
- Heatmap showing the expression pattern of the 29 RBPs during EB differentiation.
We next performed protein interactome investigations to understand the relationships between RBPs and pluripotency regulation. Here, we found that RBPs differentially expressed during EB formation predominantly interacted with pluripotency‐associated genes (PAG) (Lu et al, 2014; Fig 1C). The dynamic expression of RBPs during exit‐from‐pluripotency combined with these extensive interactions with PAGs suggests their functional importance during exit from pluripotency.
We next took advantage of our own previous PiggyBac transposon‐based screening results (Liu et al, 2017) and three reported genome‐wide screens for genes required for exit from pluripotency (Yang et al, 2012; Betschinger et al, 2013; Leeb et al, 2014). We re‐analyzed these data and found that 29 RBPs were potentially required for exit from pluripotency (Fig 1D; Dataset EV2). These RBPs mainly contribute to RNA splicing and processing, as well as RNA transport and stability (Fig 1E). When go back to their expressions during pluripotency exit, we found that 19 RBPs increased during EB formation, consistent with their necessities during exit‐from‐pluripotency (Fig 1F). These data suggest that RBPs could have a notable role in mediating ES cell exit from pluripotency.
Seven RBPs participate in ES cell exit from pluripotency
To comprehensively dissect the role of the 29 identified RBPs in regulating exit from pluripotency, we constructed individual RBP‐KO ES cells and examined their ability to exit from pluripotency through forming EB and random differentiation by LIF (leukemia inhibitory factor, used for maintenance of ES cell self‐renewal in vitro culture) withdrawal assay (Fig 2A).
Figure 2. Seven RBPs are required for ES cell exit from pluripotency.
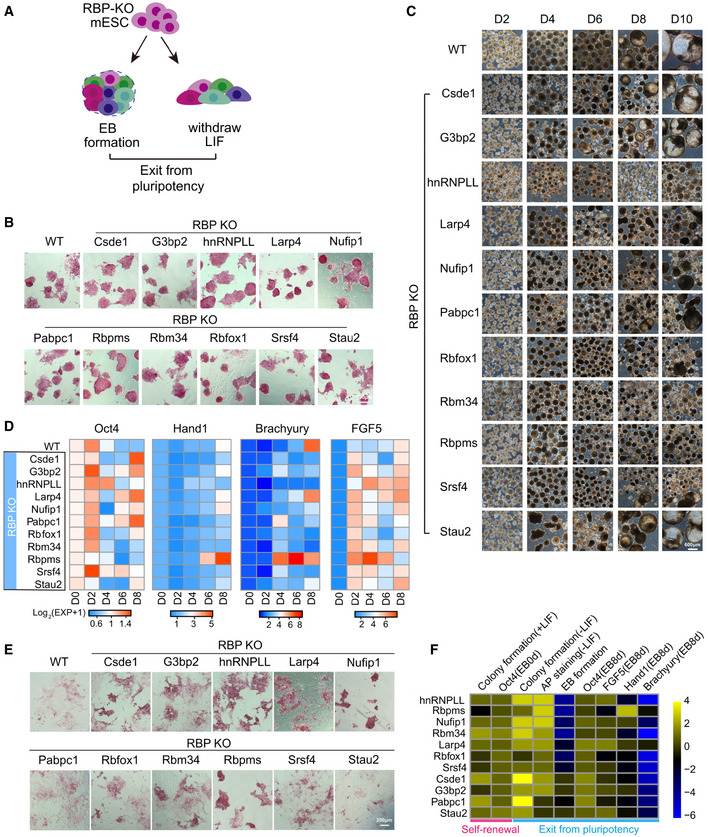
- The research strategy to dissect the role of RBPs in regulating exit from pluripotency. Firstly, we constructed individual RBP‐KO ES cells and examined their ability to exit from pluripotency through forming EB and random differentiation by LIF withdrawal assay.
- AP staining of 11 RBP‐KO ES cell lines maintained under ES cell culture conditions. Scale bar, 200 μm.
- EB formation from WT and RBP‐KO ES cell lines. WT ES cell‐derived EBs formed cystic structures by day 8 of differentiation; most of RBP‐KO ES cell‐derived EBs were more compact. Scale bar, 600 μm.
- Time‐course analysis of Oct4 expression and three lineage differentiation markers (Hand1, Brachyury, and FGF5) during EB formation. It was quantified by qRT‐PCR, the relative expression of genes was calculated by the double delta C t (ΔΔC t) method, normalized to the expression value at day 0. The relative expression data were represented as the heatmap.
- AP staining of WT and 11 RBP‐KO ES cell lines after LIF withdrawal. Scale bar, 200 μm.
- RBP ranking based on ES cell abilities to self‐renewal (determined by colony formation (±LIF) and Oct4 expression) and exit from pluripotency (determined by AP staining, EB formation, and marker genes expression) in the context of RBP‐KO. The expression value of Oct4, FGF5, Hand1, and Brachyury was represented as the relative level of these genes compared with their expressions in WT ES cells. The quantitation of other phenotypes, such as colony formation, AP staining, and EB formation was manually defined.
Of the 29 RBPs, the role of some in regulating proliferation, apoptosis, or pluripotency had already been studied (Malmegrim et al, 2002; Fathi et al, 2009; van Hoof et al, 2009; Ougland et al, 2012; Fagoonee et al, 2013; Liu et al, 2013; Leeb et al, 2014; Ye & Blelloch, 2014). KO mice were already characterized for a further three RBPs (Khdrbs3, Sfrs12, Snrpc, according to the MGI database), the phenotypes of which are normal (Dataset EV2). We thus focused on the remaining 16 RBPs and used the CRISPR/Cas9 nickase system (Shen et al, 2014) to ablate these RBPs with minimal off‐target cleavage. We successfully disrupted the open reading frames of critical exons of 11 of the candidate RBPs (Csde1, G3bp2, hnRNPLL, Larp4, Nufip1, Pabpac1, Rbpms, Rbm34, Rbfox1, Srsf4, Stau2) in ES cells. We were unable to establish RBP‐KO ES cell lines for the remaining five RBPs (Lsm6, Prpf38b, Pcbp2, Sf3b4, and Hnrnpc), likely due to either their essential contributions to cell survival or low guide RNA activities (Appendix Fig S1).
Going forward with the 11 RBP‐KO ES cells, we first assessed their self‐renewal ability. Alkaline phosphatase (AP) activity and Oct4 expression analysis showed that deletion of these RBPs did not affect ES cell self‐renewal when maintained under ES cell culture conditions (Figs 2B and EV1A). We then used EB formation from wild‐type (WT) and RBP‐KO ES cells as an in vitro model for early embryonic development (Rodda et al, 2002). During early‐stage differentiation (before day 6), all RBP‐KO ES cells aggregated to form an EB‐like structure. Most of the WT ES cell‐derived EBs formed cystic structures by day 8, while some RBP‐KO cell lines (hnRNPLL, Larp4, Nufip1, Pabpc1, Rbfox1, Rbm34, Rbpms, Srsf4) generated compact and/or small EBs (Fig 2C). Following up with the marker gene expression detection, they showed different expression pattern of self‐renewal marker Oct4, germ layer‐specific marker Hand1, Brachyury, and FGF5, compared with WT ES cells (Fig 2D; Dataset EV3). In response to LIF withdrawal, WT ES cells differentiated and lost AP activity, while some of the RBP‐KO clones (Csed1, G3bp2, hnRNPLL, Larp4, Nufip1, Rbm34, Rbpms) still showed strong colony formation and AP activity (Fig 2E), which was more obvious (except the Csed1‐KO clone) when these ES cells were maintained in 2i + LIF culture condition first and then withdrawing 2i + LIF (Fig EV1B). Both the EB formation defects and high levels of AP activity after LIF withdrawal suggest that some of these RBP‐KO ES cells can maintain their self‐renewal but cannot exit from pluripotency successfully.
Figure EV1. hnRNPLL‐KO ES cell lines show a differentiation deficiency.
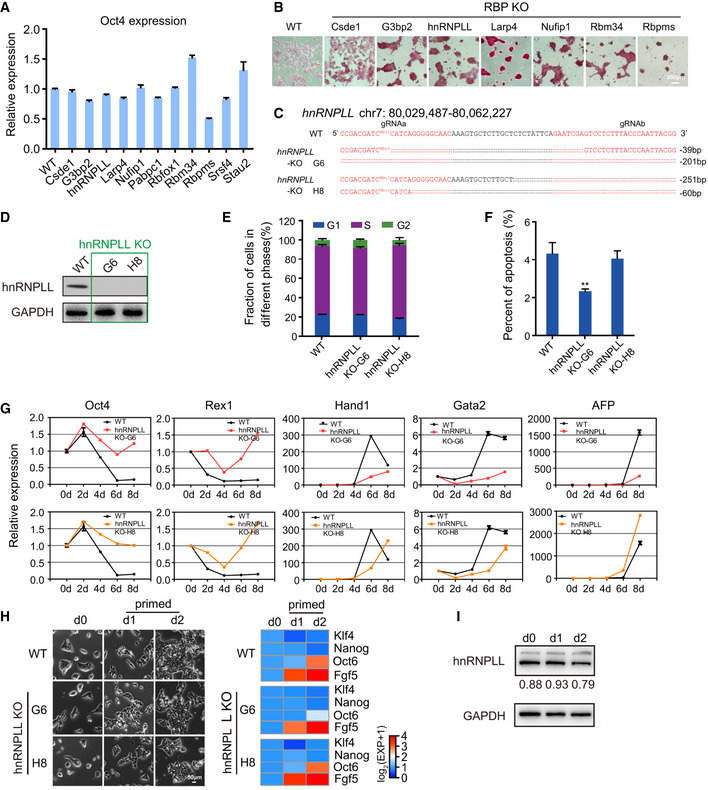
- Relative expression of Oct4 determined by qRT–PCR. The data represent the means ± SD (n = 3, from biological repeats).
- AP staining of WT and 7 RBP‐KO ES cell lines after 2i + LIF withdrawal under N2B27 culture conditions. Scale bar, 200 μm.
- The sequence of the hnRNPLL‐KO allele in ES cell lines derived from clones G6 and H8.
- Western blot analysis of hnRNPLL expression in hnRNPLL‐KO ES cell lines.
- Cell cycle phase distribution in WT and hnRNPLL‐KO ES cell lines. The data represent the means ± SD (n = 3, from biological repeats).
- Apoptosis analysis of WT and hnRNPLL‐KO ES cell lines. The data represent the means ± SD (n = 3, from biological repeats). **P < 0.01. t‐Test was used to determine the significance.
- Relative expression levels of pluripotency genes (Oct4 and Rex1) and lineage‐specific genes (Hand1, Gata2, AFP) during EB formation at the indicated days as measured by qRT–PCR. The data represent the means ± SD (n = 3, from biological repeats).
- The transition of WT and the two hnRNPLL‐KO ES cell clones from the naïve to the primed state. Cells were firstly cultured under the naïve state (2i + LIF) and then transited to the primed state for the indicated number of days. Naïve (Klf4 and Nanog) and primed (FGF5 and Oct6)‐associated gene expression during the transition process (right) was examined by qRT‐PCR. Scale bar, 50 μm.
- Western blot detection of hnRNPLL expression during the transition of WT ES cells from the naïve to the primed state.
We next ranked the RBPs according to the degree of deficiency that the corresponding RBP‐KO ES cells showed in exit from pluripotency (Fig 2F). We deemed the top seven RBPs, including hnRNPLL, Rbpms, Nufip1, Rbm34, Larp4, Rbfox1, and Srsf4 as being important for proper onset of ES cell differentiation. These RBP‐KO ES cells showed higher colony formation abilities and AP activities after LIF withdrawal, as well as higher Oct4 expression and aberrant expression of differentiation markers during EB formation compared with WT ES cells (Fig 2F; Dataset EV3). Because hnRNPLL‐KO ES cells showed the most obvious change in ES cell differentiation, we focused on this RBP for subsequent functional analyses.
hnRNPLL disruption hinders ES cells exit from pluripotency
hnRNPLL is a member of the heterogeneous nuclear RNA‐binding protein (hnRNP) family and was originally identified as a critical inducible regulator of CD45 alternative splicing (AS) in T cells (Oberdoerffer et al, 2008). To confirm its role in regulating ES cell exit from pluripotency, we picked another hnRNPLL‐KO ES cell line (clone H8) and compared the differentiation deficiency with the original hnRNPLL‐KO ES cell line (clone G6). hnRNPLL was not expressed in either clones, despite the disrupted regions were different (Figs 3A, and EV1C and D).
Figure 3. hnRNPLL disruption or overexpression influences exit from pluripotency of ES cells.
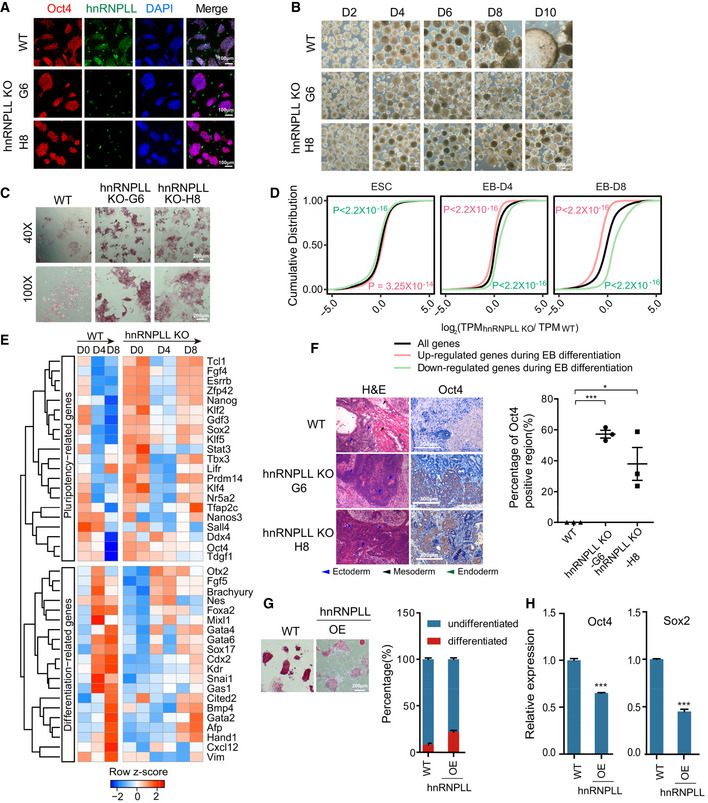
- Immunofluorescence staining of Oct4 and hnRNPLL in WT and hnRNPLL‐KO ES cells from two clones (H8 and G6). hnRNPLL was not expressed in either of the hnRNPLL‐KO clones. DAPI stained the nuclei. Scale bar, 100 μm.
- EB formation from WT and the two hnRNPLL‐KO ES cell clones. WT ES cell‐derived EBs formed cystic structures by day 8 of differentiation, while both hnRNPLL‐KO ES cell clones‐derived EBs were more compact. Scale bar, 200 μm.
- AP staining of WT and both hnRNPLL‐KO ES cell clones after LIF withdrawal. Scale bar, 200 μm.
- Cumulative distribution showing the relative expression of WT‐differentially expressed genes in hnRNPLL‐KO ES cells during EB differentiation. Red lines and green lines represent the up‐ and down‐regulated genes during EB differentiation of WT ES cells, while the black line stands for all expressed genes in WT and hnRNPLL‐KO ES cells. Kolmogorov–Smirnov test was used to determine the significance. The relative expression ratio was log2‐transformed before plotting.
- Heatmap of key pluripotency‐ and differentiation‐associated gene expression during EB formation from WT and hnRNPLL‐KO clones (H8 and G6).
- Hematoxylin and eosin labeling and Oct4 immunohistochemical staining of teratomas derived from WT and hnRNPLL‐KO ES cells (left). Representative Oct4‐positive regions are shown. The percentage of Oct4‐positive region (%) is plotted (right). The data represent the means ± SD, (n = 3, from biological repeats). *P < 0.05; ***P < 0.001. t‐Test was used to determine the significance. Scale bar, 100 μm.
- AP staining of WT and hnRNPLL overexpression ES cell lines maintained under ES cell culture conditions and colony formation assay. Scale bar, 200 μm. The data represent the means ± SD (n = 3 biological repeats).
- Relative expression of Oct4 and Sox2 after hnRNPLL overexpression. The data represent the means ± SD (n = 3, from biological repeats). ***P < 0.001. t‐Test was used to determine the significance.
Consistent with the findings from clone G6, Oct4 expression in clone H8 was not affected by hnRNPLL deletion (Fig 3A). The cell cycle profile and apoptosis ratios were as also comparable with WT ES cells, with just a slightly lower apoptotic cell ratio in ES cells derived from clone G6 (Fig EV1E and F). Clone H8 also showed a lack of cystic EB structures and marker expression, high AP activity following LIF withdrawal (Figs 3B and C, and EV1G). The similarities in deficiencies in exit from pluripotency between the two clones support that indeed hnRNPLL is involved in this process. We also tested the possible ability of hnRNPLL‐KO ES cells on transition from the naïve to the primed state in vitro (Martello & Smith, 2014; Neagu et al, 2020). Both the morphological observation and the molecular examination indicated that hnRNPLL‐KO ES cells can successfully convert to epiblast stem cells (EpiSCs) (Fig EV1H). In addition, the hnRNPLL protein level was unchanged during the conversion process (Fig EV1I). So, we concluded that hnRNPLL did not participate in the transition from the naïve to the primed state.
Going forward with the cells from clones G6 and H8, we next aimed to understand the disturbed molecular circuitry in hnRNPLL‐KO cells during EB formation. We recovered WT ES cells as well as clone G6 and H8 at the indicated days during EB formation and processed them for RNA sequencing. To undergo EB formation, ES cells must silence the self‐renewal program and simultaneously activate differentiation‐associated genes (Desbaillets et al, 2000). We first compared day 4 or day 8 differentiated cells with self‐renewing ES cells (day 0) and identified a subset of up‐regulated and down‐regulated genes that correlate with EB formation. Coincident with the abnormal EB macroscopic structure, the “up‐regulated” genes were significantly repressed in hnRNPLL‐KO cells during EB differentiation compared with all genes, while the “down‐regulated” genes were promoted (Fig 3D). These data strongly support overall general retardation on differentiation in hnRNPLL‐KO cells (Fig 3D). Pluripotency‐associated genes (e.g., Oct4, Sox2, Nanog, and Klf4) remained active in hnRNPLL‐KO ES cells, while genes associated with germ layer development (e.g., Brachyury, Foxa2, and Kdr) were relatively suppressed (Figs 3E and EV2A; Dataset EV4). We also noticed that some self‐renewal genes down‐regulated at day 4, but then elevated again at day 8, which may be due to the heterogeneous ES cell populations cultured in Serum + LIF condition (Martello & Smith, 2014; Guo et al, 2016). Under the EB differentiation, although the majority of hnRNPLL‐KO cells cannot exit the pluripotency successfully, small sub‐populations with likely formative or primed states may still transit into lineage commitments. During EB formation, an initial morphological event was the formation of an outer layer of extraembryonic endoderm cells. The improper up‐regulation of extraembryonic endoderm marker genes (Gata4, Gata6, Sox17) in differentiating hnRNPLL‐KO ES cells suggested the defect of extraembryonic endoderm differentiation. We checked the morphological differentiation event during EB formation by immunofluorescent staining. As a result, Gata4‐ or Sox17‐positive endoderm cells were mainly at the outer layer of EBs at differentiation day 4 or 8. However, in EBs generated from hnRNPLL‐KO cells, the expression of Gata4 or Sox17 was dislocated (Fig EV2B). These results further confirmed the abnormal defect of the differentiation from the naïve state to extraembryonic endoderm of hnRNPLL‐KO ES cells.
Figure EV2. hnRNPLL deletion blocks ES cells exit from pluripotency.
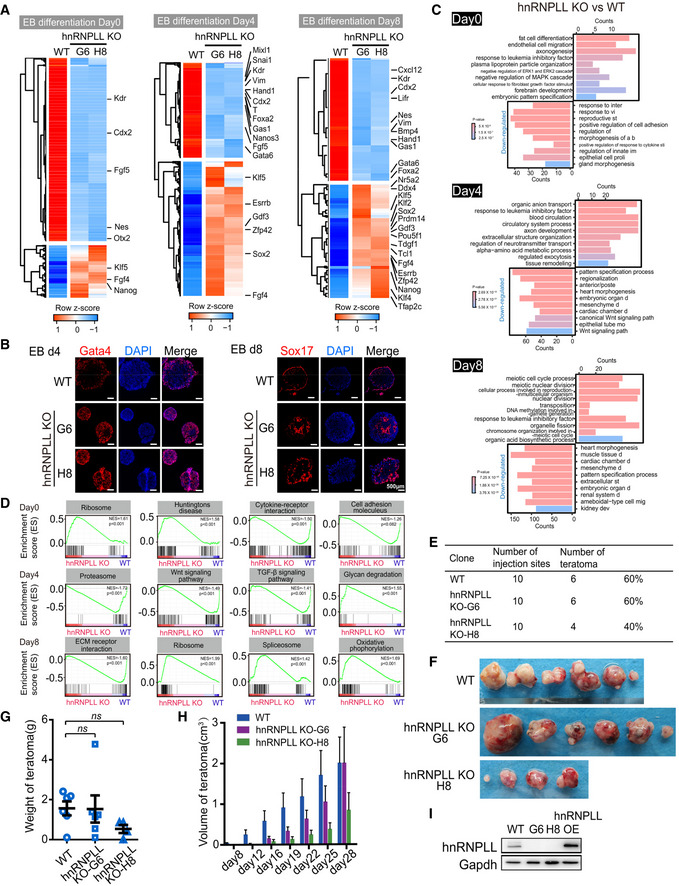
- Heatmap showing the differential expressed genes between WT and hnRNPLL‐KO ES cells during EB formation. The important pluripotency‐ and differentiation‐associated genes were indicated.
- Immunofluorescence staining of Gata4 or Sox17 in EBs generated from WT and two hnRNPLL‐KO ES cell clones (H8 and G6) at the indicated time point. Gata4‐ or Sox17‐positive endoderm cells were mainly at the outer layer of WT Ebs at differentiation day 4 or 8. Scale bar, 500 μm.
- Top 10 enriched GO terms of differentially expressed genes between WT and hnRNPLL‐KO ES cells during EB formation.
- GSEA of KEGG pathways between hnRNPLL‐KO and WT ES cells during EB formation at day 0 (upper), day 4 (middle), and day 8 (lower).
- Teratoma formation efficiency from WT and hnRNPLL‐KO ES cell lines.
- Representative images of teratomas formed from WT and hnRNPLL‐KO ES cell lines.
- Weight of teratomas derived from WT and hnRNPLL‐KO ES cells (H8 and G6). The data represent the means ± SD (n > 3 biological repeats). Ns, not significant. t‐Test was used to determine the significance.
- Volumes of teratomas at different days after injection. The data represent the means ± SD (n > 3, from biological repeats).
- Western blot validation of hnRNPLL overexpression in ES cells.
Gene Ontology (GO) analysis of the differentially expressed genes showed an enrichment in terms associated with embryonic organ development and mesenchymal development, indicative of perturbed differentiation (Fig EV2C). Furthermore, Gene Set Enrichment Analysis (GSEA) showed that ribosome and Huntingtons disease‐related genes were up‐regulated in hnRNPLL‐KO ES cells, while important cellular signaling pathways required for embryonic development, such as the Wnt and TGF‐β pathways were reduced in hnRNPLL‐KO EBs (Fig EV2D).
We next tested the ability of hnRNPLL‐KO ES cells to exit from pluripotency in vivo through teratoma formation assay. hnRNPLL‐KO ES cells showed a relatively fast proliferation rate and produced teratomas with highly variable volumes compared with WT ES cells; however, the average frequency, volume, and weight of teratomas from hnRNPLL‐KO ES cells were comparable to those from WT ES cells (Fig EV2E–H). Superficially, it seemed that teratoma formation per se was unaffected in the context of hnRNPLL‐KO ES cells. We next interrogated the teratomas composition. ES cell‐derived teratomas normally consist of a heterogeneous mix of differentiated cells representing three germ layers. We detected many large Oct4‐positive, undifferentiated regions in hnRNPLL‐KO‐derived teratomas, which by comparison were only occasional in the WT derivations (Fig 3F). Moreover, although some primitive ectoderm neuronal rosettes can be observed in some teratomas derived from hnRNPLL‐KO ES cells, mature differentiated cells such as neuroectoderm, adipocyte, cartilage, muscle cells, respiratory, and intestine‐like epithelium were distinctly lacking compared with their prevalence in teratomas derived from WT ES cells (Fig 3F). These data support that hnRNPLL‐KO ES cells exhibit a defect in exit from pluripotency and onset of differentiation both in vivo and in vitro.
Enforced expression of hnRNPLL can drive ESC differentiation
To further illustrate the role of hnRNPLL, we also tested whether the overexpression of hnRNPLL was sufficient to drive ES cell differentiation. So, we constructed the hnRNPLL overexpression vector and transfected ES cells with the vector. The results of Western blot showed that hnRNPLL was overexpressed successfully (Fig EV2I). The AP activity was reduced in ES cells and the differentiated clones increased when hnRNPLL was overexpressed (Fig 3G). Consistently, the expression of pluripotency‐associated genes (Oct4 and Sox2) was also down‐regulated in hnRNPLL‐OE ES cells (Fig 3H). All these results indicated that hnRNPLL overexpression can drive ES cell differentiation to a certain extent even under the self‐renewal culture condition.
hnRNPLL‐deficient mice are developmental impaired and growth retarded
As hnRNPLL was critical for exit from pluripotency in cultured ES cells, we would expect hnRNPLL‐KO embryos to show an abnormal developmental profile that would potentially result in lethality. To test this hypothesis, we generated hnRNPLL +/− mice, in which we deleted the critical exon 2 of hnRNPLL using CRISPR/Cas9 technology (Fig 4A). We aimed to generate hnRNPLL −/− mice from heterozygous intercrosses and confirmed the genotypes by region‐specific PCR (Fig EV3A). As expected, no hnRNPLL −/− homozygotes mice were produced from more than half (10/18) of the pregnancies; the actual birth rate of hnRNPLL −/− mice was only 12.4%, which was deviated from Mendelian genotype ratios, significantly lower than that of WT hnRNPLL +/+ mice (39.7%), suggesting that a proportion of hnRNPLL −/− littermates had embryonic lethal defects (Figs 4B and EV3B). Of the few hnRNPLL −/− homozygous mice that were born at term, they were visibly smaller and growth retarded, and all died by 3–4 weeks after birth (Fig 4C–E). Postmortem analysis showed that organ weight: body weight ratios of hnRNPLL −/− mice were abnormal for the spleen, liver, heart, and lung compared with WT or heterozygous littermates (Fig EV3C). These results indicate that hnRNPLL is important for normal ontogeny, especially embryonic development.
Figure 4. hnRNPLL‐deficient mice are developmental impaired and growth retarded.
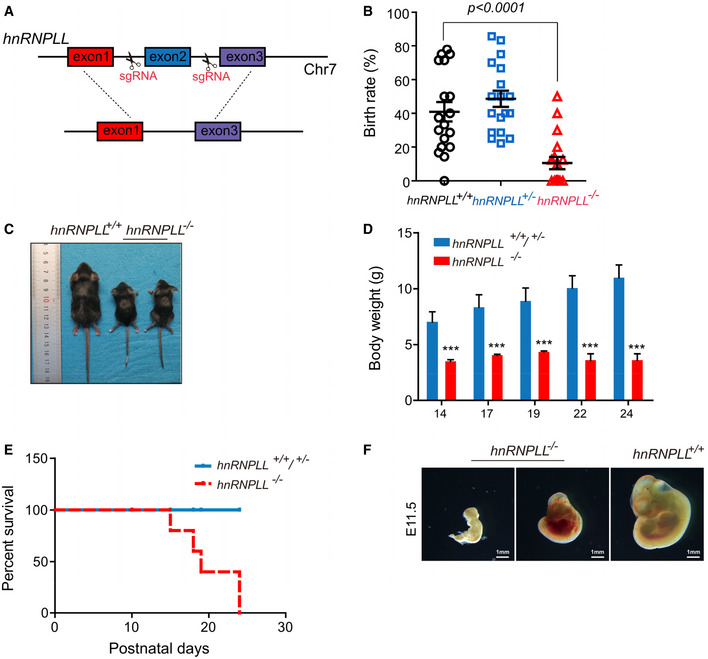
- Schematic of hnRNPLL knockout mouse, in which its exon two is deleted.
- Birth rate of the different hnRNPLL genotypes, the birth rate of hnRNPLL −/− mice was significantly lower than that of WT hnRNPLL +/+ mice. The data represent the means ± SD. t‐Test was used to determine the significance.
- Representative images of hnRNPLL +/+ and hnRNPLL −/− littermates at 3 weeks.
- Body weight growth curve of mice at different postnatal days. The data represent the means ± SD (n = 5–10). ***P < 0.001. t‐Test was used to determine the significance.
- Survival curve analysis for hnRNPLL +/+, hnRNPLL +/−, and hnRNPLL −/−mice (ten mice in hnRNPLL +/+/+/− group and seven mice in hnRNPLL −/−group).
- Genotypes and representative images of hnRNPLL +/+ embryo and retarded hnRNPLL −/− embryos at E11.5 which generated from hnRNPLL +/− intercrosses. Scale bar, 1 mm.
Figure EV3. hnRNPLL‐deficient mice exhibit a developmental delay and growth retardation.
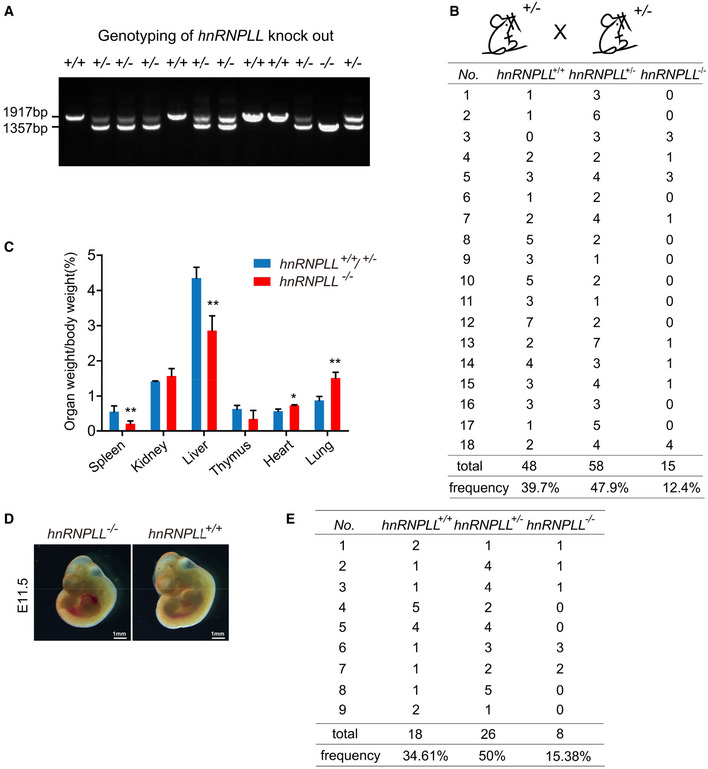
- Genotyping of littermates generated from hnRNPLL +/− intercrosses.
- Statistical analysis of the genotypes of 18 litters generated from hnRNPLL +/− intercrosses.
- The organ to body weight ratio for hnRNPLL +/+, hnRNPLL +/−, and hnRNPLL −/− mice. The data represent the means ± SD (n ≥ 3, from biological repeats). *P < 0.05; **P < 0.01. t‐Test was used to determine the significance.
- Representative images of hnRNPLL +/+ embryo and hnRNPLL −/− embryo at E11.5 which generated from hnRNPLL +/− intercrosses. The image of hnRNPLL +/+ embryo was a repeated display of Fig 4F. Scale bar, 1 mm.
- Statistical analysis of the genotypes of 9 litters at E11.5 generated from hnRNPLL +/− intercrosses.
To further determine the impact of hnRNPLL on mammalian embryo development, we collected the mouse embryos at 11.5dpc and meanwhile the corresponding genotypes were confirmed by analyzing the yolk sac samples. We found that most of the hnRNPLL −/− embryos showed serious growth retardation, exhibiting as smaller embryos and serious malformation (Fig 4F), which clearly suggested that hnRNPLL functioned earlier than E11.5. Moreover, consistent with our previous observation on the newborns (Fig 4B), some visible normal hnRNPLL −/− E11.5 embryos were also observed (Fig EV3D), although the ratio (15.38%) was significantly lower than that of WT hnRNPLL +/+ mice (34.61%) (Fig EV3E). The incomplete penetrance of hnRNPLL −/− embryos also suggested the complexity of gene regulatory network and the possible gene redundancy in vivo. Taken all the observed phenotypes from the mutant mice, we concluded that hnRNPLL played important role in early‐stage embryo development.
hnRNPLL interacts with RNA splicing proteins to regulate RNA alternative splicing in ES cells
As mentioned, hnRNPLL regulates AS in T and B cells (Oberdoerffer et al, 2008), but also seems to regulate gene expression at the transcriptional level, for example, by activating BCL2 via binding to the i‐motif in the BCL2 promoter (Roy et al, 2016). To investigate the mechanism by which hnRNPLL controls ES cell exit from pluripotency, we first performed immunoprecipitation (IP) of hnRNPLL experiments to identify any hnRNPLL interacting proteins in ES cells. We detected many specific protein bands that were enriched in the hnRNPLL IP group, compared with the IgG‐negative control (Figs 5A and EV4A). We then identified these enriched proteins by mass spectrometry to uncover the hnRNPLL interactome in ES cells. Some of these interactions were included in the STRING database, and many of the hnRNPLL partners exhibited dense interaction networks (Fig EV4B). Impressively, nearly 35% of the hnRNPLL‐interacting proteins in ES cells were associated with RNA splicing, such as hnRNPA1, hnRNPA2B1, PTBP1, and hnRNPA3; we verified these interactions by Western blotting (Fig 5B and C; Dataset EV5). The strong enrichment of RNA‐splicing proteins in the hnRNPLL interactome implies that similar to its role in immune cells, hnRNPLL function in ES cells involves the control of RNA splicing.
Figure 5. hnRNPLL regulates RNA alternative splicing in ES cells.
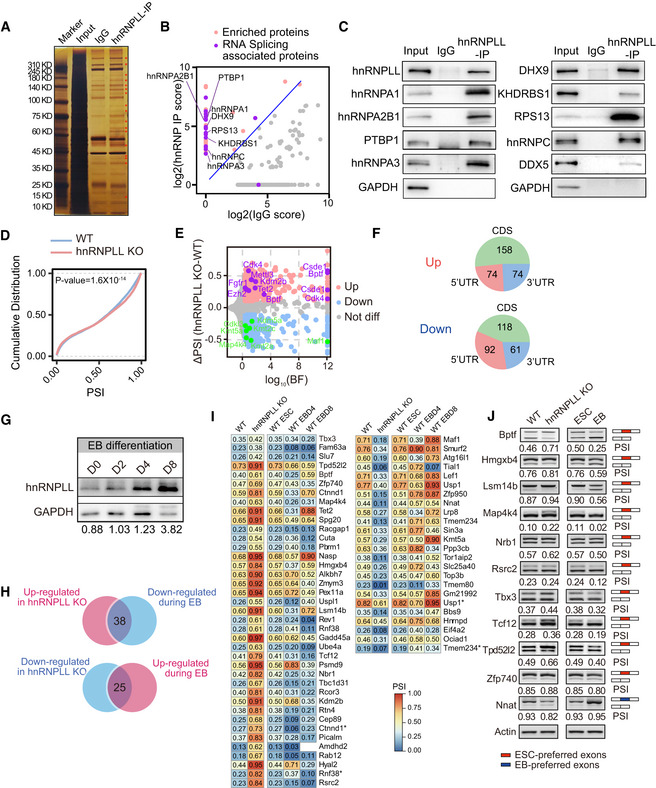
- Silver nitrate‐stained NuPAGE gel of hnRNPLL‐interacting proteins. Red star indicate hnRNPLL‐specific interacting proteins compared with IgG.
- Mass spectrum identification of the hnRNPLL interactome in ES cells. RNA splicing associated proteins are well enriched by hnRNPLL IP. The enrichment score was log2‐transformed before plotting.
- Western blot validation the interactions between hnRNPLL and the indicated splicing‐associated proteins.
- Cumulative distribution of PSI with exon skipping events between WT and hnRNPLL‐KO ES cells. Kolmogorov–Smirnov test was used to determine the significance.
- Volcano plot of differential exon skipping events between WT and hnRNPLL‐KO ES cells. Indicated AS events were labeled in purple (up‐regulated) and green (down‐regulated) with gene names.
- The regional distribution of differential exon skipping events between WT and hnRNPLL‐KO ES cells.
- Western blot analysis of hnRNPLL protein during EB formation of WT ES cells at indicated days.
- Venn plot showed the divergent exon skipping events between EB differentiation and hnRNPLL deletion. 38 exon skipping events that were inhibited by hnRNPLL but increased during EB differentiation, and 25 events that showed the opposite profile.
- Heatmap of the 38 and 25 divergent exon skipping events in Fig 5I. * indicates the different exon skipping events of the same gene.
- RT–PCR validation of partial divergent exon skipping events shown in Fig 5J. Red boxes represent ESC preferred exons, blue boxes indicate EB preferred exons.
Figure EV4. hnRNPLL regulates RNA alternative splicing in ES cells.
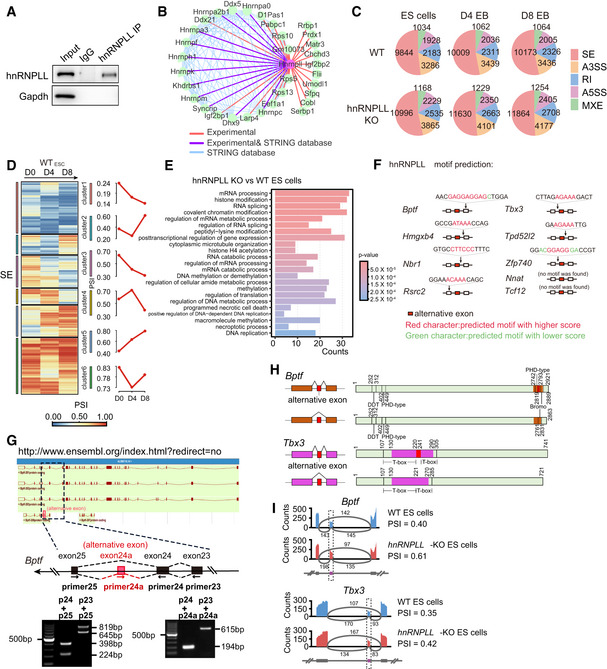
- Immunoprecipitation of hnRNPLL in ES cells.
- The protein–protein interaction network between hnRNPLL and its partners reported in the STRING database. The purple lines indicated the interactions verified by mass spectrum, while the red lines represent the interactions only reported in our mass spectrum data.
- The number of AS events of five AS types occurring at each stage during differentiation, for WT and hnRNPLL‐KO ES cell lines.
- Heatmap of the differential exon skipping events during WT EB formation (left). The AS events were classified into 6 clusters and average PSI value of each cluster were plot in red (right).
- Gene ontology enrichment based on the differentially expressed AS genes between WT and hnRNPLL‐KO cells.
- Predicted hnRNPLL‐binding motif in the indicated transcripts. Arrow indicates motif position relative to the alternatively spliced exon.
- Verification of the Bptf isoforms including the alternative exon. Images of Bptf isoforms from Ensembl database (upper).
- Position of the indicated alternative exon relative to the known Tbx3 or Bptf functional domain.
- Sashimi plots showing the read distribution of alternative spliced exons of Bptf and Tbx3 between the WT and hnRNPLL‐KO ES cells. The alternative spliced exons of the AS events were colored in purple. Numbers of exon–exon junction reads were marked above gray arc. PSI of AS events was labeled on the right.
AS of RNA generally occurs by one of five simple modes, namely exon skipping (SE), alternative 3′ splicing site (A3SS), alternative 5′ splicing site (A5SS), intron retention (RI), and mutual exon (MXE) (Chen & Weiss, 2015). We found that the proportion of each AS type was stable during EB formation and was unaffected by hnRNPLL depletion (Fig EV4C). To quantify AS, we estimated the percent spliced in (PSI)—the exon utilization rate. Proportion of each AS type including exon skipping was stable, but there were many dynamic exon skipping events, in which the PSI varied markedly during EB formation (Fig EV4D); we thus only focused on exon skipping events going forward. The PSI for exon skipping was markedly changed in hnRNPLL‐KO ES cells compared with WT ES cells (Fig 5D). We detected aberrant exon skipping of target genes in hnRNPLL‐KO ES cells, with both signs of exon skipping repression and promotion. In particular, we identified that known pluripotency genes, including Csde1, Fgfr1, Ezh2, and Bptf, were all irregularly spliced with hnRNPLL knockout (Fig 5E; Dataset EV6).
GO analysis showed that the differentially spliced genes in hnRNPLL‐KO ES cells were enriched for mRNA processing, histone modification, and RNA splicing (Fig EV4E). This finding suggests that hnRNPLL‐regulated RNA splicing elicits a cascade of effects, since AS of the important RNA processor was regulated by hnRNPLL. The alternative exons that were preferentially retained were CDS (coding sequence), followed by 5′UTR and then 3′UTR regardless as to whether the PSI was elevated or suppressed (Fig 5F).
Then, we raised the question as to which AS events hnRNPLL specifically regulated to permit ES cell exit from pluripotency. hnRNPLL is up‐regulated during EB differentiation (Fig 5G); thus, we presumed that the most significant differential exon skipping events between hnRNPLL‐KO and WT ES cells during EB differentiation might be direct targets of hnRNPLL during exit from pluripotency. We detected 38 exon skipping events that were inhibited by hnRNPLL but increased during EB differentiation and 25 events that showed the opposite profile (Fig 5H and I). Specifically, the alternative exons of Tbx3, Bptf, and Tcf12 were typically included when hnRNPLL expression was lost, but generally skipped during EB differentiation (Fig 5J and K). The converse was true for the alternative exons of Nnat, Lef1, and Kmt5a. Part of the aberrant AS events and their change during EB differentiation were validated by RT–PCR (Fig 5J). The results showed that the change of these AS events in hnRNPLL‐KO or during EB differentiation was basically consistent with the RNA‐sequencing data, although the PSI value was not exactly the same. These data suggest that the differentiation defects in hnRNPLL‐KO ES cells might be due to aberrant splicing.
hnRNPLL promotes Bptf and Tbx3 exon skipping to facilitate ES cell exit from pluripotency
We next wanted to explore whether any of the described alternative exons or nearby introns were directly bound by hnRNPLL and thus to understand the underlying molecular mechanism. To this aim, we performed hnRNPLL RNA immunoprecipitation‐PCR (RIP‐PCR) and found that of the PCR‐verified exon skipping events (Fig 5J), nine gene transcripts, including Bptf, Hmgxb4, Nbr1, Nnat, Rsrc2, Tbx3, Tcf12, Tpd52l2, and Zfp740 were highly enriched in the hnRNPLL immunoprecipitates (Fig 6A). Next, we aimed to verify these interactions with hnRNPLL using capture and ligation probe‐PCR (CLIP‐PCR), which can define the exact RBP and RNA‐binding sites by using ultraviolet (UV) cross‐linking to covalently capture close associations between an RNA and a protein (Fig 6B). We found that the potential hnRNPLL‐binding sequence was present in the flanked introns or exons of the alternative exon in seven genes, with the two exceptions being Nnat and Tcf12 (Fig EV4F). After extracting RNA from the square region indicated in Fig 6C, we found that all the predicted binding sites in the seven transcripts were enriched significantly in hnRNPLL immunoprecipitates (Fig 6C), implying that hnRNPLL binds to these sequences and promotes specific exon skipping.
Figure 6. hnRNPLL promotes Bptf and Tbx3 exon skipping to facilitate ES cell exit from pluripotency.
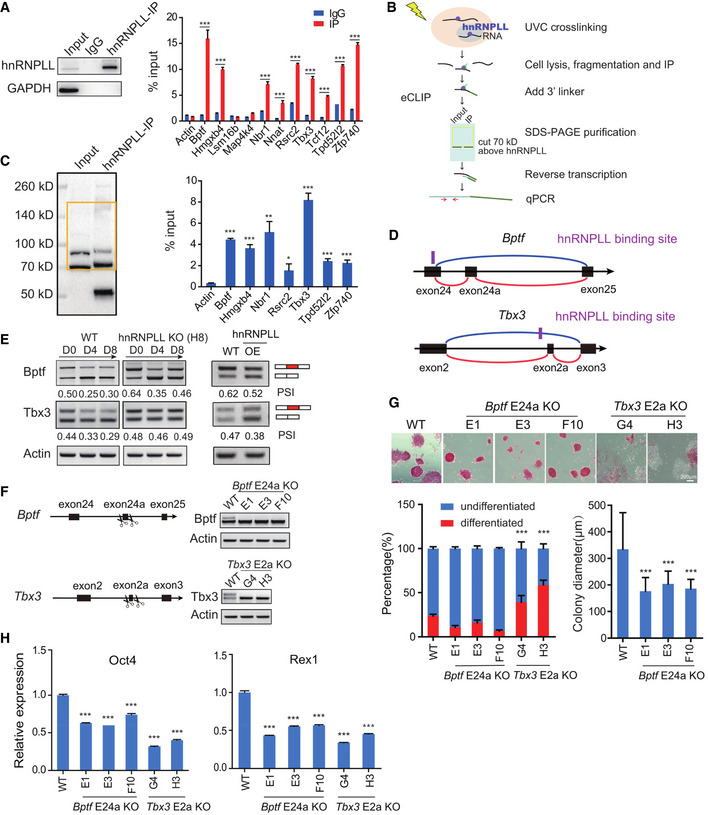
- RIP‐PCR validation of the transcripts that interact with hnRNPLL. The left panel shows the results of Western blots verification of hnRNPLL Immunoprecipitation efficiency. The data represent the means ± SD (n = 3, from biological repeats). ***P < 0.001. t‐Test was used to determine the significance.
- The CLIP‐PCR pipeline. Specifically, hnRNPLL and its target RNA are cross‐linked by UV, then enriched by immunoprecipitation. 3′linker is added to the RNA tag, then hnRNPLL–RNA complex is purified by SDS–PAGE. After protease K digestion, RNA is reversed into cDNA. qRT–PCR is performed to detect cross‐linked RNA.
- Purification of hnRNPLL–RNA complex and detection of the RNA targets in the complex by PCR. The left panel shows the membrane position labeled as the yellow square which was cut to extract the hnRNPLL–RNA complex. The bands around 70KD indicate hnRNPLL proteins, and the bands around 50KD indicate the heavy chain of antibodies used in Immunoprecipitation. The right plot indicates the enrichment of these RNA targets in the hnRNPLL–RNA complex. The data represent the means ± SD (n = 3, from biological repeats). *P < 0.05; ***P < 0.001. t‐Test was used to determine the significance.
- Bptf and Tbx3 alternative splicing model: Bptf exon24a and Tbx3 exon2a are alternatively spliced. The predicted hnRNPLL‐binding motif sites were marked in purple. The red and blue lines represented include and excluded isoforms of the AS events, respectively.
- Detection of Bptf and Tbx3 exon skipping events during EB formation in WT and hnRNPLL‐KO ES cells (left) and in hnRNPLL overexpression ES cells under ES cell culture conditions (right) by RT–PCR. OE, hnRNPLL overexpression.
- RT–PCR validation of Bptf exon24a and Tbx3 exon2a deletion in WT ES cells.
- AP staining of Bptf E24a KO or Tbx3 E2a KO ES cells maintained under normal ES cell culture conditions (upper). Scale bar, 200 μm. Colony formation assay of Bptf E24a KO or Tbx3 E2a KO ES cells under ES cell culture condition (lower left). The data represent the means ± SD (n = 3, from biological repeats). ***P < 0.001. Colony diameter of Tbx3 E2a KO ES cells (lower right). The data represent the means ± SD (n > 80). ***P < 0.001. t‐Test was used to determine the significance.
- Relative expression of Oct4 and Rex1, determined by qRT–PCR. The data represent the means ± SD (n = 3, from biological repeats). ***P < 0.001. t‐Test was used to determine the significance.
Of the seven hnRNPLL‐binding and hnRNPLL‐regulating genes, Bptf and Tbx3 are known to contribute to the pluripotency maintenance and/or the onset of differentiation in ES cells (Landry et al, 2008; Han et al, 2010; Russell et al, 2015). Although the changes in AS of Tbx3 were subtle in the RNA‐seq data, the alternative exon of Tbx3 already has been concerned (Zhao et al, 2014), and the role of the different isoforms in ES cells was unknown. So, we focused on Bptf and Tbx3 AS to determine how in this case, they might regulate ES cell exit from pluripotency. Based on Ensembl database annotation, the alternative exon of Bptf affected by hnRNPLL should be the second exon of a very short isoform (Ensembl, transcript: Bptf‐208, Fig EV4G), but our RT–PCR results verified the existence of a longer isoform containing the exon in ES cells (Fig EV4G), so we named this alternative exon as exon24a in our further work. The Bptf alternative exon24a is removed to destroy the second PHD‐type Zinc finger in the peptide (2,742–2,793 aa) (UniProt, 2019). The Tbx3 alternative exon resides between two T‐boxes (for DNA binding) but its skipping has no effect on the T‐box itself (Figs 6D and EV4H). We found that in ES cells, hnRNPLL regulates the two exon skipping events and thus the two different Bptf and Tbx3 isoforms coexist (Fig 6E). When WT ES cells exited from pluripotency, hnRNPLL up‐regulated and promoted the exon skipping, resulting in a decrease of the longer Bptf and Tbx3 isoforms. By contrast, hnRNPLL depletion led to a clear increase in expression levels of the longer Bptf and Tbx3 isoforms (Figs EV4I and 6E). The expression level of the longer isoform was maintained or did not return to normal during the attempted EB differentiation of hnRNPLL‐KO cells. Whereas, overexpression of hnRNPLL promoted the exon skipping of Bptf and Tbx3, resulting in a decrease of the longer Bptf and Tbx3 isoforms (Fig 6E). In addition, we also test the utilization of Tbx3 alternative exon in ES cells cultured under 2i + LIF condition, by analyzing published RNA‐seq data (GSE23943) (Marks et al, 2012) and RT–PCR (Fig EV5A).
Figure EV5. hnRNPLL regulates AS of Bptf and Tbx3 in ES cells.
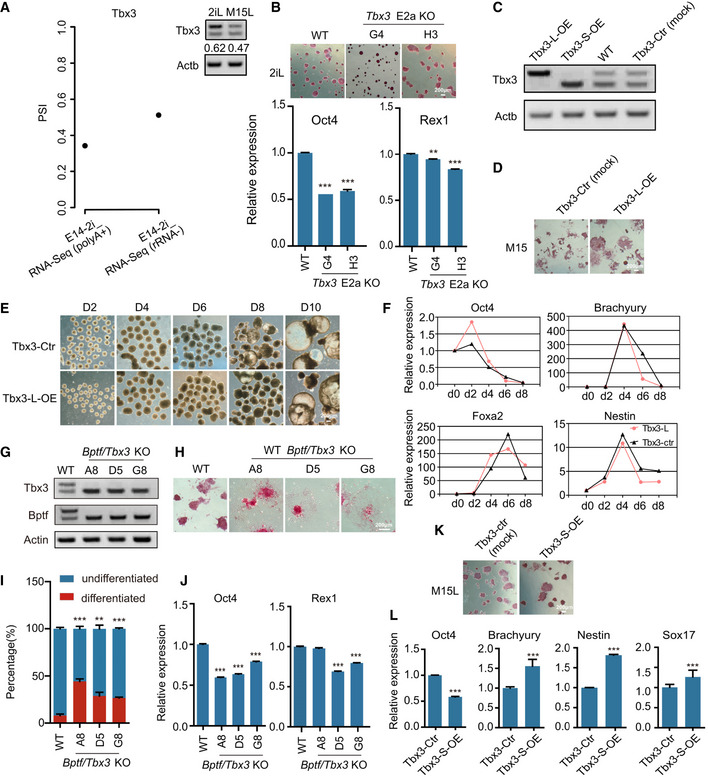
- Utilization rate of Tbx3 alternative exon in 2i + LIF culture condition in the published 2i + LIF RNA‐seq data (GSE23943) (left) and also detected by RT–PCR (right).
- AP staining and relative expression of Oct4 and Rex1 in WT or Tbx3 E2a KO ES cells maintained under 2i + LIF culture conditions. The data represent the means ± SD (n = 3, from biological repeats). **P < 0.01, ***P < 0.001. t‐Test was used to determine the significance. Scale bar, 200 μm.
- RT–PCR validation of Tbx3‐L or Tbx3‐S overexpression in ES cells. Tbx3‐L‐OE, Tbx3 long isoform overexpression. Tbx3‐S‐OE, Tbx3 short isoform overexpression. Tbx3‐Ctr, ES cells transfected with empty vector.
- AP staining of Tbx3‐Ctr and Tbx3‐L‐OE ES cell lines after LIF withdrawal. Scale bar, 200 μm.
- EB formation from Tbx3‐Ctr and Tbx3‐L‐OE ES cell clones. Scale bar, 200 μm.
- Relative expression levels of pluripotency genes (Oct4) and lineage‐specific genes (Brachyury, Nestin, Foxa2) during EB formation at the indicated days as measured by qRT–PCR. The data represent the means ± SD (n = 3, from biological repeats).
- RT–PCR validation of Bptf and Tbx3 alternative exon deletion in WT ES cells.
- AP staining of Bptf/Tbx3 KO ES cells maintained under normal ES cell culture conditions. Scale bar, 200 µm.
- Colony formation assay of Bptf/Tbx3 KO ES cells under ES cell culture condition. The data represent the means ± SD (n = 3, from biological repeats). **P < 0.01, ***P < 0.001. t‐Test was used to determine the significance.
- Relative expression of Oct4 and Rex1 in Bptf/Tbx3 KO ES cells, determined by qRT–PCR. The data represent the means ± SD (n = 3, from biological repeats). ***P < 0.001. t‐Test was used to determine the significance.
- AP staining of Tbx3‐Ctr and Tbx3‐L‐OE ES cell lines under ES cell culture condition. Scale bar, 200 μm.
- Relative expression of Oct4, Brachyury, Nestin, and Sox17 with Tbx3‐S‐OE. The data represent the means ± SD (n = 3, from biological repeats). ***P < 0.001. t‐Test was used to determine the significance.
From these results, we hypothesized that the long (exon included) Bptf and Tbx3 isoforms are important for maintaining ES cell self‐renewal and that a decline in the expression levels of these isoforms is required for ES cell exit from pluripotency. To test this hypothesis, we specifically deleted the alternative exons in these two genes in ES cells (Fig 6F) and then monitored colony formation efficiency. Although ES cell colony formation efficiency was not affected by Bptf long transcript, the colony diameter was significantly reduced (Fig 6G). Strikingly, Tbx3 long transcript depletion led to obvious differentiation and decreased colony formation ability (Fig 6G), consistent with the known role of Tbx3 in maintaining ES cell self‐renewal (Russell et al, 2015; Waghray et al, 2015). Accordingly, the expression levels of the self‐renewal markers Oct4 and Rex1 were also significantly decreased in ES cells with Bptf exon24a or Tbx3 exon2a deletion (Fig 6H). Meanwhile, Tbx3 exon2a was not required to maintain the naïve pluripotent state (Fig EV5B), which may be due to the compensations from other genes or signal transductions under 2i + LIF culture conditions. These results support that the exon‐included long Bptf and Tbx3 isoforms promote ES cell self‐renewal; abnormal expression of these isoforms as a result of hnRNPLL depletion might be the main obstacle to exit from pluripotency in hnRNPLL‐KO ES cells.
Further, we also determined the different role of Tbx3 isoforms in regulating pluripotency. We overexpressed long and short Tbx3 isoforms in ES cells, respectively. Enforced expression of the long Tbx3 isoform showed stronger colony formation and AP activity than control cells after LIF withdrawal and also can induce a little delay in Oct4 silencing during EB formation, although cannot block ES cell differentiation obviously (Fig EV5C–F). These results indicated the contribution of the long Tbx3 isoform in maintaining ES cell self‐renewal. Whereas, overexpression of the short Tbx3 isoform decreased the expression of self‐renewal marker and also a little stimulation of the differentiation markers, suggesting its potential role in facilitating ES cells exit from pluripotency (Fig EV5C, K and L).
Bptf and Tbx3 alternative exon loss can rescue defective differentiation in hnRNPLL‐KO ES cells
In our final assays, we sought to investigate whether Bptf exon24a or Tbx3 exon2a deletion can rescue the differentiation deficiency observed in hnRNPLL‐KO cells. The alternative Bptf or Tbx3 exons were uniquely or simultaneously (Bptf/Tbx3) deleted in hnRNPLL‐KO ES cells (H8) (Fig 7A) and WT ES cells (Fig EV5G). The spliced exon deletion did not alter TBX3 or BPTF protein expression significantly (Appendix Fig S2A). As the alternative splicing of Tbx3 or Bptf has little effect on their proteins’ molecular weight, Western blot cannot reflect the consequences of alternative splicing. For the WT cells, the Bptf/Tbx3 double exon‐KO led to partial differentiation and decreased colony formation ability and also the decreased expression of self‐renewal markers Oct4 and Rex1 (Fig EV5H–J). However, we observed no differences in AP activity or colony formation efficiency in these mutant hnRNPLL‐KO ES cells, indicating other events affected by hnRNPLL‐KO might compensate for Bptf or Tbx3 long isoform depletion (Appendix Fig S2B and C). When these cells were cultured without LIF, hnRNPLL‐KO H8, Bptf, and Tbx3 exon‐KO H18 ES cells still showed high AP activities and colony formation abilities, while the Bptf/Tbx3 double exon‐KO H8 ES cells differentiated normally and were comparable with WT ES cells (Fig 7B and C).
Figure 7. Bptf and Tbx3 alternative exon loss can rescue defective differentiation in hnRNPLL‐KO ES cells.
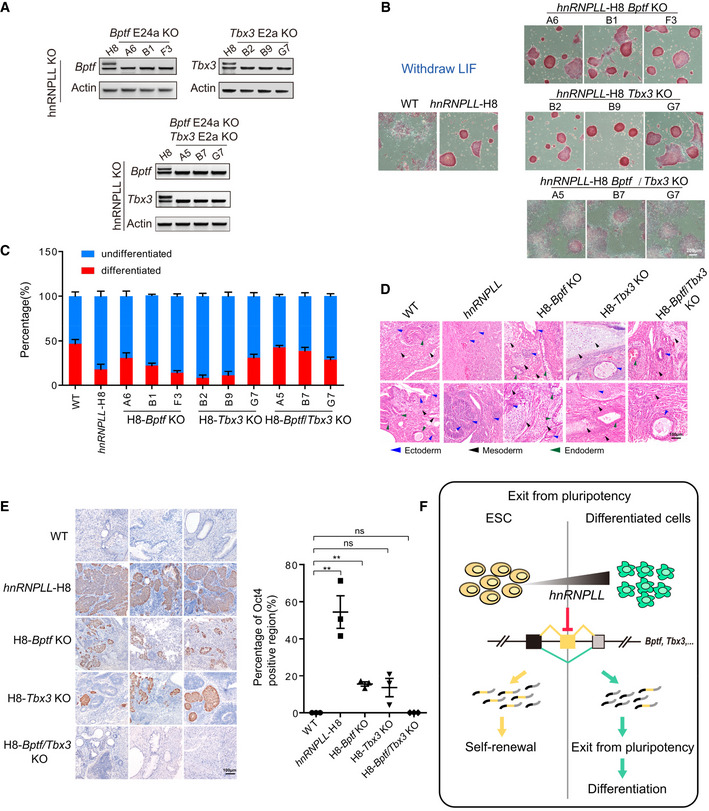
- RT–PCR validation of Bptf and Tbx3 alternative exon deletion, separately in hnRNPLL‐KO H8 cells (upper) and simultaneously in hnRNPLL‐KO H8 cells (lower).
- AP staining of H8‐Bptf or H8‐Tbx3 exon‐KO ES cells after LIF withdrawal. hnRNPLL‐H8, hnRNPLL‐KO ES cells; H8‐Bptf KO, alternative Bptf exon was deleted in hnRNPLL‐KO H8 cells; H8‐Tbx3 KO, alternative Tbx3 exon was deleted in hnRNPLL‐KO H8 cells; H8‐Bptf/Tbx3 KO, alternative Bptf or Tbx3 exons were simultaneously deleted in hnRNPLL‐KO H8 cells. Scale bar, 200 μm.
- Colony formation assay of H8‐Bptf or H8‐Tbx3 exon‐KO ES cells after LIF withdrawal. The data represent the means ± SD (n = 3, from biological repeats).
- Hematoxylin and eosin labeling of teratomas derived from WT and H8‐Bptf or H8‐Tbx3 exon‐KO ES cells. The blue triangle represent ectoderm, the dark one represent mesoderm, and the green one represent endoderm. Scale bar, 100 μm.
- Oct4 immunohistochemical staining of teratomas derived from WT and H8‐Bptf or H8‐Tbx3 exon‐KO ES cells. Representative Oct4‐positive regions are shown (left). Scale bar, 100 µm. The percentage Oct4‐positive region (%) is shown (right). The data represent the means ± SD (n = 3 biological repeats). **P < 0.01; ns, not significant. t‐Test was used to determine the significance.
- Schematic representation of hnRNPLL regulating the AS of crucial genes that have an important role in mediating exit from pluripotency.
We next repeated our EB formation and teratoma formation assay using these mutant cell lines. Morphologically, the EB formation deficiency with hnRNPLL depletion was not rescued by Bptf E24a/Tbx3 E2a KO; it might be due to the important and complicated role of Tbx3 long isoform in regulating pluripotency (Appendix Fig S2D). As before, we identified large undifferentiated regions in hnRNPLL‐H8‐derived teratomas, indicative of Oct4 expression, and only a few differentiated tissues. The extent of Oct4‐positive areas in hnRNPLL‐H8‐Bptf KO and hnRNPLL‐H8‐Tbx3 KO‐derived teratomas, however, was comparatively reduced and the hnRNPLL‐H8‐Bptf/Tbx3 KO derivations showed no overt difference with WT ES cell‐derived teratomas. Moreover, all three germ layers were visible in these hnRNPLL‐H8‐Bptf and Tbx3 KO‐derived teratomas (Fig 7D and E). These data demonstrate that the simultaneous disruption of the Bptf/Tbx3 alternative exons can rescue the differentiation deficiency observed in hnRNPLL‐KO ES cells. AS that is regulated by hnRNPLL seems to have an important role in ES cell exit from pluripotency. Mechanistically, hnRNPLL promotes multiple ES cell‐preferred exon skipping events, including exon skipping in Bptf and Tbx3. Promoting these events ultimately reduces the expression levels of ES cell‐specific isoforms and facilitates ES cells exit from pluripotency to permit differentiation (Fig 7F).
Discussion
In the present study, we combined bioinformatics analyses with newly generated ES KO cells and KO mouse models to reveal a previously unknown role for RBPs in ES cell exit‐form‐pluripotency. We found that the RBP hnRNPLL can regulate the AS of multiple genes and, in this way, repress the expression levels of ES cell‐preferred isoforms to facilitate pluripotency exit. Our data highlight the contribution of specific regulated AS during the exit from pluripotency (namely Bptf and Tbx3) and delineate the hnRNPLL‐regulated AS program responsible for dismantling pluripotency circuitry.
Detailed dissection of hnRNPLL’s role in ES cell self‐renewal and different stage of differentiation revealed that hnRNPLL played critical roles in exit from pluripotency, but did not affect the transition from naïve to primed pluripotency. Meanwhile, it may also participate in the progression to lineage commitment, as its depletion not only resulted in retarded pluripotency exit, but also the improper lineage commitment and organ developmental defect. A small part of hnRNPLL‐null embryos can survive to E11.5 even to birth which seems inconsistent with its critical role in controlling ES cell exit‐form‐pluripotency. It is most likely caused by gene redundancy, different gene expression profile, incomplete penetrance, etc., which also reflected some differences between the cultured ES cells and the epiblast in blastocyst. This is also the case for some previously defined exit from pluripotency factors, such as Flcn, Tsc2, Pum1, and Whsc1 (Betschinger et al, 2013; Leeb et al, 2014; Tian et al, 2019).
Our work supports the essential role of RBPs in mediating post‐transcriptional control during ES cell pluripotency regulation (Ye & Blelloch, 2014; You et al, 2015; Chen & Hu, 2017; Li & Izpisua Belmonte, 2018). More recently, several RBPs are also found to be required for ES cell differentiation, for example, noncanonical function of DGCR8 facilitates the splicing of Tcf7l1 (also known as Tcf3) and promotes ES cell exit from pluripotency (Cirera‐Salinas et al, 2017). RNA helicase DHX9 processes the intergenic spacer (IGS)‐rRNA and mediates nucleolar heterochromatic formation, which is required for ES cell differentiation (Leone et al, 2017). Another RNA helicase DDX6‐depleted ES cells lose P‐bodies and cannot exit pluripotent state successfully (Di Stefano et al, 2019). These studies together with our work demonstrated that RBP‐mediated RNA regulation participated in controlling ES cell pluripotency exit and the onset of differentiation. The RBP‐mediated post‐transcriptional repression of the pluripotency genes may coordinate the transcriptional repression, even earlier than the regulation at transcriptional level, as the down‐regulation of key pluripotency genes was observed before than the chromatin accessibility changes at regulatory regions of these genes (Li & Izpisua Belmonte, 2018; Di Stefano et al, 2019). The fact seems that it is necessary to shut down the transcribed pluripotency genes in ES cells through post‐transcriptional regulation when starting the differentiation program. So, RBP‐mediated post‐transcriptional regulation may play unexpected role in cell fate transition which was previously neglected and should be followed with interest. What is exciting is that repression of a single RNA‐binding polypyrimidine‐tract‐binding protein (PTBP1) is sufficient to induce the trans‐differentiation of fibroblasts into functional neurons (Xue et al, 2013). The DEAD‐box RBP DDX5 was reported to act as a reprogramming roadblock, and its knockdown can significantly enhance the efficiency of iPSC induction (Nefzger & Polo, 2017). Accumulating evidences as well as the important role of RBPs in controlling ES cell pluripotency exit uncovered here suggest that targeting RBPs holds great promise for mediating ES cell‐directed differentiation or inducing pluripotency.
AS is a versatile post‐transcriptional mechanism that ultimately expands protein diversity required for numerous cell fate transitions during development (Kalsotra & Cooper, 2011; Cieply et al, 2016; Ule & Blencowe, 2019). Accumulating evidence supports that AS is dynamically regulated during ES cell differentiation to in turn regulate ES cell pluripotency and differentiation (Salomonis et al, 2010; Chen et al, 2015; Han et al, 2017; Xu et al, 2018). Several pivotal AS events and RBP regulators (such as MBNL and SFRS2) in controlling pluripotency have been uncovered. For example, MBNL‐enhanced expression of the Foxp1 ESC‐specific isoform is critical for efficient iPSC formation, ES cell self‐renewal, and pluripotency (Han et al, 2013; Xu et al, 2018). SFRS2 regulates AS of the methyl‐CpG‐binding protein MBD2, the isoforms of which have opposing roles in ES cell maintenance and reprogramming to pluripotency (Lu et al, 2014). Transcription factor 3, TCF3 (E2A) AS is regulated by the hnRNP H/F: This splicing event results in the mutual expression of the E12 and E47 transcription regulators. TCF3‐E47 decreases E‐cadherin expression to promote human ES cell differentiation, while TCF3‐E12 contributes to pluripotency maintenance (Yamazaki et al, 2018). Here, we found that hnRNPLL can regulate a series of AS events to facilitate ES cells to exit from pluripotency. These data support that different isoforms of the same gene can have opposing roles on ES cell fate, and the switch between these isoforms can often participate in cell fate transition. Together, these data demonstrate that AS is an important layer of pluripotency regulation.
The function of Tbx3 and Bptf in ES cells has been addressed previously. Bptf is a chromatin remodeling protein that is required for ES cell differentiation and embryo development, as both EB and teratoma formation are blocked after Bptf deletion (Landry et al, 2008; Qiu et al, 2015). Here, we found that the exon24a of Bptf is alternatively spliced and the resulting two isoforms may function distinctively. The exon‐included (long) isoform participates in ES cell proliferation and colony formation, and its down‐regulation seems to be required for ES cell differentiation. By contrast, the excluded (short) isoform is up‐regulated during ES cell differentiation; whether this isoform mainly contributes to ES cell differentiation (as previously reported) is elusive and requires further investigation. The role of Tbx3 in supporting ES cell self‐renewal is also well investigated but not fully understood. Early studies demonstrated that depletion of Tbx3 in ES cells leads to differentiation (Ivanova et al, 2006; Han et al, 2010), but recent work indicates that Tbx3 plays critical role in pluripotency exit and early cell fate decision, instead of the induction and maintenance of naive pluripotency (Russell et al, 2015; Waghray et al, 2015; Zhang et al, 2019). AS of Tbx3 has been reported in ES cells (Zhao et al, 2014), but the differences in roles of the two isoforms in maintaining pluripotency were obscure. Here, we show that it is the longer transcript (including the alternative exon2a) not the shorter transcript that maintains ES cell self‐renewal, and hnRNPLL‐mediated post‐transcriptional repression of the longer transcript promotes pluripotency exit. Of course, a single AS of Tbx3 or Bptf was insufficient to drive pluripotency exit; it seems that a series of AS events regulated by hnRNPLL helped ES cells to exit from pluripotency, not just AS of Tbx3 and Bptf. Now, the underlying molecular mechanisms of the different Bptf and Tbx3 isoforms and their functions in ES cells warrant further exploration.
In conclusion, we systematically identified the roles of RBPs in controlling ES cell exit from pluripotency and uncovered an AS regulator, hnRNPLL‐mediated exon skipping was important for the exit from the pluripotent state and its absence would lead to improper lineage commitment. This study offers a valuable resource for further studying RBPs in ES cell maintenance and differentiation and also proposes the application of targeting RBPs in ES cell‐based regenerative medicine.
Materials and Methods
Cell culture
Mouse AB1 ES cells were cultured on γ‐irradiated DR4 Mouse Embryonic Fibroblast(MEF) feeder cells (neomycin, hygromycin, puromycin, and 6‐thioguanine‐resistant) in ES cell culture medium supplemented with 15% fetal bovine serum, 1,000 U/ml leukemia inhibitory factor (LIF, Millipore), 2 mM GlutaMAX (GIBCO), 1% mM MEM Non‐Essential Amino Acids Solution (GIBCO), 0.1 mM 2‐mercaptoethanol (GIBCO), 50 U/ml penicillin, and 50 U/ml streptomycin (GIBCO). The cells were cultured at 37°C with 5% CO2 in a humidified environment and passaged every other day. The naïve culture medium was N2B27 supplemented with 2i (3 μM CHIR99021, 1 μM PD0325901) and LIF (1,000 U/ml LIF). N2B27 consisted of 1:1 DMEM/F12 (Gibco) with Neurobasal medium (Gibco) supplemented with 0.5% N2 supplement (Gibco), 1% B27 supplement (Gibco), 0.033% BSA 7.5% solution (Sigma), 0.1 mM 2‐mercaptoethanol, and 2 mM Glutamax. To transit from naïve to primed state, the ES cells cultured under naïve state were trypsinized and seeded onto gelatine and FCS‐coated plates in N2B27 medium supplemented with 1,000 U/ml LIF, 2 μM IWP2 (R&D), 20 ng/ml activin A (R&D), and 12 ng/ml bFGF. All the cells used in this study are negative for Mycoplasma contamination.
CRISPR/Cas9‐mediated gene knockout in mouse ES cells
The candidate genes were disrupted as previously described (Liu et al, 2017). To minimize off‐target cleavage, we used the Cas9 nickase (D10A) (Ran et al, 2013) to disrupt candidate RBP genes with paired guide RNAs in AB1 ES cells. Briefly, the sgRNAs were cloned into the pX335 construct that expressed Cas9 nickase (D10A). Then, million cells were transfected by electroporation (230 V, 500 µF; Bio‐Rad Gene Pluser) with 30 µg paired Px335‐sgRNA and 5 µg puromycin‐selection plasmid, PB‐puro. After 48 h of puromycin‐selection, electroporated cells were trypsinized and replated to allow the growth of ES cell colonies. ES cell colonies were picked on day 7. Genomic DNA was extracted from the cells, and PCR was performed. Gene knockout clones were identified by enzyme digestion of the PCR product combined with Sanger sequencing. To disrupt the longer transcripts of target genes, sgRNAs were cloned into the pX330 construct, two pX330‐sgRNA plasmids for each gene. The target clones were first analyzed by PCR and detected the expression level by RT–PCR and real‐time PCR. The oligos and primers are listed in Dataset EV7.
Knockout mouse and embryo procedures
All the mice used in this study are C57BL/6 genetic background. hnRNPLL +/− mice were constructed in Institute of Laboratory Animal Science, CAMS&PUMC by deleting its exon2. hnRNPLL +/− mice were crossed to obtain hnRNPLL −/− mice. Genotyping was performed by genomic PCR using primers flanking hnRNPLL exon2. The birth rate, body weight, and survival time of wild‐type, hnRNPLL +/−, and hnRNPLL +/− mice were recorded and statistically analyzed. Mice were sacrificed around 2 weeks after birth to weight the organ ratio.
Embryos were obtained from crosses between hnRNPLL +/− male and female mice. Embryonic day 0.5 was recorded the morning after an observable plug was found. Embryos were collected at E11.5, and the genotypes were detected by genomic PCR. For genomic PCR detection, yolk sacs were firstly collected and then digested in mouse tail lysate at 50°C for 3–4 h, lysate was inactivated at 85°C for 45 min, and the supernatant could be used as templates. All mouse experimental protocols were approved by the Institutional Animal Care and Use Committee at Peking Union Medical College & Chinese Academy of Medical Sciences. All animal care and experimental methods were carried out in accordance with the ARRIVE guidelines for animal experiments. All the mice used in this study were fed in Specific Pathogen Free (SPF) facilities. All the mouse studies were randomly allocated, and no mouse was excluded from the analysis.
Embryoid body formation
Embryoid bodyies (EBs) were formed from wild‐type ES cells and gene disrupted ES cells as previously described (Jackson et al, 2010). We used handing drop culture method to form EBs. Briefly, cultured ES cells were dissociated with trypsin on the day of passage and sedimented for 30 min at 37°C. Plate 20µl drops containing 600 ES cells on the lid of petri dishes in regular arrays. A standard 100‐mm dish can accommodate about 30–40 drops. Invert the lid and place it over the bottom of a petri dish filled with PBS to prevent the drops from drying out. Incubate the petri dish with hanging drops in an incubator for 2 days. The ES cells aggregate into a single EB. EBs were harvested and subsequently transferred into bacterial‐grade dishes and cultivated for 2–8 days. Culture medium was containing of knockout DMEM (KO‐DMEM), 15% FBS, 2 mM GlutaMax, 1% non‐essential amino acids (NEAA), and 100 μM β‐mercaptoethanol. The cultures were replaced with fresh differentiation medium every other day.
Alkaline phosphatase staining and colony formation assay
Alkaline phosphatase activity was detected using Leukocyte Alkaline Phosphatase Kit (Sigma) following the manufacturer’s protocol. Briefly, cells were fixed with 4% paraformaldehyde, washed twice by TBST, then stained for 30 min at RT, finally washed by TBST, and suspended in PBS containing 20% glycerol for storage.
For colony formation assay, ES cells were seeded at 500 cells per well on gelatin‐coated 12‐well plate. The cells were cultured for 5 days before staining with Alkaline Phosphatase Kit, and the positive colonies were counted using an inverted microscope (Olympus, X41). For AP staining after LIF withdrawal, “undifferentiated” means colonies containing AP‐positive cells only, and “differentiated” represents colonies containing AP‐negative cells only.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, then permeabilized with 0.5% Triton X‐100 for 10 min, and blocked with 3% BSA for 1 h at room temperature (RT). Cells were incubated with primary antibody overnight at 4°C, washed three times with PBS then incubated with secondary antibody for 1 h at RT. Afterward, nuclei were labeled by DAPI and visualized by Olympus BX51.
Immunofluorescence for EBs: EBs were collected and fixed with 4% paraformaldehyde overnight. Paraffin section was prepared using standard procedures. Sections were incubated with the primary antibodies (anti‐Gata4, Santa Cruz Biotechnology, sc‐25310; anti‐Sox17, Abcam, ab191699) followed by appropriate Alexa Flour‐coupled secondary antibodies. Sections were photographed on a confocal fluorescence microscope (Olympus, #FV1000).
Teratoma formation assay
For teratoma formation assay, 5 × 105 cells were injected into one site of a SCID Beige mouse by subcutaneous injection, 10 sites (two sites per mouse) for each cell line. The size of teratoma was measured at the 9th day after injection and then measured every 2 or 3 days. Four weeks after injection, teratoma was picked and fixed in PBS containing 4% formaldehyde and embedded in paraffin. Sections were stained with hematoxylin and eosin. The percentage of Oct4‐positive region was calculated as the followings: Open the immunohistochemical picture in Photoshop and circle Oct4‐positive region using the magnetic lasso tool, then copy the layer and get the pixels using histogram. Get Oct4 negative region pixels by the same way and calculate the percentage of Oct4‐positive region using Oct4‐positive region pixel/(Oct4‐positive region pixel + Oct4 negative region pixel).
Vector construction
hnRNPLL (mouse, NM_144802) coding sequence was cloned into lentivirus vector pWPXL‐FH‐IF, which contained a drug (puromycin) selection marker. The coding sequences of the two isoforms of Tbx3 (mouse, NM_011535; NM_1980520) were cloned into lentivirus vector pLV‐hef1a‐mNeongreen‐P2A‐Puro‐WPRE‐CMV‐MCS‐3Xflag, respectively. Lentiviruses were prepared using 293T packaging cell line according to the manufacturer's instruction (Invitrogen).
Western blot
Remove the media and wash the cells by PBS for two times. Collect the cells by scraping with a cell lifter and remove PBS. Then, cells were lysed on ice for 30 min by lysis buffer (50 mM Tris–HCl, pH 7.4; 100 mM NaCl; 1% NP‐40; 0.1% SDS). Centrifuge the cell lysate at 16,000 g for 20 min at 4°C. Transfer the supernatant and quantify protein by microplate. Fifty micrograms of protein was mixed with protein loading buffer and thermal denatured. Protein samples were run on 10% SDS–PAGE for separation and transferred to PVDF membrane. The membrane was then blocked with 5% milk blocking buffer. Antibody was incubated with the membrane for antigen hybridization. All the antibodies are listed in Dataset EV7.
RNA extraction, qPCR, and RT–PCR
All ES cells and EB samples were washed by PBS and then harvested. Total RNA was extracted by TRIzol (Thermo Fisher, 15596018) and then reversed to cDNA using M‐MLV reverse transcriptase (Thermo Fisher, 28025021). We performed qRT–PCR with the SYBR Green PCR kit (Applied Biosystems, Cat# A25742). The relative expression of genes during EB differentiation was calculated by the double delta C t (ΔΔC t) method, normalized to the expression value at day 0. The relative expression data were represented as the heatmap. To analyze PSI of the alternative exon, forward and reverse primers were designed in the flanking exons, respectively. RT–PCR products were run on a 3% agarose gel. For transcripts quantification, cDNA was amplified by quantitative PCR by Roch LightCycler480. Primer sequences used are shown in Dataset EV7.
RNA immunoprecipitation (RIP)
For each reaction, 50 μl Protein A Dynabeads (Thermo Fisher, 10002D) were washed three times with RIP lysis buffer (20 mM HEPES pH 7.9, 150 mM NaCl, 0.5 mM EDTA pH 8.0, 10 mM KCl, 1.5 M MgCl2, 0.5% Np‐40, 10% glycerin, 1.5 mM DTT, 1 × Protease Inhibitor cocktail (Roche, 539134), 10 U/ml RNase Inhibitor) and then incubated with 5 μg anti‐hnRNPLL antibody (CST, 4783) and normal rabbit IgG (Millipore, 12‐370) for 4 h. Ten million ESC were resuspended in 400 μl cold RIP lysis buffer and lysed for 30 min, and then centrifuged for 30 min to separate insoluble fraction. The supernant was immunoprecipitated with antibody‐Dynabeads complex by rotating at 4°C overnight. After that, Dynabeads were washed two times, and RNA was purified by TRIzol (Thermo Fisher, 15596018) and then reversed to cDNA. qPCR was used to detect hnRNPLL–RNA interaction. Primer sequences are shown in Dataset EV7.
Cross‐linking and immunoprecipitation‐PCR
Twenty million ESC were washed by PBS and then cross‐linked with 125 mJ/cm2. 1 ml CLIP lysis buffer (50 mM Tris–HCl pH 7.4, 100 mM NaCl, 1% NP‐40, 0.1% SDS, 0.5% sodium deoxycholate) with protease inhibitor (Roche, 539134) were used to prepare cell lysate. Before centrifugation, cell lysate was treated with Turbo DNase (Thermo Fisher, AM2239) and RNase A (Qiagen, 19101) in 37°C for 10 min. Two copies of 1% supernatant were took as input and stored at 4°C. 15 μg anti‐hnRNPLL antibody was added to the cell lysate for immunoprecipitation, incubating overnight. 70 μl Protein A Dynabeads (Thermo Fisher, 10002D) were washed by CLIP lysis buffer for three times and then added to the cell lysate. After immunoprecipitation, Dynabeads were washed with high salt buffer for three times. 3′end of RNA was repaired by PNK, and then 3′linker was ligated to the RNA tag. 10% of the Dynabeads was taken for Western blot validation. CLIP sample and input sample were run on a 4–12% NuPAGE Bis‐Tris gel (Thermo Fisher, NP0335BOX) and transferred to nitrocellulose membrane for RNA–protein complex purification. The area overtop hnRNPLL proteins circled by the box (Fig 6C) was cut for RNA elution. RNA was eluted by protease K (Roch, 3115887001) digestion and purified by acid phenol/chloroform/isoamyl alcohol (pH 6.5) using Zymo RNA columns (ZYMO research, R1013). Input sample was separately processed by end repairment and linker ligation. Since then, all the samples were synchronized. First strand of cDNA was obtained by SuperScript III (Thermo Fisher, 18080044) with specific primer. hnRNPLL‐binding sites were detected by qPCR. IP efficiency was evaluated by Western blot.
CO‐immunoprecipitation
Ten million ESC were washed by PBS and lysed by lysis buffer (20 mM Tris–HCl pH 8.0, 137 mM NaCl, 1%NP‐40, 2 mM EDTA). Lysis supernatant was then incubated with anti‐hnRNPLL antibody (CST, 4783) and normal rabbit IgG (Millipore, 12‐370) overnight, and then, Protein A Dynabeads (Thermo Fisher, 10002D) was added to the mixture for 4 h. IP complex was eluted by LDS sample buffer (Thermo Fisher, NP0007), and proteins were separated by 4–12% NuPAGE Bis‐Tris gel (Thermo Fisher, NP0335BOX), then conducted to silver staining. The differential bands between hnRNPLL IP and IgG controls were cut for mass spectrum in Biomedical Testing Center of Tsinghua University. Proteins with MS score < 5 were filtered out, and proteins whose score in the hnRNPLL IP group were significantly higher than that in IgG group were selected as potential hnRNPLL interacting proteins. Some of them were further verified by Western blot combined with hnRNPLL immunoprecipitation. All the antibodies are listed in Dataset EV7.
RNA‐sequencing data processing
All ES cells and EB samples were washed by PBS and then harvested. Total RNA was extracted by Trizol (Thermo Fisher, 15596018), then conducted to RNA sequencing. All sequenced reads were trimmed with trimmomatics (v0.36) and mapped to GENCODE (vM12) mouse genome (mm10) with Hisat2 (v2.0.5). Samtools (v1.9) was used to sort and index aligned bam files. Gene expression levels were quantified as transcripts per kilobase of exon per million reads (TPM) with StringTie (v1.3.3b). Python script named “prepDE.py” from StringTie was used to calculate raw counts.
Protein–protein interaction, GSEA, and GO analysis
The original mouse protein–protein interaction (PPI) data were downloaded from STRING database (version 11.0), and the interactions with “binding” mode were remained for further analyses. Next, we extracted the interaction relationships which both interactors contain pluripotency‐associated genes (PAGs) or differential expressed RBPs during EB formation. Cytoscape (v3.8.0) was used to visualize the PPI network in Fig 1C. For hnRNPLL‐associated PPI network in Fig EV4B, we integrated mass spectrum verified PPI interactions with STRING collected PPI interactions into networks. Gene Set Enrichment Analysis (GSEA) analysis was performed between wild‐type AB1 ES cells and hnRNPLL‐KO ES cells during EB formation by using GSEA software (http://software.broadinstitute.org/gsea/index.jsp) against KEGG pathways. Gene Ontology (GO) enrichment was performed using “clusterProfiler” package with only “biological process” category. The significant enriched terms were obtained with Benjamini & Hochberg adjusted P‐value < 0.05. For GO enrichment of differential AS events, the AS events were firstly mapped to genes before enrichment. Domain information of enriched RBPs in Fig 1E was obtained from Uniprot, and Cytoscape was used to visualized the interaction network between enriched terms and RBPs.
Alternative splicing analysis
Percentage spliced in (PSI) of alternative spliced exons were quantified with aligned bam files using MISO (v0.5.4). A total of 5 types of alternative splicing events (SE, A3SS, A5SS, MXE, and RI) were identified. Differential alternative splicing events were defined as absolute value of δPSI more than 0.1, and Bayes factor ≥ 2. RBPmap (version1.1) was used to identify hnRNPLL‐binding sites of Bptf and Tbx3 in Fig 6D. The location of CDS, 5′UTR, and 3′UTR region was downloaded from ENSEMBL (v97) via Biomart. Bedtools (v2.27.1) were used to map alternative exons to gene regions. Cumulative distribution of PSI was calculated with PSI value in wild‐type and hnRNPLL‐KO ES cells. The “pheatmap” package was used to perform hierarchical clustering and draw heatmap. Especially, the gene expression level was scaled with z‐score before clustering.
Statistical analysis
All statistical analyses were performed with R software using a two‐sided Student’s t‐test, unless specified otherwise. Coefficient of variation (CV) was used to estimate the difference during EB formation, neural differentiation, and cardiac differentiation. Top 50% of genes with highest CV were firstly selected and then take intersection with all the 789 RBPs in Fig 1A. The “edgeR” package was used to evaluate the statistical significance of differential expressed genes using raw counts from StringTie. Genes with corrected P‐value (FDR) < 0.05 were considered as significant differential expressed.
The cumulative distribution of gene expression was calculated with log2‐transformed ratio between hnRNPLL‐KO and WT ES cells at day 0, day 4, and day 8 using an R function named “ecdf”. The log2‐transformed ratio of up‐regulated, down‐regulated, and all genes were calculated using TPM separately and then merged together to make comparison for each time point. Kolmogorov–Smirnov test was used to determine the statistical significance between up‐/down‐regulated genes and all genes. The “pheatmap” package was used to perform hierarchical clustering and draw heatmap. Especially, the gene expression level was scaled with z‐score before clustering.
Author contributions
Project conception and study design: YH, YM, JY, and DW; Experiment design and performance: XW, CP, PT, TL, YFH, YS, LZ, and XSW; In vitro and in vivo experiments: XW, CP, PT, SY, CS, and GL; RNA‐sequencing analysis and interpretation of the corresponding bioinformatic analyses: XW, CP, PT, FW, MZ, YJ, CS, XSW, YFH, and DW; Manuscript writing and editing: XW, CP, PT, YM, and YH; Comments on the manuscript: All authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Review Process File
Acknowledgements
We thank Dr. Allan Bradley (the Wellcome Trust Sanger Institute) for kindly providing AB1 ES cell line, and we also thank Dr. Xing Chang (Shanghai Jiao Tong University School of Medicine) for free hnRNPLL antibody, Dr. Jingyi Hui (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) for generous CLIP advising. This work was funded by National Key Research and Development Program of China [2016YFA0100103 to Y.H. and Y. M.; 2019YFA0801800 to J.Y.; 2019YFA0802600 to Y.M.]; CAMS Innovation Fund for Medical Sciences [2016‐I2M‐3‐002 to Y.H. and Y.M.; 2019‐I2M‐2‐001 to J.Y., 2017‐I2M‐3‐009 to X.W. and J.Y.]; National Natural Science Foundation of China [81530007 and 31725013 to J.Y.; 81970101 to Y.M.; 31801151 to X.W.; 82070109, 81770104, 62002153 to D.W.].
The EMBO Journal (2021) 40: e104729.
Contributor Information
Dong Wang, Email: wangdong79@smu.edu.cn.
Jia Yu, Email: j-yu@ibms.pumc.edu.cn.
Yanni Ma, Email: yanni_ma@126.com.
Yue Huang, Email: huangyue@pumc.edu.cn.
Data availability
The RNA‐sequencing data are available from the GEO database under the accession number GSE136955 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136955).
References
- Alexander J, Wamstad J (2013) Gene Expression Omnibus GSE47948 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47948) [DATASET]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A (2013) Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI (2012) Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 3: 923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Weiss WA (2015) Alternative splicing in cancer: implications for biology and therapy. Oncogene 34: 1–14 [DOI] [PubMed] [Google Scholar]
- Chen K, Dai X, Wu J (2015) Alternative splicing: an important mechanism in stem cell biology. World J Stem Cells 7: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Hu G (2017) Post‐transcriptional regulation of the pluripotent state. Curr Opin Genet Dev 46: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B, Park JW, Nakauka‐Ddamba A, Bebee TW, Guo Y, Shang X, Lengner CJ, Xing Y, Carstens RP (2016) Multiphasic and dynamic changes in alternative splicing during induction of pluripotency are coordinated by numerous RNA‐binding proteins. Cell Rep 15: 247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera‐Salinas D, Yu J, Bodak M, Ngondo RP, Herbert KM, Ciaudo C (2017) Noncanonical function of DGCR8 controls mESC exit from pluripotency. J Cell Biol 216: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR (2011) RBPDB: a database of RNA‐binding specificities. Nucleic Acids Res 39: D301–D308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbaillets I, Ziegler U, Groscurth P, Gassmann M (2000) Embryoid bodies: an in vitro model of mouse embryogenesis. Exp Physiol 85: 645–651 [PubMed] [Google Scholar]
- Di Stefano B, Luo EC, Haggerty C, Aigner S, Charlton J, Brumbaugh J, Ji F, Rabano Jimenez I, Clowers KJ, Huebner AJ et al (2019) The RNA helicase DDX6 controls cellular plasticity by modulating P‐Body homeostasis. Cell Stem Cell 25: 622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, Tolosano E, Provero P, Pandolfi PP, Silengo L et al (2013) The RNA binding protein ESRP1 fine‐tunes the expression of pluripotency‐related factors in mouse embryonic stem cells. PLoS One 8: e72300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi A, Pakzad M, Taei A, Brink TC, Pirhaji L, Ruiz G, Sharif Tabe Bordbar M, Gourabi H, Adjaye J, Baharvand H et al (2009) Comparative proteome and transcriptome analyses of embryonic stem cells during embryoid body‐based differentiation. Proteomics 9: 4859–4870 [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA‐binding proteins and post‐transcriptional gene regulation. FEBS Lett 582: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Pinello L, Han X, Lai S, Shen L, Lin TW, Zou K, Yuan GC, Orkin SH (2016). Serum‐based culture conditions provoke gene expression variability in mouse embryonic stem cells as revealed by single‐cell analysis. Cell Rep 14: 956–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T (2009) Forcing cells to change lineages. Nature 462: 587–594 [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL et al (2010) Tbx3 improves the germ‐line competency of induced pluripotent stem cells. Nature 463: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN et al (2013) MBNL proteins repress ES‐cell‐specific alternative splicing and reprogramming. Nature 498: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Braunschweig U, Gonatopoulos‐Pournatzis T, Weatheritt RJ, Hirsch CL, Ha KCH, Radovani E, Nabeel‐Shah S, Sterne‐Weiler T, Wang J et al (2017) Multilayered control of alternative splicing regulatory networks by transcription factors. Mol Cell 65: 539–553 [DOI] [PubMed] [Google Scholar]
- van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5: 214–226 [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR (2006) Dissecting self‐renewal in stem cells with RNA interference. Nature 442: 533–538 [DOI] [PubMed] [Google Scholar]
- Jackson M, Taylor AH, Jones EA, Forrester LM (2010) The culture of mouse embryonic stem cells and formation of embryoid bodies. Methods Mol Biol 633: 1–18 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA (2011) Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN (2013) The RNA‐binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20: 1122–1130 [DOI] [PubMed] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, Xie X, Wan J, Xing Y, Freudenberg JM et al (2014) Fip1 regulates mRNA alternative polyadenylation to promote stem cell self‐renewal. EMBO J 33: 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS et al (2008) Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet 4: e1000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A (2014) Genetic exploration of the exit from self‐renewal using haploid embryonic stem cells. Cell Stem Cell 14: 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone S, Bar D, Slabber CF, Dalcher D, Santoro R (2017) The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep 18: 1248–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Izpisua Belmonte JC (2018) Deconstructing the pluripotency gene regulatory network. Nat Cell Biol 20: 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QV, Dixon G, Verma N, Rosen BP, Gordillo M, Luo R, Xu C, Wang Q, Soh CL, Yang D et al (2019) Genome‐scale screens identify JNK‐JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat Genet 51: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB (2010) RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Timani K, Ou X, Broxmeyer HE, He JJ (2013) C‐MYC controlled TIP110 protein expression regulates OCT4 mRNA splicing in human embryonic stem cells. Stem Cells Dev 22: 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X et al (2014) A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol 10: 93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Wang X, Liu Y, Zhang M, Cai T, Shen Z, Jia Y, Huang Y (2017) Arrayed mutant haploid embryonic stem cell libraries facilitate phenotype‐driven genetic screens. Nucleic Acids Res 45: e180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Gaidatzis D, Sack R, Meister G, Filipowicz W (2013) The mammalian TRIM‐NHL protein TRIM71/LIN‐41 is a repressor of mRNA function. Nucleic Acids Res 41: 518–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Goke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH (2013) SON connects the splicing‐regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol 15: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ et al (2014) Alternative splicing of MBD2 supports self‐renewal in human pluripotent stem cells. Cell Stem Cell 15: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmegrim KC, Pruijn GJ, van Venrooij WJ (2002) The fate of the U1 snRNP autoantigen during apoptosis: implications for systemic autoimmunity. Isr Med Assoc J 4: 706–712 [PubMed] [Google Scholar]
- Manning KS, Cooper TA (2017) The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol 18: 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A et al (2012) The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Smith A (2014) The nature of embryonic stem cells. Annu Rev Cell Dev Biol 30: 647–675 [DOI] [PubMed] [Google Scholar]
- Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagu A, van Genderen E, Escudero I, Verwegen L, Kurek D, Lehmann J, Stel J, Dirks RAM, van Mierlo G, Maas A et al (2020) In vitro capture and characterization of embryonic rosette‐stage pluripotency between naive and primed states. Nat Cell Biol 22: 534–545 [DOI] [PubMed] [Google Scholar]
- Nefzger CM, Polo JM (2017) DEAD‐Box RNA binding protein DDX5: not a black‐box during reprogramming. Cell Stem Cell 20: 419–420 [DOI] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A (2008) Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S, Paz Yaacov N, Safran M, Moshkovitz S, Shtrichman R, Sherf O, Jacob‐Hirsch J, Keshet G, Amariglio N, Itskovitz‐Eldor J et al (2010) Alu sequences in undifferentiated human embryonic stem cells display high levels of A‐to‐I RNA editing. PLoS One 5: e11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, Nordstrand LM, Rognes T, Lee JT, Klungland A, Kouzarides T et al (2012) ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells 30: 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Song C, Malakouti N, Murray D, Hariz A, Zimmerman M, Gygax D, Alhazmi A, Landry JW (2015) Functional interactions between NURF and Ctcf regulate gene expression. Mol Cell Biol 35: 224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y et al (2013) Double nicking by RNA‐guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respuela P, Nikolic M, Tan M, Frommolt P, Zhao Y, Wysocka J, Rada‐Iglesias A (2016) Foxd3 promotes exit from naive pluripotency through enhancer decommissioning and inhibits germline specification. Cell Stem Cell 18: 118–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, Kavanagh SJ, Rathjen J, Rathjen PD (2002) Embryonic stem cell differentiation and the analysis of mammalian development. Int J Dev Biol 46: 449–458 [PubMed] [Google Scholar]
- Roy B, Talukder P, Kang HJ, Tsuen SS, Alam MP, Hurley LH, Hecht SM (2016) Interaction of individual structural domains of hnRNP LL with the BCL2 Promoter i‐Motif DNA. J Am Chem Soc 138: 10950–10962 [DOI] [PubMed] [Google Scholar]
- Russell R, Ilg M, Lin Q, Wu G, Lechel A, Bergmann W, Eiseler T, Linta L, Kumar PP, Klingenstein M et al (2015) A dynamic role of TBX3 in the pluripotency circuitry. Stem Cell Reports 5: 1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG (2009) The let‐7 target gene mouse lin‐41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 11: 1411–1420 [DOI] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS et al (2010) Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA 107: 10514–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X et al (2014) Efficient genome modification by CRISPR‐Cas9 nickase with minimal off‐target effects. Nat Methods 11: 399–402 [DOI] [PubMed] [Google Scholar]
- Tian TV, Di Stefano B, Stik G, Vila‐Casadesus M, Sardina JL, Vidal E, Dasti A, Segura‐Morales C, De Andres‐Aguayo L, Gomez A et al (2019) Whsc1 links pluripotency exit with mesendoderm specification. Nat Cell Biol 21: 824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Blencowe BJ (2019) Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell 76: 329–345 [DOI] [PubMed] [Google Scholar]
- UniProt C (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47: D506–D515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghray A, Saiz N, Jayaprakash AD, Freire AG, Papatsenko D, Pereira CF, Lee DF, Brosh R, Chang B, Darr H et al (2015) Tbx3 controls Dppa3 levels and exit from pluripotency toward mesoderm. Stem Cell Reports 5: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I et al (2013) The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self‐renewal and somatic cell reprogramming. Cell Stem Cell 13: 676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH (2016) Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 17: 155–169 [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhao W, Olson SD, Prabhakara KS, Zhou X (2018) Alternative splicing links histone modifications to stem cell fate decision. Genome Biol 19: 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y et al (2013) Direct conversion of fibroblasts to neurons by reprogramming PTB‐regulated microRNA circuits. Cell 152: 82–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Liu L, Lazarev D, Al‐Zain A, Fomin V, Yeung PL, Chambers SM, Lu CW, Studer L, Manley JL (2018) TCF3 alternative splicing controlled by hnRNP H/F regulates E‐cadherin expression and hESC pluripotency. Genes Dev 32: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Kalkan T, Morrisroe C, Smith A, Sharrocks AD (2012) A genome‐wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet 8: e1003112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Blelloch R (2014) Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 15: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA‐protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KT, Park J, Kim VN (2015) Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29: 2004–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chronis C, Chen X, Zhang H, Spalinskas R, Pardo M, Chen L, Wu G, Zhu Z, Yu Y et al (2019) The BAF and PRC2 complex subunits Dpf2 and Eed antagonistically converge on Tbx3 to control ESC differentiation. Cell Stem Cell 24: 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li G, Wei C (2013) Gene Expression Omnibus GSE44067 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44067) [DATASET]
- Zhao D, Wu Y, Chen K (2014) Tbx3 isoforms are involved in pluripotency maintaining through distinct regulation of Nanog transcriptional activity. Biochem Biophys Res Commun 444: 411–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Alexander J, Wamstad J (2013) Gene Expression Omnibus GSE47948 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47948) [DATASET]
- Zhang Y, Li G, Wei C (2013) Gene Expression Omnibus GSE44067 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44067) [DATASET]
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Review Process File
Data Availability Statement
The RNA‐sequencing data are available from the GEO database under the accession number GSE136955 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136955).


