Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread to more than 200 countries and regions globally. SARS-CoV-2 is thought to spread mainly through respiratory droplets and close contact. However, reports have shown that a notable proportion of patients with coronavirus disease 2019 (COVID-19) develop gastrointestinal symptoms and nearly half of patients confirmed to have COVID-19 have shown detectable SARS-CoV-2 RNA in their faecal samples. Moreover, SARS-CoV-2 infection reportedly alters intestinal microbiota, which correlated with the expression of inflammatory factors. Furthermore, multiple in vitro and in vivo animal studies have provided direct evidence of intestinal infection by SARS-CoV-2. These lines of evidence highlight the nature of SARS-CoV-2 gastrointestinal infection and its potential faecal–oral transmission. Here, we summarize the current findings on the gastrointestinal manifestations of COVID-19 and its possible mechanisms. We also discuss how SARS-CoV-2 gastrointestinal infection might occur and the current evidence and future studies needed to establish the occurrence of faecal–oral transmission.
Subject terms: Viral infection, Dysbiosis, SARS-CoV-2
Although COVID-19 is a respiratory disease and its causative agent, SARS-CoV-2, principally infects the respiratory tract, extrapulmonary manifestations are observed. This Perspective explores the gastrointestinal symptoms associated with COVID-19 and the putative underlying mechanisms, discussing experimental evidence on SARS-COV-2 gastrointestinal infection and the potential for faecal–oral transmission.
Introduction
Coronaviruses belong to the Coronaviridae family of the order Nidovirales1. They are positive-sense strand RNA viruses with ~30 kb RNA genomes and an envelope with surface ‘spikes’1. Historically, these viruses have not been considered highly pathogenic to humans because they cause only mild, self-limiting respiratory infections, that is, until the outbreak of severe acute respiratory syndrome (SARS) in 2002 (ref.2). In December 2019, an outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was reported in several hospitals in Wuhan (Hubei Province, China)3. Since then, the virus has infected >105 million people as of 8 February 2021, spreading to over 200 countries or regions worldwide4 and is the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic.
There are three highly pathogenic coronaviruses, namely SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV)1 and SARS-CoV-2 (ref.3), which belong to the Betacoronavirus genus. They mainly infect the respiratory tract through attachment to the angiotensin-converting enzyme 2 (ACE2) receptor (SARS-CoV and SARS-CoV-2) or dipeptidyl peptidase 4 (DPP4) receptor (MERS-CoV)1,5. After entry, SARS-CoV, MERS-CoV and SARS-CoV-2 infections only induce a low or delayed interferon response6–8. As a consequence, the rapid viral replication triggers the release of pro-inflammatory cytokines and/or chemokines and massive infiltration of inflammatory cells6,7. These events, in severe cases, lead to acute lung injury and acute respiratory distress syndrome, which are the leading causes of death during SARS-CoV-2 infection9.
According to a report of 72,314 cases in China, the overall case-fatality rate was 2.3%10. Mild to moderate disease, which was considered as non-pneumonia and mild pneumonia, was reported in 81% of cases10. Meanwhile, severe (that is, dyspnoea, respiratory frequency ≥30 breaths per minute, blood oxygen saturation ≤93%, partial pressure of arterial oxygen to fraction of inspired oxygen ratio <300, and/or lung infiltrates >50% within 24–48 hours) and critical (that is, respiratory failure, septic shock, and/or multiple organ dysfunction or failure) disease were reported in 14% and 5% of cases, respectively10. On admission, the most common symptoms were cough (67.8%) and fever (43.8%)11 and ground-glass opacity was the most common radiological finding on chest CT12.
Most studies have focused on SARS-CoV-2 infection and clinical symptoms in the respiratory tract. However, ACE2 is also highly expressed in the intestine13,14. According to a meta-analysis of data from a Hong Kong cohort published in 2020, 17.6% of patients with COVID-19 have gastrointestinal symptoms15 and 48.1% of faecal samples from patients with COVID-19 have tested positive for viral RNA15. Potential gastrointestinal infection and faecal–oral transmission should therefore be carefully considered.
In this Perspective, we summarize the gastrointestinal manifestations of COVID-19 and the possible underlying mechanisms, the current lines of evidence showing intestinal infection, and the in vitro and in vivo models that are being used to study SARS-CoV-2 intestinal infection. Additionally, we discuss the possible influence of intestinal infection on the immune response that might contribute to the so-called cytokine release syndrome (CRS), also known as ‘cytokine storm’, during viral pneumonia. Moreover, we discuss the remaining questions that need to be resolved surrounding SARS-CoV-2 intestinal infection and its potential faecal–oral transmission to stimulate the desperately needed research on this topic to improve our understanding of this pandemic.
Gastrointestinal symptoms in COVID-19
Multiple studies have reported gastrointestinal symptoms in patients with COVID-19. Diarrhoea, nausea, vomiting, anorexia and abdominal pain are described as the main gastrointestinal symptoms in most studies, although other gastrointestinal symptoms, such as acid reflux, upper gastrointestinal bleeding, haematochezia, constipation and melena, have also been reported in a few cases11,16–29 (Table 1). According to a meta-analysis that included 10,890 patients, the pooled prevalence estimates of gastrointestinal symptoms were diarrhoea (7.7%), nausea or vomiting (7.8%), and abdominal pain (2.7%)30. It is worth mentioning that different studies have adopted different criteria for what constitutes gastrointestinal symptoms. In some studies, gastrointestinal symptoms only included diarrhoea, nausea and vomiting16,19,23, whereas in others, symptoms such as loss of taste, abdominal pain and loss of appetite were also included17,24,25,29. For example, a multicentre study in China reported gastrointestinal symptoms, including diarrhoea (3.8%), nausea and vomiting (5%), as less common symptoms11, whereas one single-centre study in Baltimore, USA, reported gastrointestinal symptoms, including nausea, vomiting, diarrhoea, abdominal pain, anorexia, haematochezia, and loss of smell or taste, in more than 70% of patients with COVID-19 (ref.24). The great variation in the proportion of patients with gastrointestinal symptoms among different studies might also be related to geographical region30 and whether symptoms were reported on admission or during hospitalization25. Moreover, patients might receive different medications for COVID-19, some of which are likely to induce diarrhoea as an adverse event, for example, lopinavir or antibiotics. In addition, given the differing epidemic prevention policies between regions, the lack of data on COVID-19 in some communities, and the fact that mild cases are less reported or are not admitted to hospital in some regions, there might be deviations in the proportion of gastrointestinal symptoms reported between studies.
Table 1.
Studies reporting proportion of patients with COVID-19 with gastrointestinal symptoms
| Study | Region | Time | Total number of patients | Gastrointestinal symptom | Point at which symptoms were reported | Gastrointestinal symptoms and disease severity |
|---|---|---|---|---|---|---|
| Studies in adults | ||||||
| Lin et al.25 | Zhuhai, China | 17 Jan to 15 Feb 2020 | 95 | Diarrhoea 5.3%; nausea 3.2%; anorexia 5.3%; acid reflux 1.1% | On admission | No statistically significant difference in the clinical outcomes (remained in hospital, discharged or died) |
| Diarrhoea 18.9%; nausea 14.7%; vomiting 4.2%; anorexia 12.6%; acid reflux 1.1%; epigastric discomfort 2.1% | During hospitalization | |||||
| Papa et al.27,a | Rome, Italy | 15 Mar to 14 Apr 2020 | 34 | Any gastrointestinal symptoms 8.8% | On admission | Gastrointestinal symptoms associated with reduced mortality |
| Any gastrointestinal symptoms 32.3% | During hospitalization | |||||
| Jin et al.23 | Zhejiang, China | 17 Jan to 8 Feb 2020 | 651 | Diarrhoea 8.6%; vomiting 2.15%; nausea 2.0% | On admission | Gastrointestinal symptoms associated with severe disease |
| Zhou et al.16 | Wuhan, China | 29 Dec 2019 to 31 Jan 2020 | 191 | Diarrhoea 5%; nausea or vomiting 4% | On admission | No statistically significant difference in mortality |
| Chen et al.24,a | Baltimore, USA | 9 Mar to 15 Apr 2020 | 101 | Diarrhoea 50%; nausea 30%; vomiting 14%; abdominal pain 26%; anorexia 53%; haematochezia 1% | On admission | No correlation between gastrointestinal symptoms and increased hospitalization rate or ICU care needs |
| Ferm et al.17 | New York, USA | 14 Mar to 1 Apr 2020 | 892 | Diarrhoea 9.8%; nausea 16.6%; vomiting 10.2%; loss of taste 2.4%; loss of appetite 11.8%; abdominal pain 7.8% | On admission | No difference in ICU admission, length of stay, or mortality |
| Remes-Troche et al.20 | Veracruz, Mexico | 1 Apr to 5 May 2020 | 112 | Diarrhoea 7.8%; vomiting 7.1%; abdominal pain 9.8% | On admission | No statistically significant difference in disease severity |
| Redd et al.22 | Massachusetts, USA | Until 2 Apr 2020 | 318 | Diarrhoea 33.7%; nausea 26.4%; vomiting 15.4%; abdominal pain 14.5%; anorexia 34.8%; constipation 0.94%; melena 0.63%; reflux 0.63%; dysphagia 0.31%; odynophagia 0.31%; haematochezia 0.31% | During hospitalization | No correlation between gastrointestinal symptoms and disease severity |
| Wan et al.28 | China | 19 Jan to 6 Mar 2020 | 232 | Diarrhoea 21%; abdominal pain 1%; bloody stool 4% | During hospitalization | Gastrointestinal symptoms associated with severe symptoms of pneumonia |
| Díaz et al.18 | Chile | Until 11 Apr 2020 | 7,016 | Diarrhoea 7.3%; abdominal pain 3.7% | No distinction made | Gastrointestinal symptoms associated with a higher risk of hospitalization |
| Cholankeril et al.29 | California, USA | 4 Mar to 24 Mar 2020 | 116 | Diarrhoea 10.3%; nausea and/or vomiting 10.3%; abdominal pain 8.8%; loss of appetite 25.3% | No distinction made | No correlation between gastrointestinal symptoms and disease severity |
| Hajifathalian et al.21 | New York, USA | 4 Mar to 9 Apr 2020 | 1,059 | Diarrhoea 22%; abdominal pain 7% | No distinction made | Gastrointestinal symptoms associated with lower rates of death and ICU admission |
| Studies on paediatric patients | ||||||
| Xu et al.79 | Guangdong, China | Until 20 Feb 2020 | 10 | Diarrhoea 30%; vomiting 0 | On admission | NA |
| Cai et al.81 | China | Jan 19 to 3 Feb 2020 | 10 | Diarrhoea 0% | During hospitalization | NA |
| Lu et al.170 | Wuhan, China | 28 Jan to 26 Feb 2020 | 171 | Diarrhoea 8.8%; vomiting 6.4% | No distinction made | NA |
| Fakiri et al.171 | Marrakesh, Morocco | 2 Mar to 1 Apr 2020 | 74 | Diarrhoea 5.4% | No distinction made | NA |
| de Ceano-Vivas et al.172 | Madrid, Spain | 11 Mar to 9 Apr 2020 | 58 | Diarrhoea 12.1%; vomiting 15.5% | No distinction made | NA |
| Mahmoudi et al.173 | Tehran, Iran | 7 Mar to 30 Mar 2020 | 35 | Diarrhoea 26%; vomiting 29%; abdominal pain 11% | No distinction made | NA |
| CDC COVID-19 Response Team174 | USA | 12 Feb to 2 Apr 2020 | 291 | Diarrhoea 13%; nausea and/or vomiting 11%; abdominal pain 5.8% | No distinction made | NA |
| Parri et al.175 | Italy | 3 Mar to 27 Mar 2020 | 100 | Diarrhoea 9%; nausea or vomiting 10% | No distinction made | NA |
Table 1 contains selected studies that take sample size, region, symptom collection time point (on admission or during hospitalization) and research methods (prospective or retrospective) into consideration. In detail, studies on adult patients that clarified whether the gastrointestinal symptoms were collected before or after admission and with a sample size >100 were included; the two studies that distinguish the symptoms on admission and during hospitalization are placed but samples <100 are placed at the top of the table. Two studies in Chile and California, with a sample size of >100 but missing the symptom collection time point, were included owing to the few studies in these regions. One New York study with a large sample size was also included. The number of paediatric studies containing gastrointestinal symptoms are limited; studies in different regions with symptom collection time point or a sample size >35 were included. CDC, Centers for Disease Control and Prevention; COVID-19, coronavirus disease 2019; ICU, intensive care unit; NA, not available. aProspective study.
As a consequence, there is also disagreement among studies examining possible correlations between gastrointestinal symptoms and disease severity. According to an early meta-analysis that included 3,022 patients from 9 studies, no statistically significant difference could be identified between patients with or without gastrointestinal symptoms (20.5% versus 18.2%)31. Additionally, according to two studies in New York, patients with gastrointestinal symptoms had lower rates of death than those without gastrointestinal symptoms (8.5% versus 16.5% and 0% versus 5%, respectively)21,32. Consistent with that finding, a single-centre study in Italy reported that the presence of gastrointestinal symptoms was inversely associated with the risk of clinical deterioration19. By contrast, a later meta-analysis that included 4,243 patients from 60 studies globally revealed that the pooled prevalence of all gastrointestinal symptoms was higher in patients with severe disease than in patients with mild disease (17.1% versus 11.8%)15. Moreover, a multicentre study in China found that more patients with diarrhoea showed severe symptoms of pneumonia than did patients without diarrhoea (53% versus 19%)28. Further prospective studies with consistent standards for describing gastrointestinal symptoms and that exclude adverse effects caused by medications are required to determine whether gastrointestinal symptoms have a bona fide correlation to disease severity.
Diarrhoea is the most common gastrointestinal symptom in COVID-19; 2–50% of patients with COVID-19 have been reported to have diarrhoea11,16–29 (Table 1). According to a study of 651 patients in Zhejiang, China, among 53 patients with diarrhoea, the duration of diarrhoea lasted 1–9 days and, in most cases, the diarrhoea was self-limiting23. The adverse events related to treatments provided during hospitalization can partially explain the diarrhoea symptoms reported by some patients with COVID-19. In one open-label, phase II trial, 127 adults with COVID-19 were administered lopinavir–ritonavir33; the adverse effects included self-limited nausea and diarrhoea in 52 (41%) patients, which were mostly resolved within 3 days after drug initiation. Interestingly, a 22-year-old man with COVID-19 showed no respiratory symptoms but did present a 4-day history of diarrhoea and low-grade fever at the onset of illness34. Furthermore, according to a single-centre case series study (n = 138) in Wuhan, China, 10.1% of patients initially presented with diarrhoea and nausea 1–2 days prior to the development of fever and dyspnoea35. These studies suggest that gastrointestinal symptoms might be a symptom of onset in some patients with COVID-19.
Notably, patients with other respiratory virus-related diseases, including SARS, MERS36 (Table 2) and even severe influenza, also present with gastrointestinal symptoms37,38. For example, 20% of patients with SARS reported diarrhoea39. Furthermore, 97% (65 of 67) of patients with SARS reportedly tested positive for SARS-CoV RNA in faeces40, and an autopsy study of 5 patients showed infection in a limited number of enterocytes and lymphocytes41. As many as one-third of patients with severe MERS reported gastrointestinal symptoms42 and, in one study, 14.6% tested positive for viral RNA in faecal samples43. According to several cohort studies, 40–70% of patients infected with H5N1 had gastrointestinal symptoms38,44,45 and, in one case report, diarrhoea preceded respiratory manifestations by up to 1 week46. Interestingly, influenza is actually an enteric virus in birds47,48 and other coronaviruses, such as mouse hepatitis virus and bovine coronavirus49,50, also establish intestinal infection in animals.
Table 2.
Comparison of SARS-CoV-2, SARS-CoV and MERS-CoV infection
| Characteristics | SARS-CoV-2 (refs4,30,83) | SARS-CoV36,40 | MERS-CoV36,43 |
|---|---|---|---|
| Epidemiology | |||
| Confirmed cases | 105,658,476a | 8,096 | 2,519 |
| Mortality (%) | 2.2a | 9.6 | 34.4 |
| Incubation period (days) | 1–14 | 2–14 | 2–14 |
| Gastrointestinal characteristics | |||
| Nausea (%) | 7.8 | 20–35 | 21.0 |
| Vomiting (%) | 7.8 | 20–35 | 21.0 |
| Diarrhoea (%) | 7.7 | 20–25 | 26.0 |
| Percentage of patients with positive faecal samples | 55.0 | 97.0 | 14.5b |
MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2. aAs of 8 February 2021 according to the WHO. bPercentage of faecal samples tested positive.
The mechanism of diarrhoea in patients with COVID-19 is still largely unknown. Experimental studies from other viruses have indicated that several factors might lead to diarrhoea, including alterations in gut microbiota51,52, osmotic diarrhoea due to malabsorption or inflammation that is secondary to enterocyte damage53,54, release of virulent proteins or toxins53, and viral-induced intestinal fluid and electrolyte secretion by activation of the enteric nervous system55. Two retrospective studies of 87 patients with COVID-19 showed that patients with diarrhoea are more likely to test positive for viral RNA in stool15,18 and, in one study (n = 59), the faecal viral load was positively associated with diarrhoea (P = 0.06)15. This aspect is similar to what was observed during SARS-CoV infection56, indicating a potential correlation between intestinal infection and diarrhoea. Notably, the histological examination of both patients with SARS and patients with COVID-19 in several case studies showed that the mucosal epithelium of the duodenum and rectum had no major damage39,57–60. Considering the limited number of cases, more histological analyses are needed to determine whether there is enterocyte damage in the intestine, which could lead to diarrhoea.
Immune mechanisms have been proposed to play an important role in viral diarrhoea such as in norovirus infections61. Indeed, inflammatory immune responses in patients with COVID-19 with diarrhoea have been reported. Calprotectin, which is an inflammatory marker secreted by infiltrated neutrophils, was detected in high concentrations in the faecal samples of 22 patients with COVID-19 with diarrhoea62, highlighting the potential involvement of inflammation. Additionally, according to a mouse study, excessive levels of inflammatory factors caused by lung infection of influenza might also exacerbate diarrhoea symptoms through changes in composition of the gut microbiota, including decreased levels of segmented filamentous bacteria and Lactobacillus and/or Lactococcus as well as an increase in Enterobacteriaceae, independently of direct intestinal infection52. In several cases, patients with COVID-19 with gastrointestinal symptoms had no detectable viral RNA in stool15,63, which suggests that SARS-CoV-2 could also cause diarrhoea that is independent of intestinal infection.
Further studies are needed to investigate the specific mechanisms underlying the gastrointestinal symptoms caused by SARS-CoV-2 infection. For example, a comparison of the proportion of gastrointestinal symptoms among patients with or without detectable virus in the intestine would be useful to understand the contribution of direct intestinal infection on the occurrence of gastrointestinal symptoms. Given that ACE2 has been shown to function in gut microbial ecology64 and that alterations in the gut microbiota of patients with COVID-19 have been reported65,66, investigations are warranted to determine whether these alterations in gut microbiota are related to intestinal peristalsis and diarrhoea.
Evidence of intestinal infection
Intestinal expression of SARS-CoV-2 receptor and serine protease
Cell entry of SARS-CoV-2 is dependent upon binding of its spike (S) proteins to the cellular surface protein ACE2; priming of S proteins by host cell transmembrane serine protease 2 (TMPRSS2) is also essential67. Analyses of single-cell transcriptomics data in humans revealed that ACE2 and TMPRSS2 are co-expressed in lung alveolar type 2 cells, oesophageal upper epithelial and gland cells, and in absorptive enterocytes from the ileum and colon68. Notably, absorptive enterocytes in the small intestine were found to express the highest levels of ACE2 in the human body14. Meanwhile, a small fraction of human colon epithelial cells expressed ACE2 at moderate levels69.
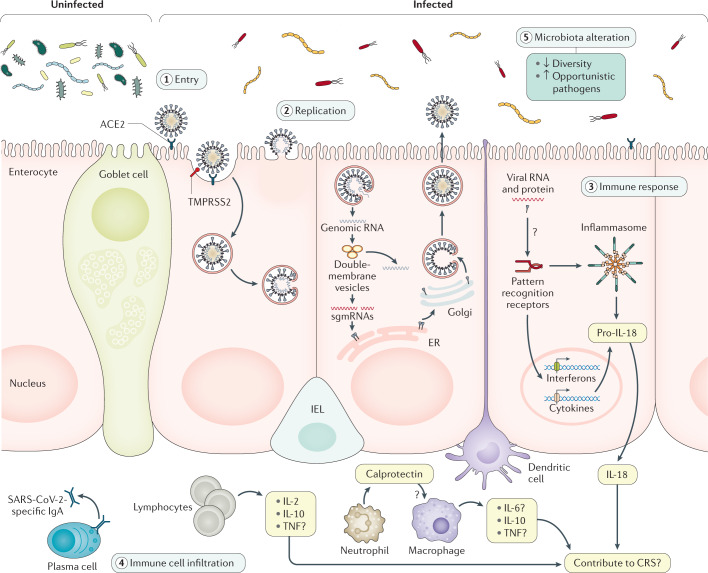
According to immunohistochemistry studies of ACE2, an abundant expression of ACE2 can be observed in the brush border of the intestinal enterocytes in the ileum70,71. ACE2 is also present in the vascular endothelium and vascular smooth muscle cells in the submucosa of the ileum70. In the colon, ACE2 is mainly present in the blood vessels and muscular layers70, consistent with single-cell transcriptomics data showing that relatively few colon epithelial cells express ACE2 (ref.69). Taken together, the expression patterns of ACE2 and TMPRSS2 indicate the potential for SARS-CoV-2 intestinal infection (Fig. 1).
Fig. 1. Evidence of SARS-CoV-2 intestinal infection.
This figure shows the putative mechanisms for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) intestinal infection. Angiotensin-converting enzyme 2 (ACE2) is mainly expressed on the brush border of enterocytes in the ileum and colon. Cell entry by SARS-CoV-2 (1) begins with the binding of spike (S) proteins to ACE2. Host cell transmembrane serine protease 2 (TMPRSS2) cleaves the S protein. Subsequently, the cell membrane fuses with the viral membrane and SARS-CoV-2 genomic RNA is released into the cytoplasm. Based on human intestinal organoid studies, SARS-CoV-2 primarily infects enterocytes but not goblet cells. The double membrane structure produced by virus replication (2) can be observed in the infected cells and the virus protein can be detected in the endoplasmic reticulum (ER). Newly assembled viral particles were released predominantly from the apical side into the lumen. The detection of subgenomic mRNA (sgmRNA) can serve as evidence of active viral replication in the intestine. SARS-CoV-2 infection activates an interferon-mediated immune response (3) in human organoids. Levels of the intestinal epithelial cell-specific inflammatory factor IL-18, which is activated by inflammasomes, have been shown to increase in patients with severe coronavirus disease 2019 (COVID-19). However, how SARS-CoV-2 triggers immune response in the gut in humans is not yet well understood, including the role of inflammatory factors caused by intestinal infection and their contribution to cytokine release syndrome (CRS), and requires further investigation. The histological examination of human intestinal samples revealed that lymphocytes and inflammatory cells infiltrated the lamina propria (4). Patients with diarrhoea exhibited increased faecal calprotectin levels, released mainly by infiltrated neutrophils. However, whether intestinal infiltrations of T cells, B cells, macrophages and neutrophils as well as of their secreted cytokine and IgA are correlated with disease severity is still unknown. SARS-CoV-2 infection altered the gut microbiota community structure (5). The enrichment of opportunistic pathogens and the depletion of beneficial commensals was observed in patients with COVID-19. These changes were correlated with the expression of inflammatory factors in the serum of these patients. However, whether the microbiota profile can predict the occurrence of CRS and whether modulation of the microbiota can resolve CRS need further study. IEL, intraepithelial lymphocyte.
Detection and isolation of SARS-CoV-2 from faeces
Multiple studies have reported the positive detection of SARS-CoV-2 RNA in faecal samples or rectal swabs from patients with COVID-19 (refs15,59,72–85) (Table 3). According to a retrospective cohort study in Zhejiang, China, viral RNA was detected in the stool of 59% (55 of 93) of patients86. The median duration of viral RNA in stool was 22 days86. Viral loads from stool samples were found to peak later in the disease, generally 2–3 weeks after symptom onset86,87. The viral RNA load in stool samples apparently reflected the course in sputum in 86% (6 of 7) of cases in a study in Germany88. However, in some patients, faecal samples remained positive for virus even after the respiratory and/or sputum samples exhibited no detectable virus79,88. In some cases, the viral load in faeces reached 107 copies/g (ref.88), even higher than that in pharyngeal swabs. The presence and persistence of such large amounts of viral RNA in faeces is unlikely to be explained by only the swallowing of virus particles replicated in the throat but rather suggests the potential for enteric infection of SARS-CoV-2.
Table 3.
Summary of patients with SARS-CoV-2-positive faecal or rectal swabs
| Author | Region | Number of patients with positive stool/rectal swab sample | Duration of positive infection (days) | Patients with positive stool/rectal sample after negative respiratory system samples | Duration after positive respiratory system samples (days) |
|---|---|---|---|---|---|
| Adult patients | |||||
| Lin et al.72 | Guangzhou, China | 46/217 (21.2%) | 3–18 | 30/46 (65.2%) | 3–15 |
| Ling et al.73 | Shanghai, China | 54/66 (81.8%) | 9–16a | 43/55 (78.2%) | 1–4 |
| Cheung et al.15 | Hong Kong, China | 9/59 (15.3%) | Data collection on presentation | NA | NA |
| Kujawski et al.74 | USA | 7/12 (58.3%) | 1–12 | 1/7 (14.3%) | 1 |
| Lo et al.75 | Macau, China | 9/9 (100%) | 1–18 | 1/9 (11.1%) | 6 |
| Young et al.76 | Singapore | 4/8 (50%) | 1–7 | 1/4 (25%) | 5 |
| Paediatric patients | |||||
| Hua et al.77 | Zhejiang, China | 32/35 (91.4%) | NA | 18/35 (51.4%) on discharge | >70 days in one child since illness onset |
| Han et al.78 | Seoul, Korea | 11/12 (91.6%) | 80% positive >3 weeks | NA | NA |
| Xu et al.79 | Guangzhou, China | 8/10 (80%) | 3–28a | 8/8 (100%) | 3–30a |
| Liu et al.80 | Shanghai, China | 8/9 (89%) | 28–66 | 8/8 (100%) | 14–52 |
| Cai et al.81 | China | 5/6 (83.3%) | 18–30a | 5/5 (100%) | 11–18 |
| Xing et al.82 | Qingdao, China | 3/3 (100%) | 6–30 | 3/3 (100%) | 8–20 |
| No distinction made | |||||
| Wu et al.83 | Zhuhai, China | 41/74 (55.4%) | 1–39 | 32/41 (78%) | 1–33 |
| Xiao et al.59 | Guangzhou, China | 39/73 (53.4%) | 1–12a | 17/39 (43.6%) | NA |
| Chen et al.84 | Wuhan, China | 28/42 (66.7%) | 1–21a | 18/28 (64.3%) | 6–10 |
| Kim et al.85 | Korea | 8/15 (53.3%) | 1–7 | 2/8 (25%) | 3–9 |
This is a table with selected studies; studies that were published before 1 Aug 2020, sample size ≥3, and proportion and duration of positive virus in the stool of the patients with COVID-19 were selected. NA, not available; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2. aSome patients remain positive at last follow-up.
The viral SARS-CoV-2 subgenomic mRNA (sgmRNA) is transcribed in actively replicating cells and SARS-CoV-2 sgmRNA has been detected in faecal samples of one patient, indicating the presence of actively infected cells in the gastrointestinal tract88. As sgmRNAs are not packaged into virions, it is therefore unlikely that they can pass through the gastrointestinal tract following their digestion by RNAse. Consistently, sgmRNAs last longer in faeces than in throat swabs, indicating that they are unlikely to be derived from the respiratory tract88. In addition, infectious virus particles have been successfully isolated from patients’ faeces89–92. The combination of very high concentrations of viral RNA, the detection of sgmRNA and the prolonged detectability in faecal samples compared with respiratory samples strongly suggests that SARS-CoV-2 actively replicates in the gastrointestinal tract (Fig. 1).
Endoscopic and histological examination in COVID-19
Endoscopy and biopsy samples of the oesophagus, stomach, duodenum and colon were taken from a 78-year-old patient with COVID-19 in Guangdong Province, China, who showed symptoms of upper gastrointestinal bleeding59. Staining for nucleocapsid proteins of SARS-CoV-2 revealed their presence in the gastrointestinal epithelium but not in the oesophageal epithelium in this patient. However, no substantial damage to the mucosal epithelium of the oesophagus, stomach, duodenum or rectum was observed. Numerous infiltrating plasma cells and lymphocytes with interstitial oedema were found in the lamina propria of the stomach, duodenum and rectum of this patient, which indicated an inflammatory response in the digestive system caused by SARS-CoV-2 infection (Fig. 1).
A similar phenomenon was also found in one patient with rectal adenocarcinoma who developed fever and cough on day 3 postoperatively and was diagnosed with COVID-19 on day 7 (ref.60). SARS-CoV-2 RNA was detected in surgical rectal tissues and virions were observed under electron microscopy of the surgical tissues on day 0. These results implied that the virus might have survived and replicated in the intestine even before respiratory symptoms appeared. SARS-CoV-2 antigens were detected in intestinal epithelial cells as well as in lymphocytes and macrophages of the lamina propria60, similar to what has been described for SARS-CoV41.
Furthermore, a study that included pathological analysis of colonoscopy samples from one patient with COVID-19 confirmed SARS-CoV-2 infection in the colon by in situ hybridization for viral RNA and by electron microscopy93. Importantly, substantial damage to the colon mucosa marked by injury to the luminal epithelial cells and goblet cell depletion was detected in colonoscopy samples93, which was not observed in two previous studies59,60. Taken together, although more histological analyses might be needed to determine whether there is enterocyte damage in the intestine, the endoscopic and histological examination in patients with COVID-19 provides direct evidence of active SARS-CoV-2 replication in the intestine.
Increased intestinal levels of pro-inflammatory cytokines and calprotectin
CRS is considered a leading cause of severe pneumonia and even death during SARS-CoV-2 infection9. Following an early increase in type 1 (antiviral, interferons) and type 3 (antibacterial or antifungal, IL-17 and IL-22) immune responses, patients with moderate COVID-19 (n = 80) displayed a progressive reduction in plasma cytokine levels94. However, patients with severe disease (n = 33) maintained these elevated responses for approximately 20 days after the onset of symptoms94. Higher plasma levels of both pro-inflammatory and anti-inflammatory cytokines IL-2, IL-6, IL-7, IL-10, GM-CSF, IP10, CCL2 (also known as MCP1), MIP1α and TNF have been detected in patients with severe disease compared with those with moderate disease94,95. Moreover, histological examination of autopsy samples from a patient with severe COVID-19 showed bilateral diffuse alveolar damage and mononuclear inflammatory lymphocytes in both lungs96. Together, these studies indicated that CRS probably plays a major role in causing acute respiratory distress syndrome and death94–96.
Increasing evidence of enteric infections with SARS-CoV-2 highlights the possible influence of intestinal inflammation on gastrointestinal symptoms and even on lung symptoms associated with COVID-19. Faecal calprotectin, which is largely expressed by neutrophil granulocytes, has been widely adopted as a reliable faecal biomarker of intestinal inflammation97. In a study including 40 Australian patients, patients with COVID-19 with diarrhoea (n = 22), and especially 9 patients with ongoing diarrhoea, had elevated faecal calprotectin concentrations compared with patients with COVID-19 without diarrhoea (n = 18)62. Moreover, faecal calprotectin production was significantly positively correlated with serum IL-6 concentration (P < 0.001)62. The intestine produces high levels of IL-6 (refs98–100), indicating a potential contribution of intestinal calprotectin and/or IL-6 in the increased serum IL-6 concentrations observed in patients with severe COVID-19 (ref.94). Intriguingly, tocilizumab, a recombinant humanized anti-human IL-6 receptor monoclonal antibody, immediately improved the clinical outcomes of 21 patients with severe and critical COVID-19 in a single arm trial101,102. However, in a randomized, double-blind, placebo-controlled phase III trial of 243 patients, tocilizumab was not effective in preventing intubation or death in moderately ill hospitalized patients with COVID-19 within 28 days (17 of 161 (10.6%) versus 10 of 80 (12.5%); P = 0.64), although there were fewer severe cases in the tocilizumab-treated group than in the placebo-treated group (13 of 161 (8.1%) versus 14 of 81 (17.3%); P = 0.03)103. In another phase III trial of 389 hospitalized patients with COVID-19 pneumonia who were not receiving mechanical ventilation, tocilizumab reduced the composite outcome of mechanical ventilation or death (12% versus 19.3%; P = 0.04) by day 28 (ref.104). However, when death from any cause alone was evaluated as a secondary outcome, no statistically significant difference was observed between the tocilizumab group and the placebo control group (10.4% versus 8.6%)104. Moreover, a randomized clinical trial of 131 patients with moderate-to-severe COVID-19 comparing the effect of tocilizumab versus standard of care (antibiotics, antiviral agents, corticosteroids, vasopressor support and anticoagulants) showed that tocilizumab might reduce the need for mechanical and non-invasive ventilation or death by day 14 (17% versus 27%) but not mortality by day 28 (ref.105). Further studies are necessary to confirm these observations, especially testing the effects of tocilizumab on patients with COVID-19 at different stages across the disease course and with different severities of the disease.
Interestingly, the level of another inflammatory cytokine mainly expressed in intestinal epithelial cells, IL-18 (refs106–108), was found to increase upon fever onset and remain highly elevated in the acute phase of SARS-CoV infection in 88 patients109. Similarly, three studies respectively found that IL-18 levels were markedly increased in the serum94,110 or faeces65 of patients with COVID-19 and that increased levels of IL-18 were positively correlated with severe disease94. IL-18 maturation is induced by the activation of the intestinal inflammasome106,108,111. IL-18 participates in several inflammatory diseases (for example, inflammatory bowel disease, metabolic syndrome and type 2 diabetes mellitus) and lung diseases (for example, allergic asthma, chronic obstructive pulmonary disease and acute lung injury)112–114. Based on experimental and clinical evidence, neutralizing IL-18 by anti-IL-18 antibody or its natural inhibitor, soluble IL-18 binding protein (IL-18BP), can attenuate the inflammatory conditions112,115,116. Interestingly, IL-18 is elevated in patients with COVID-19 but not in patients with seasonal influenza65. Additionally, the IL-18 levels were higher in the faecal supernatants obtained from patients with COVID-19 who tested positive for SARS-CoV-2 RNA than in those faecal samples that tested negative, suggesting that IL-18 can potentially serve as an indicator for intestinal infection in COVID-19 (ref.65). We postulate that intestinal release of IL-18 during SARS-CoV-2 enteric infection might also be involved in CRS in COVID-19. Further investigations into the correlation between IL-18 expression and disease severity could facilitate the potential application of IL-18 blockade in patients with severe COVID-19 with intestinal infection (Fig. 1).
Alterations in the gut microbiota in COVID-19
Previous studies have shown that respiratory viral infections are associated with altered gut microbiota composition52,117,118. Several groups have investigated changes in the faecal microbiota of patients with SARS-CoV-2 infection during hospitalization66,119. Consistently, an enrichment of opportunistic pathogens (such as Clostridium hathewayi, Actinomyces viscosus, Bacteroides nordii and Streptococcus spp.) concurrent with the depletion of beneficial commensals (including members of the Ruminococcaceae and Lachnospiraceae families) was observed in 15 patients with COVID-19 compared with healthy controls in a pilot study119. Specifically, in one patient, gut dysbiosis persisted for as long as 17 days after discharge119. The genus Coprobacillus, which contains species that strongly upregulate colonic expression of ACE2 in mice120, were the top bacteria positively associated with COVID-19 severity119. In addition, four Bacteroides species, which were associated with downregulation of ACE2 expression in the murine colon120, showed a statistically significant inverse correlation with faecal SARS-CoV-2 load119. Moreover, another study compared alterations in gut microbiota among 62 patients with COVID-19, patients with seasonal influenza and healthy individuals as controls65. An analysis of alpha diversity showed a statistically significant decrease in mean species diversity (chao1 index) in patients with SARS-CoV-2 compared with both patients with influenza and healthy controls. Moreover, patients with COVID-19 had a relatively higher abundance of Streptococcus, Clostridium, Lactobacillus, and Bifidobacterium and lower levels of Bacteroidetes, Roseburia, Faecalibacterium, Coprococcus and Parabacteroides compared with healthy individuals as controls, with IL-18 expression strongly correlating with several bacteria genera such as Peptostreptococcus, Fusobacterium and Citrobacter65. These observations bring a potential connection between microbiota dysbiosis and infection and/or inflammation caused by SARS-CoV-2 in the intestine. It is worth noting that the patients with COVID-19 involved in those studies were under medical care; further studies on treatment-naive patients would be of benefit to better characterize the connection between SARS-CoV-2 infection and gut microbiota.
Given that SARS-CoV-2 infection has been shown to lead to specific alterations in the gut microbiota, further study of the relationship between shifts in gut microbial communities and cytokine release will help to determine whether these changes directly or indirectly affect serum cytokine levels in patients. In addition, as gut microbiota-derived metabolites play important roles in the maintenance of intestinal homeostasis121–123, and given that several metabolites, such as acetate and butyrate, reportedly protect against respiratory virus infection in mice studies124,125, analyses of gut metabolism in patients with COVID-19 would also be informative of whether alterations in microbiota can cause aberrant immune responses.
Intestinal infection models
In vitro models for SARS-CoV-2 intestinal infection
Several studies based on human cell lines and organoids have confirmed that SARS-CoV-2 can infect intestinal cells in vitro69,92,126–128 (Table 4). SARS-CoV-2 was reported to infect cells of the colon carcinoma-derived lines T84 and Caco-2 (refs69,126), both of which were found to express ACE2 and TMPRSS2, as confirmed by quantitative PCR69. Interestingly, much higher amounts of infectious viral particles were produced in Caco-2 cells than in the human lung adenocarcinoma cell line Calu-3 (the virus titres were about 104 median tissue culture infectious dose (TCID50)/ml produced by Calu-3 cells versus 108 TCID50/ml produced by Caco-2 cells when infected with SARS-CoV-2 at a multiplicity of infection of 3)69.
Table 4.
Current evidence on SARS-CoV-2 replication in vitro and in vivo
| Model | Phenotypes during SARS-CoV-2 infection |
|---|---|
| In vitro studies | |
| SARS-CoV-2-infected human intestinal-derived cell line69,126 | Efficiently infected Caco-2 cells; partial infection T84 cells |
| Bat organoids92 | Susceptible to SARS-CoV-2 infection |
| Human small intestine organoids127,128 | Susceptible to SARS-CoV-2 infection; induction of ISGs |
| Human colon-derived organoids69,92,93 | Infection of 10% of colon organoid cells; induction of type III interferons and ISGs |
| Animal modela | |
| hACE2 transgenic mice138 | Viral RNA in the intestine on day 1 post-infection; no histological changes in gastrointestinal tract |
| hACE2 knock-in mice133 | Viral RNA in faeces of aged mice; intragastric infection led to lung inflammation |
| Golden Syrian hamster137,142 | Continuous viral RNA shedding in faeces; viral antigens in the intestine; successfully infected via fomites |
| Ferret134,139,141 | Continuous viral RNA shedding in faeces; viral antigens in the intestine; isolation of infectious particles from nasal swabs after intragastric transfer of faecal supernatant |
| Cat139 | Positive rectal swabs; viral RNA in the intestine |
| Dog139 | Positive rectal swabs |
| Rhesus macaques135,136 | Prolonged faecal viral shedding after being negative in respiratory samples; viral RNA in the intestinal tissue; inflammatory infiltration in the intestine; viral antigens in the intestine |
| Cynomolgus macaques144 | Viral RNA in faeces; viral RNA in ileum |
hACE2, human angiotensin-converting enzyme 2; ISGs, interferon-stimulated genes; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2. aAnimals were infected via intranasal route unless otherwise noted.
Intestinal organoids can also be used as a convenient model to study intestinal viral infections such as rotaviral infection106,129. Indeed, SARS-CoV-2 can readily infect human small intestine organoids127,128. Infection of proliferative enterocyte progenitors or ApoA1+ enterocytes, but not goblet cells, has been reported. Mature viral particles were released from the basolateral and apical cells of the lumen128. Transcriptomic or quantitative PCR analysis of SARS-CoV-2-infected intestinal organoids revealed a strong induction of interferon-stimulated genes, although the ability of SARS-CoV-2 to induce a type I and/or type III interferon response varies among different studies92,127,128. SARS-CoV-2 was also reported to infect human colon-derived organoids69,92,93. Quantification revealed that ~10% of cells were infected in colon organoids, consistent with the low percentage of colon epithelial cells found to express ACE2 in single-cell transcriptomics data69. The infection of colon organoids did not lead to type I interferon (IFNβ1) production69,92 but did result in a strong upregulation of type III interferon (IFNλ)69. Interestingly, both human lung organoids and colonic organoids were cultured in one study93 and, although only lung organoids were used to screen drug candidates that are capable of inhibiting SARS-CoV-2 entry, colonic organoids were used for drug efficacy testing and showed similar results as lung organoids, suggesting that intestinal organoids can also serve as a highly relevant infection model to test viable candidate therapeutics93.
As SARS-CoV-2 showed 96% identity to the bat coronavirus BatCoV RaTG13 (ref.3) and the intestine of Chinese horseshoe bat is considered the reservoir of many related coronaviruses130,131, bat intestinal organoids have been cultured to explore SARS-CoV-2 replication in the bat gut92. Indeed, bat organoids were fully susceptible to human SARS-CoV-2 infection and sustained robust viral replication92. These findings indicated that bat intestinal organoids could therefore also be used for mechanistic studies of SARS-CoV-2 intestinal infection. Taken together, these in vitro studies strongly suggest that SARS-CoV-2 can enter and replicate in intestinal epithelial cells.
Evidence of intestinal SARS-CoV-2 infection in animal models
SARS-CoV-2 cannot infect wild-type mice132 as the entry of SARS-CoV-2 relies on the binding of human ACE2 (hACE2) but not of mouse ACE2. Thus, many efforts have been made to establish suitable animal models for mimicking specific aspects of SARS-CoV-2 infection in humans133–139 (Table 4).
Mice expressing hACE2 have been widely used for SARS-CoV-2 infection133,138,140; the adenovirus-mediated expression of hACE2 in the lung of the mice, for example, can serve as a useful model for COVID-19 pathogenesis, vaccination and treatment140. However, as hACE2 is transiently expressed in the lung via intranasal adenovirus inoculation, virtually no viral RNA was detected in the intestines of AdV-hACE2-transduced mice140. The targeted infection of the intestine by hACE2-expressed adenovirus might be needed to test intestinal infection of SARS-CoV-2. Transgenic mice that express hACE2 have also been explored to mimic mild SARS-CoV-2 infection138. However, upon intranasal inoculation of SARS-CoV-2 in hACE2 transgenic mice, viral RNA was only transiently detected at the first day post-infection and no histopathological lesions of SARS-CoV-2 were observed. The failure to establish intestinal infection in this model could be related to suboptimal virus replication in transgenic mice, as less than 104 TCID50 per ml virus was detected in the lung and, moreover, pathological changes in lung tissue were minimal138.
For more physiological exploration of the infection process, knock-in mice expressing hACE2 under the control of the mouse ACE2 promoter can serve as a more appropriate infection model than transgenic lines. The peak viral RNA titre in the lungs of hACE2 knock-in mice reached up to 108 copies/g (ref.133). Interestingly, intranasal inoculation of aged mice (30 weeks old) with 4 × 105 plaque-forming units of SARS-CoV-2 resulted in the detection of high levels of viral RNA in faeces (2.9 × 105 copies/g)133. However, as many factors could result in the intestinal detection of viral RNA, such as swallowing of the virus, virus replication in the intestine and the fact that mice, being coprophagic, could be ‘inoculating’ themselves with faecal matter, the source of the viral RNA in faeces warrants further investigation. Notably, intragastric SARS-CoV-2 infection in hACE2 knock-in mice led to interstitial inflammation with alveolar septal thickening as well as the accumulation of detectable levels of viral RNA and antigens in lung tissue133, suggesting that intragastric administration of SARS-CoV-2 could also establish productive infection and lead to pulmonary pathological changes in hACE2 mice. This result also highlights the possibility of faecal–oral transmission of SARS-CoV-2. Notably, viral RNA was not detectable in either intestinal samples or serum via intranasal or intragastric infection in this mouse model133, which is probably due to sample collection at late time points of infection (5–6 days post infection (dpi)), when the weight loss of mice was already recovered133. Further measurements on whether there is an increase of faecal virus shedding and intestinal histopathology are needed to further prove the establishment of intestinal infections.
In addition to mouse models, evidence from several other animal models also supports intestinal viral infection. SARS-CoV-2 has been reported to infect ferrets134,139,141. Viral RNA was detected in some of the rectal swabs of virus-inoculated ferrets134,139,141, some of which continued to test positive for 4–8 days (blood (for 4 dpi), nasal washes (for 8 dpi), urine (for 8 dpi) and in faeces (for 8 dpi)). Similarly, viral antigens were detectable in the intestines of these animals134. Moreover, when naive ferrets were inoculated with faecal supernatants of infected specimens, infectious virions could be isolated from subsequent nasal washes, thereby providing direct evidence of the faecal–oral transmission of SARS-CoV-2 in ferrets134.
Furthermore, the intranasal infection of golden Syrian hamsters with SARS-CoV-2 showed intestinal infection: viral RNA was continuously detectable in the faecal samples of infected hamsters for 14 days and viral antigen was detected in intestinal epithelial cells137,142. Severe intestinal epithelial cell necrosis, damaged and deformed intestinal villi, and increased lamina propria mononuclear cell infiltration were observed142, thereby providing direct evidence for intestinal infection of SARS-CoV-2 in hamsters. Interestingly, naive golden Syrian hamsters could be infected via fomites127, objects or substances capable of carrying infectious organisms. However, it is unclear whether infection via fomites was possible via the faecal–oral route as the isolation of infectious virus particles in faecal samples failed, probably owing to the toxicity of the faecal extracts on the Vero cells used for the detection of infectious virus134,143. Further in vivo experiments by intragastric inoculation of faecal supernatants into naive animals might be needed to verify the feasibility of faecal–oral transmission.
SARS-CoV-2 has been shown to infect non-human primates, such as Rhesus macaques135,136 and cynomolgus macaques144, providing the most genetically relevant infection model to mimic human infection. Upon intranasal inoculation of SARS-CoV-2 to seven Rhesus macaques at 106 TCID50, viral RNA-positive anal swabs were observed at 3 dpi in all infected monkeys, followed by a continuous decline of the positive numbers of anal swabs until viral RNA was undetectable at 14 dpi (ref.136). Virus-positive cells and inflammatory cell infiltration were both observed in the jejunum and colon of these animals. In another study, viral RNA-positive anal swabs were reported in two of eight Rhesus macaques infected with SARS-CoV-2 via a combination of intranasal (0.5 ml per nostril), intratracheal (4 ml), oral (1 ml) and ocular (0.25 ml per eye) inoculation of a 4 × 105 TCID50/ml virus dilution135. At 3 dpi, small numbers of antigen-positive lymphocytes and macrophages in the lamina propria of the intestinal tract of all four macaques were detected. Viral mRNA, which indicates active viral replication, was detected in the respiratory tract and lymphoid tissues of all four Rhesus macaques and detected in the stomach tissue of one of four Rhesus macaques at 3 dpi (ref.135). Similar to clinical studies in patients, prolonged shedding of viral RNA in rectal swabs was observed even after nose and throat swabs returned negative from one of four Rhesus macaques that were monitored until 21 dpi (ref.135). In another study, Rhesus macaques were infected with an intragastric (n = 5) or intranasal (n = 5) challenge with 1 ml of 1 × 107 plaque-forming units of SARS-CoV-2 (ref.145). Notably, both intranasal and intragastric inoculation caused the infiltration of inflammatory cells and loss of mucosal epithelium in gastrointestinal tissues. The immunohistochemistry staining of cleaved caspase 3 was observed in the stomach, jejunum, colon and rectum at 1 dpi in both the intranasal and intragastric groups145, highlighting the apoptosis caused by SARS-CoV-2 infection in gastrointestinal epithelial cells. A decreased number of mucin-containing goblet cells, the hallmark of ulcerative colitis146, was observed in the small intestine at 4 dpi in the intranasal group and as early as 1 dpi in the intragastric group145. Consistently, a decrease in goblet cell numbers has also been reported in patients with COVID-19 (ref.93), indicating the induced intestinal inflammation by SARS-CoV-2. The expression of CD68, a marker for macrophages, substantially increased in the intestine at the earlier stage (at 1, 4 and 7 dpi) of infection by either intranasal or intragastric challenge, which is consistent with the expression of inflammatory cytokines produced by macrophages such as IL-1, IL-6, IL-12 and TNF145. Interestingly, in this study, intragastric inoculation with SARS-CoV-2 led to pneumonia; however, the viral RNAs in the lung were unable to be detected145. These results led to a hypothesis that activated macrophages in the gastrointestinal tract might induce tissue damage in the digestive tract, and even in the lung, by secreting inflammatory cytokines.
In addition, viral RNA was also found in the anal swabs of SARS-CoV-2-infected cats and dogs139. Taken together, results from a variety of animal models (mice, ferrets, hamsters and non-human primates) strongly support intestinal infection of SARS-CoV-2 and the potential for faecal–oral transmission.
Potential faecal–oral transmission
Respiratory droplets and close contact have been shown to be the main routes of SARS-CoV-2 transmission147,148. However, in one study including 96 patients in China, 59% of patients shed viral RNA in stool86, which raised concerns of a possible faecal–oral transmission route for SARS-CoV-2 in humans (Fig. 2). In 2003, a large community outbreak of SARS, affecting more than 300 residents in Amoy Gardens in Hong Kong, was presumed to be related to faecal–oral transmission149. A patient with SARS with diarrhoea visited one building in Amoy Gardens and used the toilet; subsequently, 321 cases of SARS were located in clusters within this building. The floor drain traps, which are designed as a barrier between the floor and the drain, had dried out at the time of the outbreak; however, swabs taken from the affected apartments and the sewer manholes found no genetic material of SARS149. Thus, direct evidence of faecal–oral transmission is missing in that outbreak. According to a report, based on circumstantial evidence, faecal–aerosol transmission might have caused the community outbreak of COVID-19 in a high-rise building in Guangzhou, China150. Nine patients infected with SARS-CoV-2 in three families lived in vertically aligned flats connected by drainage pipes in the master bathrooms. The first family infected had a travel history to the COVID-19 outbreak epicentre of Wuhan in January 2020, whilst the other two families had no travel history and showed a later onset of symptoms in February 2020. No evidence was found for transmission via the elevator or elsewhere. Both the confirmed time points of infections and the locations of positive environmental samples are consistent with the vertical spread of virus-laden aerosols via stacks and vents150.
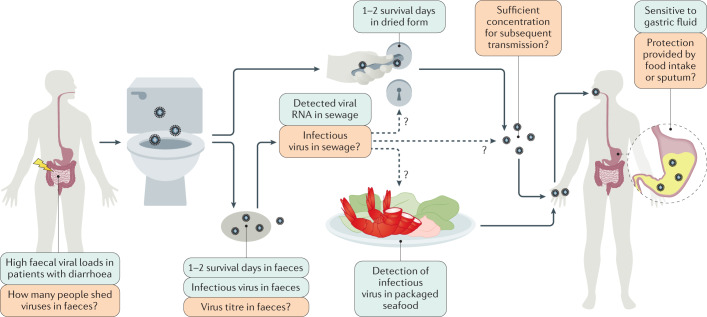
Fig. 2. The potential faecal–oral transmission of SARS-CoV-2.
The exact faecal–oral transmission route is not yet established for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); green boxes show confirmed findings, whereas orange boxes depict open questions. High viral loads were found in the faecal samples of patients with coronavirus disease 2019 (COVID-19) and diarrhoea. However, the number of patients with COVID-19 that shed infectious viruses (but not viral RNA) in faeces remains unknown. In several cases, infectious virus was isolated in faeces and an in vitro study reported 1–2 survival days of SARS-CoV-2 in faeces. Thus, the virus might possibly contaminate sewage, water, and food supplies and possibly contaminate bathroom sites via faecal–aerosol transmission. Consistent with this hypothesis, viral RNA was detected in sewage and on toilet seats, flush buttons and door handles. Moreover, the detection of infectious virus in packaged seafood was reported in China. However, the research on virus titre in faecal fomites is still lacking. Whether the virus titres in faecal fomites are of sufficient concentration and infectivity for subsequent transmission remains unknown. Moreover, SARS-CoV-2 can tolerate human small intestinal fluid but rapidly loses infectivity in gastric fluid within 10 minutes. It remains unclear whether the virus can survive during food intake or whether it is protected by sputum, which is a previously reported by-pass mechanism of Middle East respiratory syndrome coronavirus and influenza.
Three key issues should be discussed to determine whether SARS-CoV-2 can establish faecal–oral transmission. First, whether SARS-CoV-2 can tolerate exposure to gastric acid to subsequently establish an intestinal infection. Second, it remains unknown whether infectious virus particles can tolerate intestinal fluid and can then be shed through faeces. Finally, whether the virus particles in fomites are of sufficient concentration and infectivity for the subsequent transmission needs to be determined.
Can SARS-CoV-2 tolerate gastric acid and survive passage into the gut?
Enteric viruses, such as rotaviruses and noroviruses, which are characterized by an absence of a lipid envelope, are well known to be transmitted by the faecal–oral route151, whereas enveloped respiratory viruses, such as influenza virus, are thought to be cleared by the digestive juice and mucus layer in the gastrointestinal tract. SARS-CoV-2 has been found to rapidly lose infectivity in simulated gastric fluid in vitro127. As evidence of intestinal infection of SARS-CoV-2 is well reported, several protection mechanisms previously reported in influenza viruses and other respiratory coronavirus are proposed to help SARS-CoV-2 retain its infectivity in the intestine152. Hirose et al. showed that, although influenza virions are vulnerable to simulated digestive juices (that is, gastric acid and bile or pancreatic juice) in vitro, highly viscous mucus, such as viscous sputum and nasal mucus, protects the virions, thereby allowing the virus to retain its infectivity152. Moreover, both SARS-CoV and SARS-CoV-2 were reported to have infected a small number of lymphocytes41,60 and viral RNA was detected in the blood of patients infected with SARS-CoV-2 (ref.90). Whether SARS-CoV-2 could spread from the lungs to the gut carried by immune cells warrants further investigation. Notably, these protective mechanisms are typically used to explain how SARS-CoV-2 might spread from the respiratory system to the intestine in already-infected patients. However, the viral ability to establish an intestinal infection does not equate to its ability to transmit via a faecal–oral route.
The stomach environment varies over the course of gastric residence of a meal, which might affect the tolerance of respiratory viruses in gastric fluid153. Based on human clinical studies, the fed-state gastric fluid has a hyperosmolar content and higher pH compared with fasting-state gastric fluid153. In vitro, the fasting-state-simulated gastric fluid is often a salt solution that contains sodium taurocholate, lecithin and pepsin at a pH of 1.6, whereas the fed-state-simulated gastric fluid is a milk-based medium to simulate the carbohydrate to protein to fat ratio observed in the stomach after the administration of meals and has a pH of 5 (ref.154). Coronavirus MERS-CoV and HCoV-229E (an alphacoronavirus that causes mild respiratory infection in humans) were found to rapidly lose most of their infectivity in fasting-state-simulated gastric fluid143. However, by contrast, the infectivity of MERS-CoV and HCoV-229E were unaffected after 2 h of exposure to fed-state-simulated gastric fluid143. As the research on SARS-CoV-2 infectivity is based on fasting-state-simulated gastric fluid, study of the SARS-CoV-2 infection process in fed-state-simulated gastric fluid is essential to determine whether food and eating can influence the survival of SARS-CoV-2 through passage into the gut.
Can infectious SARS-CoV-2 be shed in faeces?
Shedding through faeces is another essential characteristic of faecal–oral transmission. A study in human duodenum enteroids found that the virus was released predominantly from the apical side into the lumen, suggesting the possibility of viral shedding and accumulation in the faeces of patients with COVID-19 (ref.127). Previous studies have shown that different coronaviruses have different tolerances to small intestinal fluid. Nearly half of MERS-CoV particles were found to remain viable after 2 h of exposure to fed-state-simulated intestinal fluid, whereas HCoV-229E was shown to be rapidly inactivated in the same conditions143. According to another report, residual SARS-CoV-2 virus can be found in fasting-state intestinal fluid after 24 h of incubation127. By contrast, only 20% of SARS-CoV-2 virions remained infective after 1 h of incubation in simulated human colonic fluids in the fasting state127. A clear understanding of the capacity for SARS-CoV-2 survival in fed-state colon fluid is still lacking. However, based on current research, SARS-CoV-2 could plausibly remain infectious in the stool, especially when the patient has diarrhoea symptoms127. Consistent with these findings, several reports have shown successful isolation of SARS-CoV-2 from faeces in humans89–91. For example, intact virus was isolated from the stool specimen of one patient with laboratory-confirmed COVID-19 severe pneumonia in Heilongjiang, China89, and from one patient in Guangzhou, China, who later died91. The failure of other studies to isolate infectious SARS-CoV-2 from stool might be owing to the mild disease courses of these cases, mild gastrointestinal symptoms, low virus titre or the toxicity of intestine homogenates towards the cultured cells88,143.
Can SARS-CoV-2 maintain concentration and infectivity in faecal fomites?
Several studies have investigated the infectivity of SARS-CoV-2 under various environmental conditions. One study reported that SARS-CoV-2 was able to retain viability for 3–5 days in dried glass surfaces and for 7 days in solution at room temperature155. Moreover, SARS-CoV-2 RNA was detected in sewage in Australia, USA and France156–159, and the maximum concentrations of viral RNA reach to 106 copies per litre (ref.156). Although studies on isolating infectious virus in sewage are lacking, the ability to maintain infectivity in liquids makes it possible that SARS-CoV-2 can be transmitted through sewage. In addition, SARS-CoV-2 can survive longer at low temperatures (14 days at 4°C)155; therefore, the detection of SARS-CoV-2 viral RNA and infectious virus in packaged seafood has highlighted the risk of faecal–oral transmission160.
Moreover, an in vitro study that added infectious virus (106.5 TCID50/ml) to watery stool derived from human samples reported that SARS-CoV-2 retained viability but showed a 5-log reduction in infectivity in stool at room temperature for 1–2 days155. However, as the viral titre of SARS-CoV-2 in the faeces of patients with COVID-19 and those asymptomatic but confirmed as SARS-CoV-2 positive have not been well studied, future research is needed to examine the infectious virus titre in faeces, the viability of SARS-CoV-2 in sewage and the minimum infectious viral dose in animal models to explore whether SARS-CoV-2 can be transmitted via the faecal–oral route.
Conclusions
Current research provides strong evidence for the intestinal infection of SARS-CoV-2 (Table 5). Further research is still needed to determine the mechanism of intestinal infection, the correlation with CRS and the possibility of faecal–oral transmission (Table 5). Many studies have reported the detection of viral RNA in the stool and gastrointestinal tracts of patients with COVID-19. As some patients with COVID-19 were reported to have prolonged, persistent viral RNA in rectal swabs or faeces82,83, further studies are needed to accurately quantify the proportion of patients that positively shed viral RNA in faeces and have active viral replication in the intestine. It also remains unknown for how long active intestinal infection by SARS-CoV-2 can persist. Furthermore, comparing the proportion of gastrointestinal symptoms among patients with or without intestinal infection by measuring viral genomic RNA and subgenomic viral RNA transcripts shed in the faeces can be informative on the contribution of direct intestinal infection to these gastrointestinal symptoms.
Table 5.
Current evidence and outlook on the intestinal infection and potential faecal–oral transmission of SARS-CoV-2
| Category | Current evidence | Further questions |
|---|---|---|
| Intestinal infection by SARS-CoV-2 | ||
| Clinical evidence | Nearly half of patients with COVID-19 are positive for SARS-CoV-2 RNA detection in faecal samples15 | How many patients with positive faecal tests have active viral replication as measured by viral subgenomic mRNA? |
| Persistence of viral RNA in faecal compared with respiratory samples for as long as a month after discharge77,80,83 |
Can intestinal infection serve as a reservoir for re-infection in the lung? Does intestinal infection of SARS-CoV-2 enhance its mutation rate? |
|
| Approximately 5–70% of patients with COVID-19 reported gastrointestinal symptoms11,24 |
How to unify the definition of gastrointestinal symptoms in COVID-19 among different studies? Clear distinctions should be established between gastrointestinal symptoms presented on admission and the gastrointestinal symptoms caused by medications A correlation is not yet established between gastrointestinal symptoms, the presence of faecal SARS-CoV-2 RNA and/or active viral replication |
|
| Endoscopic and histological examination of patients with COVID-19 revealed virions and inflammatory cell infiltration in the duodenum and rectum59,60 | More comprehensive autopsy or surgical specimens are needed to provide histological evidence of intestinal infection | |
|
SARS-CoV-2 infection altered gut microbiota, correlated with elevated expression of inflammatory cytokines such as IL-2 and IL-18 (refs65,66,161) High levels of faecal calprotectin in patients with COVID-19 with diarrhoea, which were positively correlated with IL-6 levels in serum62 |
Does intestinal infection lead to increased expression of inflammatory cytokines in the intestines and/or serum? If so, do intestinal infection and elevated cytokine levels contribute to cytokine release syndrome or correlate with disease severity? |
|
| IgA dominated in the early stage of SARS-CoV-2-specific humoral responses and was more potent in neutralization than IgG165 |
Does intestinal mucosa contribute to IgA production during SARS-CoV-2 infection? Will oral administration of SARS-CoV-2 vaccines achieve better efficacy? |
|
| In vitro evidence | SARS-CoV-2 infects intestinal cell lines and human intestinal organoids, thereby mediating the production of ISGs69,92,93,127,128 | Can human intestinal organoids serve as a highly relevant infection model to characterize the complete SARS-CoV-2 life cycle and test viable candidate therapeutics? |
| SARS-CoV-2 can establish an intestinal infection in hACE2 knock-in mice, hamsters, ferrets and non-human primates133–139 | More careful virological and histological examination of intestinal infection in animal models can provide evidence not easily observed in humans | |
| Potential faecal–oral transmission routes | ||
| Clinical/environmental evidence |
Viral RNA detected in the sewage156–159 Infectious virions were isolated from faecal samples of patients with COVID-19 (refs89–92) |
How long can SARS-CoV-2 survive in sewage or food surfaces? Can SARS-CoV-2 maintain sufficient concentration and infectivity in fomites for subsequent transmission? |
| In vivo evidence |
Hamsters can be infected through SARS-CoV-2 fomites137 ACE2 knock-in mice can be infected by intragastric SARS-CoV-2 (ref.133) Naive ferrets can be infected by intragastric faecal supernatant from infected ferrets134 |
Exploration of the exact route and timelines for faecal–oral infection in animal models; systematic characterization of the host response for lung infection and intestinal infection in animal models |
| Prolonged shedding of viral RNA in rectal swabs was observed from one infected Rhesus macaque even after nose and throat swabs returned negative135 | More evidence in humans on whether SARS-CoV-2 can infect the next host via the faecal–oral route is needed | |
ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019; hACE2, human ACE2; ISGs, interferon-stimulated genes; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
According to endoscopic and histological examination of patients with COVID-19, intestinal infection with SARS-CoV-2 caused inflammatory infiltration59,60. SARS-CoV-2 infection also altered the composition of the gut microbiota, which correlated with the elevated expression of inflammatory cytokines such as IL-2, IL-4, IL-6, IL-10 and IL-18 (refs65,66,161). Furthermore, faecal calprotectin was found to accumulate in high levels in the stools of patients with COVID-19 with diarrhoea, which was also correlated with serum levels of the inflammatory cytokine IL-6 (ref.62). It is therefore worth further scrutiny to determine whether higher intestinal inflammatory infiltration or more severe dysbiosis of microbiota are correlated with higher cytokine serum levels, the occurrence of CRS and/or disease severity in patients with COVID-19. In addition, further studies in animal models can help illustrate the relative contributions of intestinal inflammation and gut microbiota to CRS and disease severity.
IgA is mainly produced in mucosal surfaces. Humans produce more IgA (40–60 mg/kg per day) than all other immunoglobulin isotypes combined and at least 80% of all plasma cells are located in the intestinal lamina propria162,163. A study found that 33% (2 of 6) of patients with COVID-19 showed an IgA-dominant serum immunoglobin pattern164. According to a study including 159 patients with COVID-19, SARS-CoV-2-specific IgA is likely to be detected earlier than IgG and IgA contributed to virus neutralization to a greater extent than IgG165. Other work showed that IgA detection exhibited the highest positive diagnostic rate compared with IgM and IgG detection at 4–10 days after symptom onset166. Consistently, in a case report, one patient with IgA nephropathy showed an IgA-dominant pattern during SARS-CoV-2 infection and had reduced renal function during and after recovery from SARS-CoV-2 infection owing to the production of high levels of IgA in both serum and faeces167. Together, these reports suggest that IgA is the main type of immunoglobin induced by mucosal infection of SARS-CoV-2 and that IgA-mediated mucosal immunity plays a vital role in anti-SARS-CoV-2 infection. Notably, oral vaccination has also been discussed to prevent subsequent respiratory diseases by activating gut mucosal immunity168,169. Thus, whether oral administration of SARS-CoV-2 vaccines might achieve better efficacy and longer protection would be interesting to test.
In vivo animal studies have shown that gavage with faecal supernatant or exposure to faecal fomites can infect a proportion of animals134,137. Thus, more evidence is clearly required to unequivocally demonstrate that SARS-CoV-2 can establish a faecal–oral transmission route. First, it remains unknown what proportion of people who are infected shed infectious virus particles in faeces. Second, the virus titre in faeces and the minimum infectious dose of SARS-CoV-2 are both still unresolved. A rigorous and systematic determination of whether the virus titre in faecal fomites can exceed the minimum infection dose is essential to accurately quantify the true risk of SARS-CoV-2 transmission through a faecal–oral route.
Acknowledgements
The authors thank Z. G. Tian, H. M. Wei, R. B. Zhou, T. C. Jin, X. L. Ma, K. G. Zhang, J. P. Weng, Y. C. Wang and T. Xue from the University of Science and Technology of China, China, for discussion and comments. The authors are supported by a grant from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29030101, S.Z.), National Key R&D Program of China (2018YFA0508000, S.Z.) and National Natural Science Foundation of China (81822021, 82061148013, 91842105, 31770990, 81821001, S.Z.). R.A.F. is an investigator of the Howard Hughes Medical Institute.
Author contributions
All authors contributed equally to discussion of content. M.G., S.Z. and W.T. wrote the article. S.Z. and R.A.F. reviewed/edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks J. F. W. Chan, S. Perlman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meng Guo, Wanyin Tao.
Contributor Information
Richard A. Flavell, Email: richard.flavell@yale.edu
Shu Zhu, Email: zhushu@ustc.edu.cn.
References
- 1.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J, Li F, Shi Z. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. WHO coronavirus disease (COVID-19) dashboard. WHOhttps://covid19.who.int/ (2020).
- 5.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 6.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Channappanavar R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindler E, et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio. 2013;4:e00611-12. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 11.Guan WJ, et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang W, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 14.Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung KS, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong Cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferm S, et al. Analysis of gastrointestinal and hepatic manifestations of SARS-CoV-2 infection in 892 patients in Queens, NY. Clin. Gastroenterol. Hepatol. 2020;18:2378–2379.e1. doi: 10.1016/j.cgh.2020.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Díaz LA, et al. Symptom profiles and risk factors for hospitalization in patients with SARS-CoV-2 and COVID-19: a large cohort from South America. Gastroenterology. 2020;159:1148–1150. doi: 10.1053/j.gastro.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aghemo A, et al. COVID-19 digestive system involvement and clinical outcomes in a large academic hospital in Milan, Italy. Clin. Gastroenterol. Hepatol. 2020;18:2366–2368.e3. doi: 10.1016/j.cgh.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remes-Troche JM, et al. Initial gastrointestinal manifestations in patients with SARS-CoV-2 in 112 patients from Veracruz (Southeastern Mexico) Gastroenterology. 2020;159:1179–1181. doi: 10.1053/j.gastro.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajifathalian K, et al. Gastrointestinal and hepatic manifestations of 2019 novel coronavirus disease in a large cohort of infected patients from New York: clinical implications. Gastroenterology. 2020;159:1137–1140.e2. doi: 10.1053/j.gastro.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redd WD, et al. Prevalence and characteristics of gastrointestinal symptoms in patients with severe acute respiratory syndrome coronavirus 2 infection in the United States: a multicenter cohort study. Gastroenterology. 2020;159:765–767.e2. doi: 10.1053/j.gastro.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin X, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen A, et al. Are gastrointestinal symptoms specific for COVID-19 infection? A prospective case-control study from the United States. Gastroenterology. 2020;159:1161–1163.e2. doi: 10.1053/j.gastro.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, et al. Epidemiological and clinical predictors of COVID-19. Clin. Infect. Dis. 2020;71:786–792. doi: 10.1093/cid/ciaa322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papa A, et al. Gastrointestinal symptoms and digestive comorbidities in an Italian cohort of patients with COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2020;24:7506–7511. doi: 10.26355/eurrev_202007_21923. [DOI] [PubMed] [Google Scholar]
- 28.Wan Y, et al. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. Lancet Gastroenterol. Hepatol. 2020;5:534–535. doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cholankeril G, et al. High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology. 2020;159:775–777. doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sultan S, et al. AGA institute rapid review of the gastrointestinal and liver manifestations of COVID-19, meta-analysis of international data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology. 2020;159:320–334.e27. doi: 10.1053/j.gastro.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Cui M, Yang T, Yao P. Correlation between gastrointestinal symptoms and disease severity in patients with COVID-19: a systematic review and meta-analysis. BMJ Open Gastroenterol. 2020;7:e000437. doi: 10.1136/bmjgast-2020-000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobel YR, et al. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology. 2020;159:373–375.e2. doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung IF, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Y, et al. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69:1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wootton SH, et al. Detection of NH1N1 influenza virus in nonrespiratory sites among children. Pediatric Infect. Dis. J. 2014;33:95–96. doi: 10.1097/INF.0b013e3182a09de7. [DOI] [PubMed] [Google Scholar]
- 38.Yuen KY, et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 39.Leung WK, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peiris JS, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi X, et al. Severe acute respiratory syndrome associated coronavirus is detected in intestinal tissues of fatal cases. Am. J. Gastroenterol. 2005;100:169–176. doi: 10.1111/j.1572-0241.2005.40377.x. [DOI] [PubMed] [Google Scholar]
- 42.Arabi YM, et al. Middle East respiratory syndrome. N. Engl. J. Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corman VM, et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chotpitayasunondh T, et al. Human disease from influenza A (H5N1), Thailand, 2004. Emerg. Infect. Dis. 2005;11:201–209. doi: 10.3201/eid1102.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hien TT, et al. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 2004;350:1179–1188. doi: 10.1056/NEJMoa040419. [DOI] [PubMed] [Google Scholar]
- 46.Beigel JH, et al. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 47.Humberd J, Guan Y, Webster RG. Comparison of the replication of influenza A viruses in Chinese ring-necked pheasants and chukar partridges. J. Virol. 2006;80:2151–2161. doi: 10.1128/JVI.80.5.2151-2161.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 49.Körner RW, Majjouti M, Alcazar MAA, Mahabir E. Of mice and men: the coronavirus MHV and mouse models as a translational approach to understand SARS-CoV-2. Viruses. 2020;12:880. doi: 10.3390/v12080880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saif LJ, Jung K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J. Clin. Microbiol. 2020;58:e01355–20. doi: 10.1128/JCM.01355-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DuPont HL. Acute infectious diarrhea in immunocompetent adults. N. Engl. J. Med. 2014;370:1532–1540. doi: 10.1056/NEJMra1301069. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, et al. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014;211:2397–2410. doi: 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crawford SE, et al. Rotavirus infection. Nat. Rev. Dis. Prim. 2017;3:17083. doi: 10.1038/nrdp.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundgren O, et al. Role of the enteric nervous system in the fluid and electrolyte secretion of rotavirus diarrhea. Science. 2000;287:491–495. doi: 10.1126/science.287.5452.491. [DOI] [PubMed] [Google Scholar]
- 56.Hung IF, et al. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.To KF, et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lang Z-W, et al. A clinicopathological study of three cases of severe acute respiratory syndrome (SARS) Pathology. 2003;35:526–531. doi: 10.1080/00313020310001619118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao F, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qian Q, et al. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hassan E, Baldridge MT. Norovirus encounters in the gut: multifaceted interactions and disease outcomes. Mucosal Immunol. 2019;12:1259–1267. doi: 10.1038/s41385-019-0199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Effenberger M, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park SK, et al. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimoto T, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao W, et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020;5:100023. doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gu S, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H, et al. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69:1010–1018. [Google Scholar]
- 69.Stanifer ML, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamming I, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.To KF, Lo AW. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2) J. Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin W, et al. Association between detectable SARS-COV-2 RNA in anal swabs and disease severity in patients with coronavirus disease 2019. J. Med. Virol. 2021;93:794–802. doi: 10.1002/jmv.26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ling Y, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133:1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kujawski SA, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat. Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- 75.Lo IL, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young BE, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hua CZ, et al. Epidemiological features and viral shedding in children with SARS-CoV-2 infection. J. Med. Virol. 2020;92:2804–2812. doi: 10.1002/jmv.26180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han MS, et al. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul. Emerg. Infect. Dis. J. 2020;26:2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu P, et al. Dynamic surveillance of SARS-CoV-2 shedding and neutralizing antibody in children with COVID-19. Emerg. Microbes Infect. 2020;9:1254–1258. doi: 10.1080/22221751.2020.1772677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai J, et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020;71:1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xing YH, et al. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020;53:473–480. doi: 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Y, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 85.Kim SE, et al. Viral load kinetics of SARS-CoV-2 infection in saliva in Korean patients: a prospective multi-center comparative study. J. Korean Med. Sci. 2020;35:e287. doi: 10.3346/jkms.2020.35.e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng S, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walsh KA, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wölfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y, et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- 90.Wang W, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao F, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou J, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 93.Han Y, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Magro F, et al. Comparison of different histological indexes in the assessment of UC activity and their accuracy regarding endoscopic outcomes and faecal calprotectin levels. Gut. 2019;68:594–603. doi: 10.1136/gutjnl-2017-315545. [DOI] [PubMed] [Google Scholar]
- 98.Jeffery V, Goldson AJ, Dainty JR, Chieppa M, Sobolewski A. IL-6 signaling regulates small intestinal crypt homeostasis. J. Immunol. 2017;199:304–311. doi: 10.4049/jimmunol.1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dann SM, et al. IL-6-dependent mucosal protection prevents establishment of a microbial niche for attaching/effacing lesion-forming enteric bacterial pathogens. J. Immunol. 2008;180:6816–6826. doi: 10.4049/jimmunol.180.10.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu X, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl Acad. Sci. USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luo P, et al. Tocilizumab treatment in COVID-19: a single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stone JH, et al. Efficacy of tocilizumab in patients hospitalized with covid-19. N. Engl. J. Med. 2020;383:2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salama C, et al. Tocilizumab in patients hospitalized with covid-19 pneumonia. N. Engl. J. Med. 2021;384:20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hermine O, et al. Effect of tocilizumab vs usual care in adults hospitalized with COVID-19 and moderate or severe pneumonia: a randomized clinical trial. JAMA Intern. Med. 2021;181:32–40. doi: 10.1001/jamainternmed.2020.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu S, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546:667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang P, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang KJ, et al. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang Y, et al. Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. Preprint at. medRxiv. 2020 doi: 10.1101/2020.03.02.20029975. [DOI] [Google Scholar]
- 111.Pu Q, et al. Interaction among inflammasome, autophagy and non-coding RNAs: new horizons for drug. Precis. Clin. Med. 2019;2:166–182. doi: 10.1093/pcmedi/pbz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Siegmund B, et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-gamma and TNF-alpha production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;281:R1264–R1273. doi: 10.1152/ajpregu.2001.281.4.R1264. [DOI] [PubMed] [Google Scholar]
- 113.Monteleone G, et al. Bioactive IL-18 expression is up-regulated in Crohn’s disease. J. Immunol. 1999;163:143–147. [PubMed] [Google Scholar]
- 114.Pizarro TT, et al. IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn’s disease: expression and localization in intestinal mucosal cells. J. Immunol. 1999;162:6829–6835. [PubMed] [Google Scholar]
- 115.Nold MF, et al. IL-37 is a fundamental inhibitor of innate immunity. Nat. Immunol. 2010;11:1014–1022. doi: 10.1038/ni.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gabay C, et al. Open-label, multicentre, dose-escalating phase II clinical trial on the safety and efficacy of tadekinig alfa (IL-18BP) in adult-onset Still’s disease. Ann. Rheum. Dis. 2018;77:840–847. doi: 10.1136/annrheumdis-2017-212608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, Schmolke M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. 2018;6:9. doi: 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hanada S, Pirzadeh M, Carver KY, Deng JC. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front. Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zuo T, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Geva-Zatorsky N, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943.e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang W, Zhu S. Gut metabolites: make orphans adopted. Precis. Clin. Med. 2019;2:87–89. doi: 10.1093/pcmedi/pbz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bilotta AJ, Cong Y. Gut microbiota metabolite regulation of host defenses at mucosal surfaces: implication in precision medicine. Precis. Clin. Med. 2019;2:110–119. doi: 10.1093/pcmedi/pbz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haase S, Haghikia A, Wilck N, Müller DN, Linker RA. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. 2018;154:230–238. doi: 10.1111/imm.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Antunes KH, et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019;10:3273. doi: 10.1038/s41467-019-11152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Trompette A, et al. Dietary fiber confers protection against flu by shaping Ly6c- patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity. 2018;48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 126.Chu H, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zang R, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ding S, et al. STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat. Commun. 2018;9:1485. doi: 10.1038/s41467-018-03782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lau SK, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ge XY, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun SH, et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim YI, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Munster VJ, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Deng W, et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sia SF, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 139.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hassan AO, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Richard M, et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chan JF, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou J, et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3:eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rockx B, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiao L, et al. The gastrointestinal tract is an alternative route for SARS-CoV-2 infection in a nonhuman primate model. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Conrad K, Roggenbuck D, Laass MW. Diagnosis and classification of ulcerative colitis. Autoimmun. Rev. 2014;13:463–466. doi: 10.1016/j.autrev.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 147.CDC. How COVID-19 spreads. CDChttps://www.cdc.gov/coronavirus/2019-ncov/about/transmission.html (2020).
- 148.Offord, C. How COVID-19 is spread. The Scientisthttps://www.the-scientist.com/news-opinion/how-covid-19-is-spread-67143 (2020).
- 149.Hong Kong Government. WHO Environmental Health Team reports on Amoy Gardens. Hong Kong Governmenthttps://www.info.gov.hk/gia/general/200305/16/0516114.htm (2003).
- 150.Kang M, et al. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann. Int. Med. 2020;173:974–980. doi: 10.7326/M20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bushman FD, McCormick K, Sherrill-Mix S. Virus structures constrain transmission modes. Nat. Microbiol. 2019;4:1778–1780. doi: 10.1038/s41564-019-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hirose R, et al. Mechanism of human influenza virus RNA persistence and virion survival in feces: mucus protects virions from acid and digestive juices. J. Infect. Dis. 2017;216:105–109. doi: 10.1093/infdis/jix224. [DOI] [PubMed] [Google Scholar]
- 153.Kalantzi L, et al. Characterization of the human upper gastrointestinal contents under conditions simulating bioavailability/bioequivalence studies. Pharm. Res. 2006;23:165–176. doi: 10.1007/s11095-005-8476-1. [DOI] [PubMed] [Google Scholar]
- 154.Jantratid E, Janssen N, Reppas C, Dressman JB. Dissolution media simulating conditions in the proximal human gastrointestinal tract: an update. Pharm. Res. 2008;25:1663–1676. doi: 10.1007/s11095-008-9569-4. [DOI] [PubMed] [Google Scholar]
- 155.Chan KH, et al. Factors affecting stability and infectivity of SARS-CoV-2. J. Hosp. Infect. 2020;106:226–231. doi: 10.1016/j.jhin.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wurtzer S, et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. Preprint at. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu F, et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614-20. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nemudryi A, et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1:100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ahmed W, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total. Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pang X, et al. Cold-chain food contamination as the possible origin of COVID-19 resurgence in Beijing. Natl Sci. Rev. 2020;7:1864–1864. doi: 10.1093/nsr/nwaa264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gou W, et al. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. Preprint at. medRxiv. 2020 doi: 10.1101/2020.04.22.20076091. [DOI] [Google Scholar]
- 162.Fagarasan S, Honjo T. Intestinal IgA synthesis: regulation of front-line body defences. Nat. Rev. Immunol. 2003;3:63–72. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- 163.Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol. Rev. 2014;260:76–85. doi: 10.1111/imr.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tao W, et al. Re-detectable positive SARS-CoV-2 RNA tests in patients who recovered from COVID-19 with intestinal infection. Protein Cell. 2020 doi: 10.1007/s13238-020-00778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sterlin D, et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma H, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang Z, et al. Potential role of aberrant mucosal immune response to SARS-CoV-2 in pathogenesis of IgA nephropathy. Preprint at. medRxiv. 2020 doi: 10.1101/2020.12.11.20247668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mehan A, Venkatesh A, Girish M. COVID-19: should oral vaccination strategies be given more consideration? Ther. Adv. Vaccines Immunother. 2020;8:2515135520946503. doi: 10.1177/2515135520946503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Moreno-Fierros L, García-Silva I, Rosales-Mendoza S. Development of SARS-CoV-2 vaccines: should we focus on mucosal immunity? Expert Opin. Biol. Ther. 2020;20:831–836. doi: 10.1080/14712598.2020.1767062. [DOI] [PubMed] [Google Scholar]
- 170.Lu X, et al. SARS-CoV-2 infection in children. N. Engl. J. Med. 2020;382:1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fakiri KE, Nassih H, Sab IA, Draiss G, Bouskraoui M. Epidemiology and clinical features of coronavirus disease 2019 in Moroccan children. Indian Pediatr. 2020;57:808–810. doi: 10.1007/s13312-020-1958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.de Ceano-Vivas M, et al. SARS-CoV-2 infection in ambulatory and hospitalised Spanish children. Arch. Dis. Child. 2020;105:808–809. doi: 10.1136/archdischild-2020-319366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mahmoudi S, et al. The coronavirus disease 2019 (COVID-19) in children: a study in an Iranian Children’s Referral Hospital. Infect. Drug Resist. 2020;13:2649–2655. doi: 10.2147/IDR.S259064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.CDC COVID-19 Response Team Coronavirus disease 2019 in children - United States, February 12-April 2, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:422–426. doi: 10.15585/mmwr.mm6914e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Parri N, Lenge M, Buonsenso D. Children with COVID-19 in pediatric emergency departments in Italy. N. Engl. J. Med. 2020;383:187–190. doi: 10.1056/NEJMc2007617. [DOI] [PMC free article] [PubMed] [Google Scholar]