Abstract
Background
Rb-pathway disruption is of great clinical interest, as it has been shown to predict outcomes in multiple cancers. We sought to develop a transcriptomic signature for detecting bi-allelic RB1 loss (RBS) that could be used to assess the clinical implications of RB1 loss on a pan-cancer scale.
Methods
We utilized data from the Cancer Cell Line Encyclopedia (N=995) to develop the first pan-cancer transcriptomic signature for predicting bi-allelic RB1 loss (RBS). Model accuracy was validated using the TCGA Pan-Cancer dataset (N=11,007). RBS was then used to assess the clinical relevance of bi-allelic RB1 loss in TCGA Pan-Cancer and in an additional metastatic castration-resistant prostate cancer (mCRPC) cohort.
Results
RBS outperformed the leading existing signature for detecting RB1 bi-allelic loss across all cancer types in TCGA Pan-Cancer (AUC: 0.89 vs. 0.66). High RBS (RB1 bi-allelic loss) was associated with promoter hypermethylation (P=0.008) and gene body hypomethylation (P=0.002), suggesting RBS could detect epigenetic gene silencing. TCGA Pan-Cancer clinical analyses revealed that high RBS was associated with short progression-free (P<0.00001), overall (P=0.0004), and disease-specific (P<0.00001) survival. On multivariable analyses, high RBS was predictive of shorter progression-free survival in TCGA Pan-Cancer (P=0.03) and of shorter overall survival in mCRPC (P=0.004) independently of the number of DNA alterations in RB1.
Conclusions
Our study provides the first validated tool to assess RB1 bi-allelic loss across cancer types based on gene expression. RBS can be useful for analyzing datasets with or without DNA-seq results to investigate the emerging prognostic and treatment implications of Rb-pathway disruption.
Introduction
RB1 is a tumor suppressor that has been implicated in the pathogenesis of numerous cancer types. In addition to causing pediatric retinoblastoma, RB1 alterations have been shown to play a major role in the progression of osteosarcoma1, lymphoma2, and breast3–5, lung6,7, and prostate8,9 malignancies. Moreover, recent studies have highlighted RB1 loss as an important clinical prognostic factor in specific cancer types. For example, RB1 loss has been shown to be associated with poor overall survival in osteosarcoma1, glioblastoma10, and lung cancers11 and has been shown to predict resistance or sensitivity to various small cell lung cancer7, pancreatic cancer12, and breast cancer therapies3,13.
In order to study the clinical implications of RB-pathway disruption, one must first be able to confidently assess RB1 status. Next-generation DNA-sequencing (NGS) approaches are well suited for identifying mutations, copy number alterations, and structural variants. However, there is often uncertainty as to whether a DNA alteration truly inactivates the affected allele. Moreover, other mechanisms of gene inactivation exist that may not be captured by DNA sequencing techniques (e.g. epigenetic, post-transcriptional, or post-translational modifications). An alternative approach to assessing gene inactivation is to examine the sequelae of genomic alterations by assessing the resulting expression of related, downstream genes.
There exist a few RB1 gene sets (genes theorized to be collectively indicative of RB1 status) and two gene signatures (combinatorial expression pattern of the genes in a gene set) for predicting RB1 loss14,15. However, they all share the key limitation that they consist largely of cell cycle genes (whose expression is not specific to RB1 loss). Moreover, since these gene sets and signatures were primarily developed using breast cancer data, their generalizability to different cancer types has not been validated. Our first aim was to develop a novel pan-cancer RB1 bi-allelic loss gene signature (RBS) that outperformed existing RB1-loss signatures and accurately predicted bi-allelic RB1 loss across cancer types.
After generating and validating RBS, we then sought to use it to assess RB1 loss as a prognostic factor across all major cancer types using the TCGA Pan-Cancer database (N=11,007). Since RB1 loss was known to be clinically important in metastatic prostate cancer (not included in the TCGA Pan-Cancer dataset), we examined the prognostic significance of RBS in an independent metastatic castration-resistant prostate cancer (mCRPC) cohort.
Methods
Variable definitions
We defined “RB1 loss” in our manuscript as predicted bi-allelic loss of RB1. For the purposes of training and testing our RB1-loss classifier (RBS), ground-truth labels of RB1 status for each tumor were assigned based on the number of DNA alterations (i.e., non-silent exonic mutations, copy number loss, and inactivating structural variants) observed in RB1. For these ground-truth labels, RB1 loss was defined as presence of at least two DNA alterations in RB1.
RB1-loss gene signature (RBS) development and validation using the CCLE and TCGA pan-cancer datasets
Taking an unbiased approach to selecting genes indicative of RB1 loss, we leveraged microarray log2-normalized RPKM gene expression data of 951 pan-cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE)16. We extracted GISTC2.017 and whole-exome sequencing (WES)-based mutation calls from UCSC Xena Browser to annotate RB1 copy number (CN) and mutation calls18. Cell lines with GISTC score < −0.8 were annotated as deep (two-copy) deletion (CN-2) and cell lines with GISTC score between −0.8 and −0.4 were annotated as shallow (single-copy) deletion (CN-1). The remaining cell lines were annotated as two-copy intact (CN-0). To build an mRNA classifier to predict RB1 functional loss, we defined the tumor cell lines with predicted bi-allelic loss (i.e. deep deletion, shallow deletion with additional DNA mutation, or 2+ DNA mutations) as the RB1-loss group and remaining cell lines as the RB1-intact group. To identify differentially expressed genes between the two groups, we used the Wilcoxon Mann-Whitney test with an adjusted P-value threshold of P < 1×10-10.
We then used a nearest shrunken centroid approach (PAM)19 to generate our gene signature based on the expression pattern of the genes selected as described above. We trained the model by applying PAM to CCLE expression data, using posterior class probabilities for RB1 loss class predictions. The model was trained using 10-fold cross validation to optimize the PAM shrinkage parameter.
RBS was then validated on the TCGA Pan-Cancer RNA-seq expression dataset of 11,007 tumor samples spanning 33 cancer types, downloaded from UCSC Xena Browser using the Synapse platform (syn4976369). RB1 copy number calls and mutation data for these samples were obtained from UCSC Xena Browser, and the same GISTIC2.0 copy number thresholds and mutation criteria as used in the CCLE training set were applied to the validation set. Final RB1-loss annotations were defined based on the number of RB1 DNA alterations observed: 2-alterations (deep deletion, shallow deletion with one mutation, or 2+ mutations), 1-alteration (shallow deletion with no mutations or one mutation with no deletion), 0-alterations (no deletions or mutations). Model accuracy was assessed based on area under the ROC curve (AUC), benchmarked against the leading existing RB1-loss signature14.
RBS pathway enrichment analysis
The EnrichR web tool was used to identify genomic pathways enriched in the RBS gene set. Candidate gene sets were defined as all pathways in the KEGG, Reactome, WikiPathways, and BioCarta databases. Pathways were considered significantly enriched if their adjusted P-values were less than a predetermined significance level of 0.05.
Differential expression analysis of RB1 loss due to two or more RB1 mutations
Differential expression analysis between CN-0 tumors with no mutations and CN-0 tumors with two or more mutations was performed to identify genes that were differentially expressed in tumors with 2+ RB1 mutations. Given that there were far fewer tumors with 2+ mutations than there were with no mutations, we randomly subsampled a set of CN-0 tumors with no mutations equal in size to the subset of tumors with 2+ mutations. We then performed a differential expression analysis between the tumors with 2+ mutations and the tumors with no mutations using the Wilcoxon Mann-Whitney test with an adjusted P-value threshold of P < 0.001. For statistical robustness, we performed a boostrapped analysis with 1,000 different subsamples. Genes were considered significantly differentially expressed if, in >95% of all comparisons, they demonstrated the same directionality of over- vs. under-expression and had adjusted P-values below the predetermined significance level of 0.001.
Promoter and gene body methylation analysis
To assess the utility of RBS in detecting gene silencing due to methylation, we downloaded TCGA Pan-Cancer methylation data for 49 RB1 methylation probes from the UCSC Xena Browser. We first filtered out probes that were previously identified to be of low quality20. We then computed Spearman correlation coefficients between RBS score and Illumina DNA methylation 450K array beta values for each RB1 methylation probe. To test whether the correlations between RBS score and methylation probe values were significant in the RB1 promoter and gene body regions, we generated a null model by computing the correlation between RBS score and methylation in the promoter and gene body regions of 20 other random tumor suppressors not known to be related to RB1. For this analysis, a large set of tumor suppressors (N=1,217) was downloaded from the Tumor Suppressor Gene Database21 and those not located on the same chromosome as RB1 (i.e., not on chromosome 13) and not included in RBS were used as candidate genes for the null model. Spearman correlation coefficients computed between RBS and each methylation probe in the promoter region of a gene (defined as +/− 1.5kb of the transcription start site22) were then modeled as a distribution. The distribution of correlations between RBS and RB1 promoter methylation probes was compared to the distribution of correlations between RBS and non-RB1 promoter methylation probes using the Kolmogorov-Smirnov test with a two-sided significance level of 0.05. Analogous analyses were performed for the gene body region, where gene body was defined as the region 1.5kb downstream of the transcription start site to the transcription terminator. Transcription start sites and terminators were defined using the ‘biomaRt’ R package23.
Characterizing the prognostic value of RB1 loss across cancer types
Clinical outcomes data (progression-free, overall and disease-specific survival) were obtained from the TCGA Pan-Cancer Clinical Data Resource24. All patients with available log2-normalized RPKM RNA-seq data and clinical outcomes data were included in survival analyses. Microarray expression data were log2-normalized and scaled prior to RBS analysis. Data for the metastatic castration-resistant prostate cancer (mCRPC) cohort were obtained from a previously published study25. This cohort consisted of 101 patients with deep whole-genome sequencing, whole-transcriptome RNA-seq, and clinical outcomes data available. The mCRPC RNA-seq data were log2-normalized FPKM values. The clinical endpoint examined was overall survival, with time of study entry defined as date of mCRPC diagnosis.
The threshold of RBS score used to assign binary RB1-impaired vs. RB1-intact status in both cohorts was determined by using the Youden index (computed using the ‘OptimalCutpoints’ R package26) to select a threshold that maximized prediction accuracy in the CCLE training dataset. Cox proportional hazard models were used to model time-to-event data. All survival analyses were performed using R version 3.5.0.
Results
RB1-loss gene signature development and validation using CCLE and TCGA Pan-Cancer data
To define our RB1-loss gene set, we identified genes that were differentially expressed between CCLE cell lines that demonstrated RB1 loss and cell lines that had intact RB1. 951 of the 995 total cell lines had both copy number and microarray expression data available. Of these 951, 126 were identified as having bi-allelic RB1 loss (99 harbored two-copy deletions, 23 harbored single-copy deletions with an additional mutation, and 11 harbored 2+ mutations) and 797 were identified as RB1 intact. Our unbiased approach to defining an RB1-loss gene set using CCLE data identified a final set of 186 genes that were indicative of RB1 loss (Supplementary Table 1A). Of note, only 7 of the 186 genes overlapped with genes in the existing RB1-loss signature14.
To assess the potential utility of our 186-gene signature for predicting RB1 loss, we first performed t-SNE dimensionality reduction on our CCLE training data (N=951). Visualization of the t-SNE embedding revealed that cell lines with 2+ DNA alterations in RB1 tended to map to similar parts of the embedding, suggesting that these cell lines had similar 186-gene expression profiles (Figure 1A). This finding supported the hypothesis that the 186 genes were useful in differentiating between RB1-loss and RB1-intact samples.
Figure 1:
Training a classifier for detecting RB1-impaired tumors using Cancer Cell Line Encyclopedia (CCLE) data. A) t-SNE embedding of CCLE cell lines colored by number of DNA alterations in RB1. Embedding was constructed based on expression levels of the 186 genes found to be differentially expressed between RB1-impaired and RB1-intact cell lines. Cell lines with 2 DNA alterations in RB1 map to similar parts of the embedding, suggesting these 186 genes in aggregate are useful for differentiating between RB1-impaired and RB1-intact cancers. B) Heatmap visualizing expression values of 186 genes (rows) in 951 CCLE cell lines (columns). Cell lines are ordered from left to right in terms of increasing RBS score, where high RBS score denotes impaired RB1. Orange represents high expression and blue represents low expression.
The expression values of the 186 genes nominated as described above were then used in a supervised learning approach (PAM) to compute a RBS score for predicting RB1 loss. The model was trained using the gene expression profiles of CCLE cell lines with known RB1 status (i.e., RB1-loss vs. RB1-intact). The model identified 144 genes whose expression values were most predictive of RB1 status – these genes were used compute the final RBS score (Figure 1B, Supplementary Table 1B). RB1 and CCND1 were among the genes expressed at relatively low levels in RB1-loss samples, while CDKN2A was among the genes expressed at relatively high levels in RB1-loss samples. This was consistent with prior studies which found a high ratio of CDKN2A to CCND1 expression to be associated with RB1-loss in multiple cancer types27,28. Since we noticed that prior RB1 gene sets and gene signatures largely consisted of cell proliferation genes, we assessed the association between RBS and a previously published cell proliferation activity score29. While a previously published RB1 loss signature14 was found to be strongly correlated with the cell proliferation score (r=0.93), we found that RBS was only weakly correlated with the cell proliferation score (r=0.03). These findings suggested that RBS was not a surrogate marker for cell proliferation and was potentially more specific to RB1 loss than existing signatures. Moreover, Enrichr pathway enrichment analysis revealed that RBS was enriched for genes not only in the cyclin D – CDK4/6 and cell cycle-related pathways but also in the DNA damage response and TP53 signaling pathways (Supplementary Table 2). Altogether, these results were consistent with recent literature that suggests RB1 may play a role in processes other than cell-cycle control30.
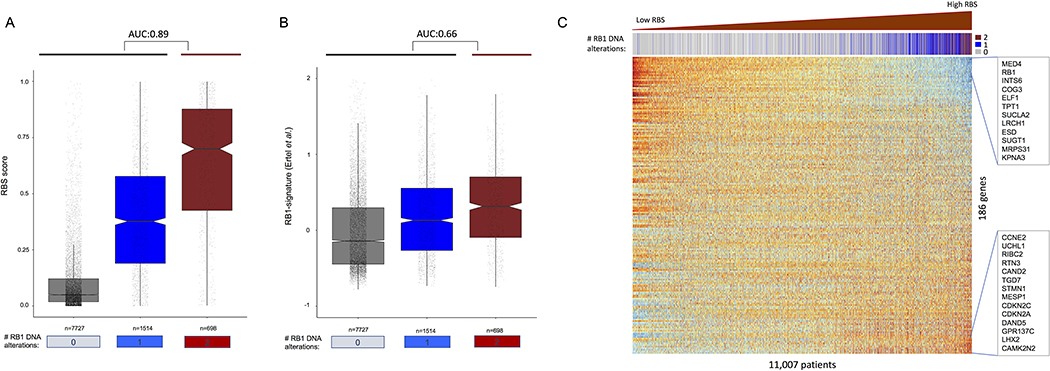
To validate RBS as an accurate model for predicting bi-allelic RB1 loss, we used the TCGA Pan-Cancer atlas expression dataset containing RNA expression data for 11,007 tumors spanning 33 cancer types with known mutation and copy number annotations. 698 of these samples were annotated as having two or more RB1 DNA alterations [559 had deep deletion (CN-2), 89 had shallow deletion with mutation (CN-1/mut), and 50 had two or more mutations with no deletions], 1,514 as having one RB1 alteration [1,332 with shallow deletion and no mutation (CN-1/no-mut), and 182 with one mutation and no deletions (CN-0/mut)] and 7,727 as having no RB1 DNA alterations. RBS achieved an AUC of 0.89 for predicting RB1 bi-allelic loss in this validation set – far superior to an AUC of 0.66 achieved by applying the leading existing RB1-loss signature14 to the same dataset (Figure 2A–B). RBS also outperformed a predictive model based solely on the ratio of CDKN2A to CCND1 expression (AUC=0.72), which was previously reported to be associated with RB1 loss. Genes including CAMK2N2, CDKN2A, and GPR137C were positively correlated with RBS score (i.e., high expression in RB1 loss) while genes including MED4 and RB1 were most negatively correlated with RBS score (Figure 2C).
Figure 2:
TCGA Pan-Cancer data validates accuracy of RBS in predicting bi-allelic RB1 loss. Boxplots showing accuracy of A) RBS in predicting bi-allelic RB1 loss compared to B) the leading existing model. C) Heatmap of TCGA Pan-Cancer data showing mRNA expression profiles of 186 genes (rows) in 11,007 patients (columns). Patients are ordered from left to right in terms of increasing RBS score, where high RBS score denotes impaired RB1. Orange represents high expression and blue represents low expression.
RBS was highly accurate at identifying RB1 loss due to deep deletion and due to shallow-deletion with additional mutation, which comprised the large majority of RB1-loss tumors. However, RBS was less effective at detecting the few RB1-loss tumors with 2+ RB1 mutations, suggesting that these tumors may have a distinct gene expression profile. To investigate this further, we performed a bootstrapped differential expression analysis to identify genes over- and under-expressed in CN-0 tumors with two or more RB1 mutations compared to tumors with no RB1 mutations (Methods). We identified 448 genes significantly overexpressed and 245 genes significantly underexpressed in the tumors with two or more RB1 mutations (Supplementary Table 3). Of these, 16 overexpressed genes (including CCNE2 and CDKN2A) and three underexpressed genes (most notably RB1) were also found in RBS. Additionally, several known regulators or effectors of RB1 such as CCNE1, CDK2, EZH2, HOXB7, and select E2F-family genes were not in RBS but were differentially expressed in the tumors with two or more mutations in RB130–34. Altogether, these findings suggested that there are some transcriptomic similarities but also notable differences between RB1-loss due to deletion and due to bi-allelic RB1 mutations.
RBS can be useful for capturing the effects of gene inactivation due to epigenetic modification
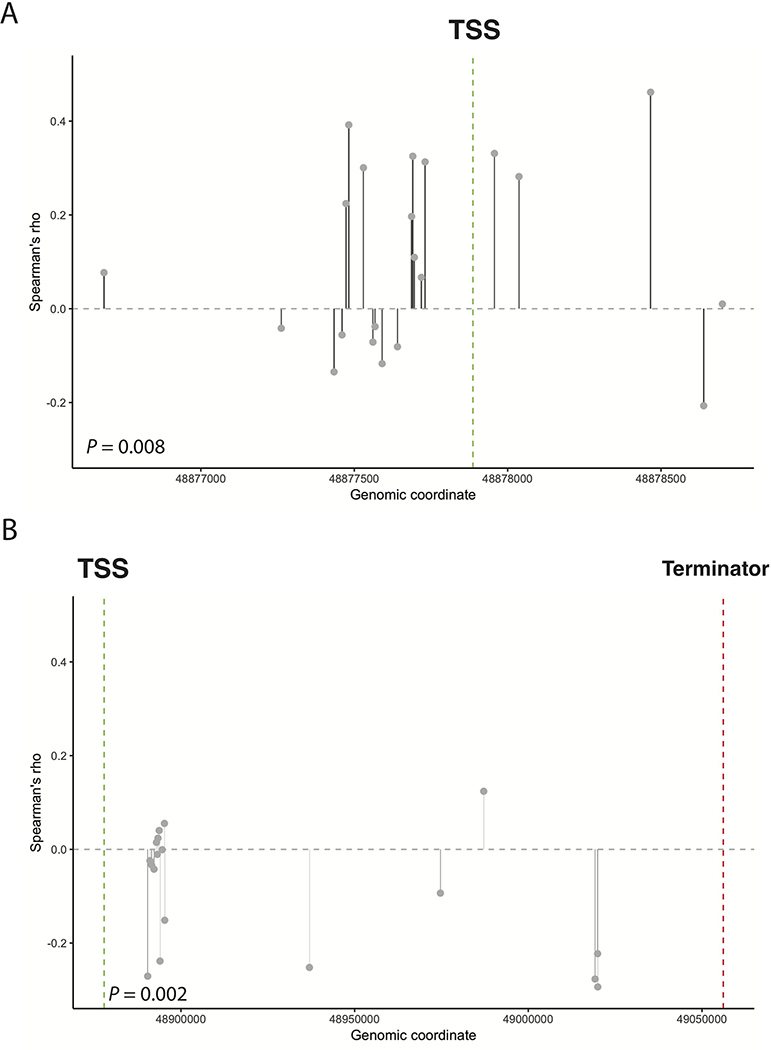
To assess the utility of RBS in capturing the effects of epigenetic events on gene expression, we examined the correlation between RBS score and the methylation scores of 39 methylation probes in the Pan-Cancer cohort (Figure 3). To test whether the pattern of correlation between RBS and methylation probe values was significant in the RB1 promoter and gene body regions, we compared our results with the correlation between RBS score and methylation in the promoter and gene body regions of 20 other random tumor suppressors unrelated to RB1 (Supplementary Table 4). We found that the positive correlation between RBS score and RB1 promoter methylation and negative correlation between RBS score and RB1 gene body methylation were significant (P=0.0077 and P=0.0016 respectively). The directionality of correlation was also consistent with existing literature, which suggests that promoter methylation is associated with decreased gene expression and gene body methylation is associated with increased gene expression in tumor suppressors22. These findings supported the hypothesis that RBS could detect the downstream effects of RB1 loss due to multiple etiologies, including those (such as methylation) that may not be captured using DNA-sequencing techniques.
Figure 3:
High RBS (impaired RB1) is A) positively correlated with methylation of CpGs in the RB1 promoter region and B) negatively correlated with methylation of CpGs in the RB1 gene body. Given prior reports of promoter hypermethylation and gene body hypomethylation being associated with gene inactivation, these results suggest RBS may detect tumors with impaired RB1 due to methylation-based gene silencing.
RBS highlights RB1 loss as a recurrent genomic event and prognostic factor across cancer types
After assessing the accuracy of RBS for predicting RB1 loss, we sought to use RBS to investigate the prognostic significance of RB1 loss across cancer types. For this analysis, we included patients in the TCGA Pan-Cancer dataset with available clinical follow-up. High RBS was defined as scores above a threshold of 0.6, determined based on the Youden Index approach applied to the CCLE training dataset. Of note, we found that the majority of cancer types had an RB1 2-hit prevalence of greater than 5%, suggesting that RB1 loss was common and potentially important in many cancer types. In our pooled analysis of all patient samples across cancer types, we found that RB1 loss defined using RBS was predictive of short progression free survival (median PFS: 36 vs. 56 months, HR:1.3[95%CI,1.18–1.44], P<0.0001; Figure 4A), short disease specific survival (median DSS: 88 vs. 219 months, HR:1.34[95%CI,1.17–1.55], P<0.0001; Figure 4B), and short overall survival (median OS: 70 vs. 94 months, HR:1.23[95%CI,1.09–1.38], P=0.0004; Figure 4C). In a multivariable survival model including both RBS and cancer type, high RBS was found to be independently prognostic of short PFS (HR:1.12[95%CI,1.02–1.26], P=0.04). These findings supported the hypothesis that RB1 loss is clinically important across cancer types and may indicate more advanced or aggressive disease in general.
Figure 4:
High RBS (RNA-seq profile consistent with impaired RB1) is associated with short A) progression free survival (PFS), B) disease-specific survival (DSS), and C) overall survival (OS). RB1 Similarly, presence of 2+ DNA alterations in RB1 is associated with short D) PFS, E) DSS, and F) OS.
We additionally assessed the prognostic significance of a DNA-sequencing based definition of RB1 loss, namely, having at least two DNA alterations in RB1. We found that similarly to high-RBS, presence of 2+ DNA alterations in RB1 was associated with short OS, PFS, and DFS compared to presence of 0 or 1 DNA alterations in RB1 (Figure 4D–F). These findings suggested that our definition of “RB1 loss” as predicted bi-allelic loss of RB1 was clinically meaningful.
RBS is predictive of poor clinical outcomes independently of the number of DNA alterations in RB1
In our methylation analysis, we showed that RBS could potentially be used to detect RB1 loss through mechanisms that could not be detected by DNA sequencing. Additionally, it is known that not all DNA mutations and copy number loss events in a gene have the same effect on the affected allele (i.e., resulting protein may still be partly or completely functional). Since RBS measures the downstream effects of DNA and non-DNA alterations at the gene expression level, we hypothesized that RBS may offer information on RB-pathway disruption that is independent of DNA-sequencing results.
To explore this hypothesis, we assessed the prognostic significance of high-RBS for predicting survival in the TCGA Pan-Cancer cohort independently of number of observed DNA alterations. For these analyses, we focused on PFS as our clinical endpoint of interest due to a prior study that found that PFS was generally the most accurate endpoint collected across all cancer types in the TCGA Pan-Cancer dataset24. On multivariable analysis adjusting for number of DNA alterations in RB1, high RBS was independently predictive of short PFS (HR:1.14[95%CI,1.02–1.29], P=0.03). This suggested that RBS may help distinguish patients with a more pronounced RB1-impaired clinical phenotype from those with a less pronounced phenotype independently of the number of DNA alterations observed in the gene. Moreover, using a criteria of high RBS or 2+ DNA alterations in RB1 to select RB1-impaired patients resulted in a 73% increase in group size as compared to using the criteria of just 2+ DNA alterations (Supplementary Figure 1A). Thus, RBS may be useful for identifying a more comprehensive group of patients with RB-pathway disruption than can be recovered using DNA sequencing alone.
To explore this concept further, we examined a previously published cohort of patients with metastatic castration-resistant prostate cancer (mCRPC)25 – the lethal subtype of prostate cancer not represented in the TCGA Pan-Cancer cohort. RB1 loss (as defined based on detected DNA alterations in RB1) has been shown to be associated with short survival in mCRPC35. Interrogating the mCRPC cohort of 101 patients with both whole-genome sequencing and RNA-seq data available, we aimed to assess whether high-RBS might be predictive of short OS independently of the number of DNA alterations present. First, we examined the degree of concordance between RB1 status as defined based on number of DNA alterations observed and as defined based on RBS score. We found that while RBS score was strongly related to the number of DNA alterations observed (AUC=0.90), not all tumors with high RBS score harbored 2+ DNA alterations and vice versa (Figure 5A). By expanding the DNAseq-based definition of RB1-loss (2+ RB1 DNA alterations) to include tumors with fewer than 2 DNA alterations in RB1 but with high RBS, one could recover 50% more tumors with RB1-impaired status (Supplementary Figure 1B). Next, we examined the prognostic significance of high-RBS in the mCRPC cohort. We found that RB1 loss as defined by high-RBS was predictive of short OS in mCRPC (median OS 15.0 vs. 42.0 months, HR:2.93[95%CI[1.47–5.83], P=0.001; Figure 5B]). Finally, to assess whether the RNA-seq (high-RBS) and DNA-seq (number of DNA alterations in RB1) results were independently predictive of survival outcomes, we performed a multivariable analysis including both the RNA-seq and DNA-seq definitions of predicted RB1 loss. We found that both the RNA-seq and DNA-seq definitions were independently predictive of short OS (P=0.0036 and P=0.046 respectively), suggesting that both RNA-seq and DNA-seq offered unique information on RB1 status that could be used to detect a clinical phenotype of RB1-impaired, clinically aggressive mCRPC.
Figure 5:
A) RBS is associated with number of DNA alterations in RB1, and high RBS is predictive of bi-allelic RB1 loss, as defined using whole genome sequencing results (AUC=0.90). B) High RBS (impaired RB1) is associated with shorter overall survival in mCRPC.
Discussion
In order to assess the clinical implications of RB1 loss across cancer types, we developed a pan-cancer RB1-loss signature (RBS) that predicted bi-allelic loss of RB1 based on gene expression data. We found that RBS was highly accurate at predicting RB1 loss across cancer types compared to existing RB1 gene signatures. Moreover, RBS was able to capture RB1 inactivation due to both DNA and epigenetic changes. Using pan-cancer (N=10,486) and metastatic prostate cancer (N=101) cohorts, we demonstrated that high-RBS was predictive of poor clinical outcomes independently of the number of DNA alterations in RB1.
There are several possible explanations as to why RBS was much more accurate than the leading existing RB1 signature at predicting bi-allelic loss of RB1 (AUC of 0.89 vs. 0.66). For one, RBS was the only RB1-loss signature that was designed to be applied across cancer types. Since RBS was trained on CCLE cell-line data derived from many different primary tissue types, it was well-suited to assess RB1 loss in the TCGA Pan-Cancer validation set, which also included patient samples from many different disease sites. Moreover, in contrast to existing RB1 loss signatures, which included genes largely or exclusively based on prior annotations, the RBS gene set was selected in an unbiased, unsupervised manner. Our approach nominated genes from the set of all existing genes that were most differentially expressed in our pan-cancer, RB1-loss training set samples. A final methodological strength of RBS was that it was trained on a very large dataset (N=995) including many samples with known RB1 loss (N=133) that could be collectively used to represent a distinct RB1-loss expression pattern.
It is important to note that that the “accuracy” of our model for AUC analyses was defined as concordance between (RBS-based) RB1-loss calls and DNA sequencing-based variant calls (i.e. mutation, copy number, and structural variant data when available). This was because DNA-sequencing results are commonly used to predict gene functional status and were the only data available for comparison. However, DNA-sequencing calls do not capture certain forms of gene inactivation such as DNA methylation of the RB1 promoter. While RBS demonstrated high concordance with DNA sequencing calls in our pan-cancer and mCRPC-specific analyses (AUCs of 0.89 and 0.87 respectively), the differences in RB1 loss assignments may not be due to error but rather improved identification of RB1 gene inactivation.
This study is not without limitations. We evaluated RBS as a potential tool to identify RB1 loss due to DNA-sequence alterations and DNA methylation at the RB1 locus. However, still other mechanisms of RB1 inactivation exist, such as CDK phosphorylation of the Rb protein36,37. It is unclear whether these mechanisms of RB1 inactivation result in a similar pattern of gene expression and whether RBS can be used to identify these Rb-inactivated tumors. Future work may involve collecting and integrating phosphoproteomic data with DNA-seq and RNA-seq data to study these additional cases of tumors with RB1 gene inactivation. Additionally, since our analysis was conducted primarily using the CCLE and TCGA Pan-Cancer databases (which focus on primary cancers), an extension to metastatic cancers is needed. In particular, as RB1 loss and RB1 under-expression have been implicated as predictors of more advanced disease in various cancers38–40, future disease-specific studies with a range of indolent and aggressive tumors may leverage RBS to study RB1 loss in the context of disease progression.
The data presented here offer several novel insights and contributions. First, our study is the first to examine the clinical implications of RB1 loss on a pan-cancer scale. We found that RB1 loss was associated with shorter progression-free survival, overall survival and disease-specific survival, highlighting the widespread clinical importance of the genomic event. Second, our novel transcriptomic signature (RBS) is highly accurate at predicting RB1 loss and can be used as a tool in future studies to shed new light on the biological and clinical impact of RB1 loss. This is especially relevant in light of recent studies which suggest that RB1 loss may associated with response to various cancer therapies including radiotherapy3,41, platinum-based chemotherapy3,7, and CDK4/6 inhibitors13,15 in breast, prostate, and small-cell lung cancers. RBS may be useful for detecting differential response to specific cancer therapies for an even broader range of therapies and cancer types than has been already studied. Third, RBS is specific to RB1-loss and not strongly correlated with cell proliferation scores (in contrast to existing RB1-loss signatures). Altogether, our study along with others suggest RB1 may have important functions aside from regulating cell proliferation, such as DNA damage repair41–43. Additional studies are needed to assess this in greater detail. Fourth, our transcriptomic signature may be used to identify RB1-impaired tumors that may not be detected using standard DNA sequencing-based definitions of predicted RB1 loss. The results of our multivariable analyses on two independent cohorts suggest that both RNA-seq and DNA-seq results may be useful to identify a more complete set of RB1-impaired patients.
Our approach to developing an RB1-loss signature is generalizable to studying a wide range of genomic alterations and may serve as a paradigm for generating expression-based gene signatures in an unbiased manner. Since RBS is an expression-based signature, it is complementary to and potentially more holistic than DNA sequencing-based approaches, which may fail to capture the full spectrum of genomic events that can result in a specific gene expression profile or phenotype. Given the plethora of studies highlighting RB1 loss as a driver event in a number of cancer types, the potential clinical implications, and the increasing availability of gene expression data for both retrospective and prospective cohorts, RBS is an immediately useful tool that can be used to assess RB1 loss in a variety of settings. Our analyses and the findings of others suggest that RB1 loss may be predictive not only of survival but also of response to cytotoxic and targeted therapies. RBS may be invaluable for investigating these relationships further with the broader goal of developing personalized cancer treatment regimens.
Supplementary Material
Translational Relevance.
RB1 loss is a recurrent genomic alteration that has been shown to predict response to various treatments including radiotherapy, platinum-based chemotherapy, and CDK4/6 inhibitors in multiple cancer types. Leveraging the transcriptomic and DNA sequencing data of over 11,000 cancer cell lines and clinical tumor samples, we identified a novel pan-cancer transcriptomic signature for identifying RB1 loss (RBS). RBS is more accurate than existing transcriptomic signatures in detecting RB1 loss and can be used alongside DNA sequencing to identify Rb-loss tumors more comprehensively. Using RBS, we found that RB1 loss was associated with impaired survival across cancer types, supporting the notion that RB1 loss constitutes a biologically and clinically distinct subgroup of cancers. Our novel transcriptomic signature can be used to further investigate the clinical implications of RB1 loss and may be coupled with treatment response data to help develop personalized cancer treatment regimens.
Acknowledgements
S.G. Zhao, B.A. Mahal, D.A. Quigley, E.J. Small and F.Y. Feng are supported by the Prostate Cancer Foundation.
Disclosure of Potential Conflicts of Interest
S.G. Zhao reports receiving commercial research support from and holds ownership interest (including patents) in GenomeDx Biosciences. Y. Liu and E. Davicioni are employees of and hold ownership interest (including patents) in GenomeDx Biosciences. E.J. Small reports receiving commercial research grants from Janssen, and is a consultant/advisory board member for Janssen and Fortis Therapeutics. P.L. Nguyen reports receiving commercial research grants from Janssen, Astellas, and Bayer; holds ownership interest (including patents) in Augmenix; and is a consultant/advisory board member for Augmenix, Ferring, Blue Earth, Bayer, Cota, Dendreon, GenomeDx, and Nanobiotix. F.Y. Feng is an employee of PFS Genomics, and is a consultant/advisory board member for Sanofi, Janssen, Medivation/Astellas, Dandreon, Ferring, EMD Serono, Bayer, and Clovis.
References
- 1.Ren W & Gu G Prognostic implications of RB1 tumour suppressor gene alterations in the clinical outcome of human osteosarcoma: a meta-analysis. European Journal of Cancer Care 26, e12401 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Pinyol M et al. Inactivation of RB1 in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood 109, 5422–5429 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Robinson TJW et al. RB1 Status in Triple Negative Breast Cancer Cells Dictates Response to Radiation Treatment and Selective Therapeutic Drugs. PLoS ONE 8, e78641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones RA et al. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. Journal of Clinical Investigation 126, 3739–3757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witkiewicz AK & Knudsen ES Retinoblastoma tumor suppressor pathway in breast cancer: prognosis, precision medicine, and therapeutic interventions. Breast Cancer Research 16, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meuwissen R et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4, 181–189 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Dowlati A et al. Clinical correlation of extensive-stage small-cell lung cancer genomics. Annals of Oncology 27, 642–647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ku SY et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu P et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53 - and RB1 -deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bäcklund LM et al. Short postoperative survival for glioblastoma patients with a dysfunctional Rb1 pathway in combination with no wild-type PTEN. Clin. Cancer Res. 9, 4151–4158 (2003). [PubMed] [Google Scholar]
- 11.Clinical Lung Cancer Genome Project (CLCGP) & Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med 5, 209ra153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hijioka S et al. Rb Loss and KRAS Mutation Are Predictors of the Response to Platinum-Based Chemotherapy in Pancreatic Neuroendocrine Neoplasm with Grade 3: A Japanese Multicenter Pancreatic NEN-G3 Study. Clinical Cancer Research 23, 4625–4632 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Condorelli R et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Annals of Oncology 29, 640–645 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Ertel A et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle 9, 4153–4163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malorni L et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget 7, 68012–68022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mermel CH et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman M et al. The UCSC Xena Platform for cancer genomics data visualization and interpretation. (2018). doi: 10.1101/326470 [DOI]
- 19.Tibshirani R, Hastie T, Narasimhan B & Chu G Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proceedings of the National Academy of Sciences 99, 6567–6572 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naeem H et al. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics 15, 51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao M, Kim P, Mitra R, Zhao J & Zhao Z TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Research 44, D1023–D1031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X et al. Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell 26, 577–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durinck S, Spellman PT, Birney E & Huber W Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–416.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quigley DA et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 174, 758–769.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Ratón M, Rodríguez-Álvarez MX, Suárez CC & Sampedro FG OptimalCutpoints : An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. Journal of Statistical Software 61, (2014). [Google Scholar]
- 27.Mizuarai S et al. Expression ratio of CCND1 to CDKN2A mRNA predicts RB1 status of cultured cancer cell lines and clinical tumor samples. Molecular Cancer 10, 31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai H et al. Cyclin D1 Loss Distinguishes Prostatic Small-Cell Carcinoma from Most Prostatic Adenocarcinomas. Clinical Cancer Research 21, 5619–5629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuzick J et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 12, 245–255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNair C et al. Differential impact of RB status on E2F1 reprogramming in human cancer. Journal of Clinical Investigation 128, 341–358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siu KT, Rosner MR & Minella AC An integrated view of cyclin E function and regulation. Cell Cycle 11, 57–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishak CA et al. An RB-EZH2 Complex Mediates Silencing of Repetitive DNA Sequences. Mol. Cell 64, 1074–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chile T et al. HOXB7 mRNA is overexpressed in pancreatic ductal adenocarcinomas and its knockdown induces cell cycle arrest and apoptosis. BMC Cancer 13, 451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent LN et al. Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. Journal of Clinical Investigation 127, 830–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt AW et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. JAMA Oncology 2, 1598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin SM Deciphering the retinoblastoma protein phosphorylation code. Trends in Biochemical Sciences 38, 12–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyson NJ RB1 : a prototype tumor suppressor and an enigma. Genes & Development 30, 1492–1502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltran H et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature Medicine 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macleod KF The RB tumor suppressor: a gatekeeper to hormone independence in prostate cancer? Journal of Clinical Investigation 120, 4179–4182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldhoff P et al. Clinical Stratification of Glioblastoma Based on Alterations in Retinoblastoma Tumor Suppressor Protein (RB1) and Association With the Proneural Subtype. Journal of Neuropathology & Experimental Neurology 71, 83–89 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thangavel C et al. The Retinoblastoma Tumor Suppressor Modulates DNA Repair and Radioresponsiveness. Clinical Cancer Research 20, 5468–5482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook R et al. Direct Involvement of Retinoblastoma Family Proteins in DNA Repair by Non-homologous End-Joining. Cell Reports 10, 2006–2018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang PH, Cook R & Mittnacht S RB in DNA repair. Oncotarget 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.