Abstract
The base and nucleotide excision repair (BER and NER, respectively) pathways are two major mechanisms that remove DNA lesions formed by the reactions of genotoxic intermediates with cellular DNA. We have demonstrated earlier that the oxidatively generated guanine lesions spiroiminodihydantoin (Sp) and 5-guanidinohydantoin (Gh) are excised from double-stranded DNA by competing BER and NER in whole cell extracts [Shafirovich, V., et al. (2016) J. Biol. Chem. 321, 5309-5319]. In this work we compared the NER and BER yields with single Gh and Sp lesions embedded at the same sites in covalently closed circular pUC19NN plasmid DNA (cccDNA), and in the same, but linearized form (linDNA) of this plasmid. The kinetics of the Sp and Gh BER and NER incisions were monitored in HeLa cell extracts. The yield of NER products is ~ 5 times greater in covalently closed circular DNA than in the linearized form, while the BER yield is smaller by ~ 20-30% depending on the guanine lesion. Control BER experiments with 8-oxo-7,8-dihydroguanine (8-oxoG) show that the BER yield is increased by a factor of only 1.4 ± 0.2 in cccDNA relative to linDNA. These surprising differences in BER and NER activities are discussed in terms of the lack of termini in covalently closed circular DNA, and the DNA lesion search dynamics of the NER DNA damage sensor XPC-RAD23B and the BER enzyme OGG1 that recognises and excises 8-oxoG.
Graphical Abstract
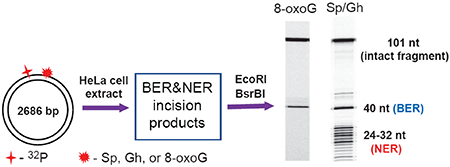
INTRODUCTION
It is well established that exposure of cellular DNA to endogenous reactive oxygen species, free radicals, metabolically activated reactive intermediates, UV light or ionizing radiation gives rise to a variety of DNA lesions.1–3 If not repaired, error-prone replication of such lesions can lead to mutations and genomic instability.4, 5 Indeed, the repair of oxidatively generated lesions is critical for maintaining genomic stability during oxidative stress.6 Existing paradigms suggest that non-bulky oxidatively generated DNA lesions are removed by base excision repair (BER) mechanisms,7 that specifically excises the damaged base, and is highly conserved from bacteria to humans.8 However, DNA helix-distorting bulky lesions are repaired by the nucleotide excision repair (NER) pathway9 that is initiated by the DNA-damage-sensing NER factor XPC-RAD23B (abbreviated as XPC).10 This initial lesion-recognition step is followed by the sequential binding of the ten-protein factor TFIIH to the XPC-DNA complex and the NER factors XPA, RPA, XPF and XPG.11 The endonucleases XPF and XPG incise the damaged strand on both sides of the lesion that generates the characteristic NER ladder of oligonucleotide excision products ~ 24 – 30 nucleotides (nt) in lengths.9–11
The oxidatively generated hydantoin DNA lesions 5-guanidinohydantoin (Gh) and diastereomeric spiroiminodihydantoin (S-Sp and R-Sp) are substrates of prokaryotic BER and NER mechanisms,12 in human cell extracts13, 14 and in human cells.15 Both hydantoin lesions are generated by the oxidation of 8-oxo-7,8-dihydroguanine (8-oxoG), itself a ubiquitous DNA lesion produced in cellular environments under conditions of oxidative stress.16
More recently, we demonstrated that the NER excision of a single bulky benzo[a]pyrene-derived DNA lesion/DNA molecule was excised with a ~ six-fold greater efficiency when it was embedded in covalently closed circular pUC19NN plasmid (contour length 2686 base pairs) than in the same but linearized form of the same plasmid.17 The first NER factor that initiates the multi-step NER process is the DNA damage sensor XPC-RAD23B that searches DNA lesions by a constrained one-dimensional hopping mechanism.18 The remarkable enhanced NER efficiency in circular DNA was attributed to the absence of blunt ends in the circular DNA plasmids. The simplest hypothesis is that in linear DNA at or near the termini, XPC molecules can dissociate from the DNA molecules (by hopping or sliding off) and have a lower probability of re-associating with DNA than in the regions more distant from the termini. Such sliding-off phenomena have been observed in the case of the p53 transcription factor.19
In this work, we compared the excision of the hydantoin lesions Gh and Sp by competing BER and NER pathways with the lesions embedded either in circular or in the linear forms of the pUC19NN plasmid with a contour length of 2686 base pairs. We have previously shown that BER and NER pathways occur in ratios of ~ NER/BER ~ 0.5 – 2.0 depending on the cell extract preparation and, presumably, the relative concentrations of relevant BER and NER proteins that initiate these repair pathways when the hydantoin lesions are embedded in short 147-mer duplexes 14. Here we elucidated how the excision of the hydantoin lesions by the two competing NER and BER pathways are affected by the topology of DNA molecules. We demonstrate that in the case of the non-bulky Sp and Gh DNA lesions, the efficiencies of the NER mechanisms are significantly enhanced in covalently closed circular DNA plasmids relative to the same but linearized DNA, while minor differences are observed in the case of base excision repair of the same lesions.
EXPERIMENTAL PROCEDURES
DNA Repair Substrates.
We designed covalently closed circular plasmids containing single Gh or S-Sp lesions at the same site harboring site-specifically guanine lesions (Figure 1A) that exclude the effects of DNA termini on DNA repair.
Figure 1.
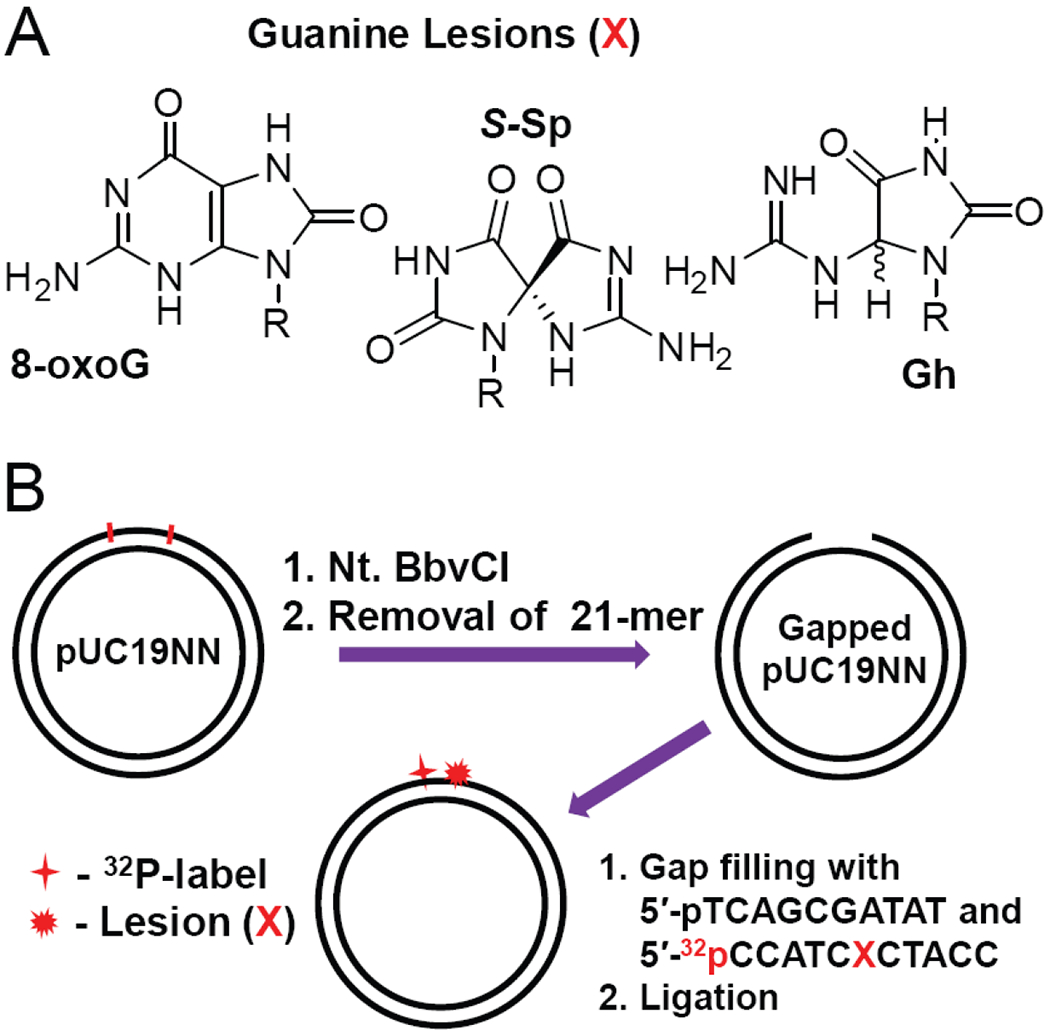
Covalently closed circular plasmid substrates (2686 bp) harboring site-specifically positioned guanine lesions (X = 8-oxoG, Gh and S-Sp); the substrates were 32P-internally labeled (*p)]. The preparation of the circular plasmid substrates were generated by a gapped-vector technology20 employing pUC19NN plasmids containing two Nt. BbvCI restriction sites.
A gapped-vector technology20 was used to generate the circular plasmid substrates from pUC19NN plasmids as shown in Figure 1B. The latter was cloned from the well-known pUC19 plasmids by inserting a fragment containing two Nt. BbvCI restriction sites.17 The gap created by tandem nicking with the Nt. BbvCI restriction enzyme was filled with 32P-labeled oligonucleotides containing site-specifically positioned Sp- or Gh-lesions, or by 8-oxoG, a classical substrate of BER proteins that was employed as a positive control of BER activity.21 The kinetics of the BER and NER incisions of these plasmid substrates were monitored in HeLa cell extracts.
Materials.
The pUC19 plasmid and restriction enzymes were obtained from New England Biolabs (Ipswich, MA). The T4-kinase, T4 ligase and proteinase K were purchased from ThermoFisher Scientific; E. coli DNA gyrase was from Millipore Sigma. The 2′-oligodeoxynucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA) and purified by denaturing polyacrylamide gel electrophoresis.
Construction of Plasmid Substrates Containing the Sp and Gh Lesions.
The pUC19NN plasmid (Figure 1B) was cloned from the pUC19 plasmid by inserting a 32-mer oligonucleotide fragment between the BamHI and HindIII restriction sites in the multiple cloning site of pUC19.17 Aliquots (50 μL) of pUC19NN (5 pmol) were treated with Nt. BbvCI restriction enzyme (50 U) at 37 °C for 1.5 h. The double-nicked pUC19NN was mixed with an excess of the complementary strand 5′-GGTAGCGATGGATATCGCTGA (500 pmol), heated to 80 °C for 5 min and cooled on ice for 2 min (this cycle was repeated 3 times). The gapped plasmid was purified by diafiltration on Amicon Ultra-0.5 spin column (100 kDa cut-off), annealed to the 5′-phosphorylated oligonucleotides 5′-pTCAGCGATAT and 32P-endlabeled 5′-32pCCATCXCTACC, the latter containing the lesion (X = Gh, S-Sp, or 8-oxoG), and finally ligated by T4 ligase at 16 °C for 16 h. The reaction products were treated with T5 endonuclease to digest linear and nicked plasmids.22 The covalently closed circular plasmids were purified by diafiltration on Amicon Ultra-0.5 spin columns (100 kDa cut-off). The nicks in circular plasmids were sealed by ligase thus generating covalently closed plasmids in their covalently closed, relaxed forms.23, 24 The supercoiled forms of the circular plasmids were generated by treatment of the relaxed plasmids with E. coli DNA gyrase using the manufacturer’s suggested protocol. The linear plasmid substrates were prepared by treatment of the covalently closed circular plasmids with ScaI restriction enzyme. The integrity of the plasmid substrates was confirmed by 1% agarose gel electrophoresis using both 32P and fluorescence (Ethidium Bromide) detection.
DNA Repair Assays in HeLa Cell Extracts.
The BER and NER-competent whole-cell extracts were prepared from HeLaS3 cells (Cell Culture Company, Minneapolis, MN) as described in detail elsewhere.25, 26 The extracts (protein concentration: 12 – 14 mg/mL by Bradford assay) in the dialysis buffer (25 mM HEPES, pH 7.9, 70 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 12% glycerol) were aliquoted, frozen in liquid nitrogen and stored at – 80 °C. The DNA repair reactions were performed in the reaction solution (total volume 100 μL) containing 25 mM HEPES, pH 7.9, 70 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 4 mM ATP, 200 μg/μL bovine serum albumin, 4% glycerol, 20 fmol of 32P-labeled DNA substrate and 30 μL HeLa cell extract. After incubation at 30 °C for fixed period of time the reactions were stopped by addition of 40 μg proteinase K in 0.3% SDS and incubated for 30 min at 37 °C. After phenol/chloroform extraction, the DNA was ethanol precipitated, treated with EcoRI and BsrBI restriction enzymes, and subjected to denaturing 12% polyacrylamide gel electrophoresis.
Analysis of Gel autoradiographs.
The polyacrylamide gels were dried, exposed to a Molecular Dynamics Storage Phosphor Screen, and then scanned with a Typhoon FLA 9000 laser scanner. The gel autoradiographs thus obtained were analysed using the Total Lab TL120 1D software.
RESULTS AND DISCUSSION
Interplay between BER and NER pathways in covalently closed circular plasmids.
The plasmid substrates harboring guanine lesions were generated from the pUC19NN plasmid by a gapped-vector technology.20 The single-stranded gap was created by removing the 21-mer fragment generated by tandem nicking with the Nt. BbvCI restriction enzyme, which cleaves only one strand in the plasmid (Figure 1B). The resulting gap was filled with the 10-mer 5′-pTCAGCGATAT and the 32P-labeled 11-mer 5′-32pCCATCXCTACC that contains the guanine lesions X = Gh, S-Sp, and 8-oxoG. The Gh/Sp lesions are efficient substrates of both BER and NER pathways,13, 27 whereas 8-oxoG is a well-known substrate of BER.21 After incubation with HeLa cell extracts, the DNA samples were isolated from the reaction mixture and treated with the EcoRI and BsrBI restriction enzymes to excise 32P-labeled 40-mer fragments, the products of successful BER activity, or the 101-mer intact fragments in the absence of BER and NER activities (Figure 2).
Figure 2.
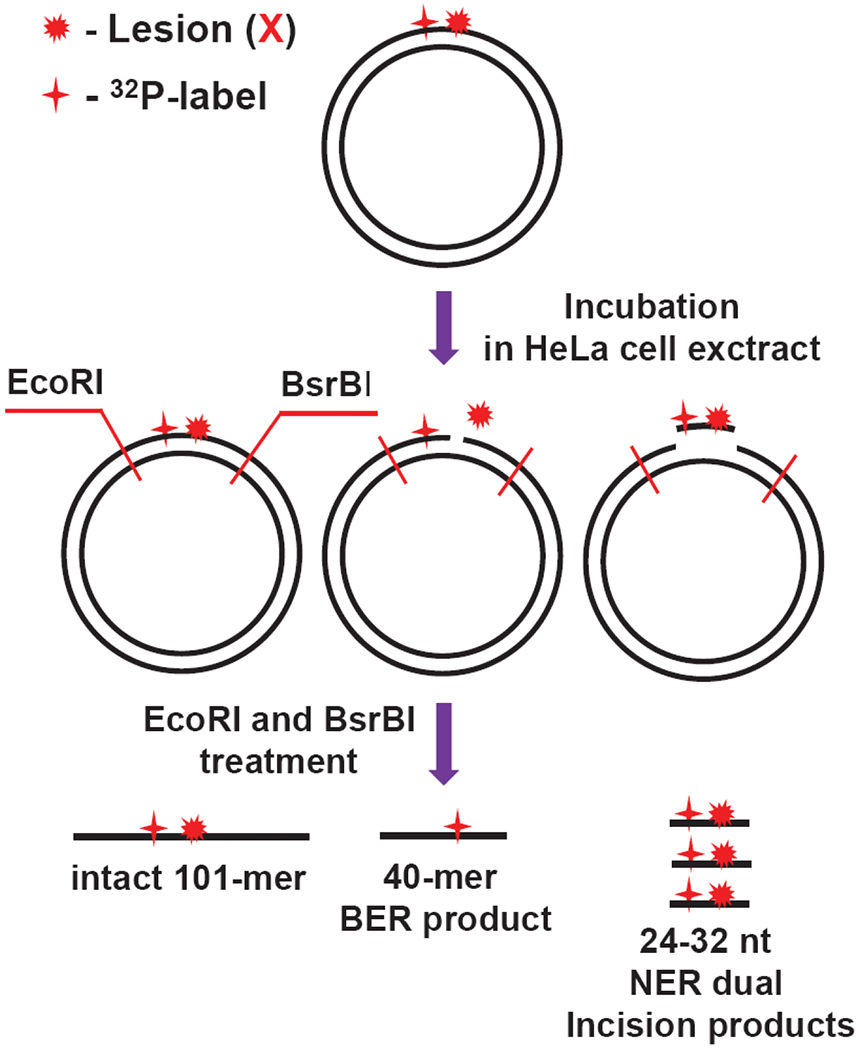
Schematic summary of the analysis of products derived from the incisions of covalently closed circular plasmids by BER and NER mechanisms after incubation in HeLa cell extracts.
In the case of the NER, treatment of DNA with EcoRI and BsrBI enzymes is not required, because successful NER activity generates the dual incision products ~24 – 32 nucleotides in lengths containing the 32P-label and the lesion.28, 29
Typical results of DNA repair assays in HeLa cell extracts obtained with the different lesions S-Sp, Gh and 8-oxoG embedded at the same site in the same plasmid at identical 0.2 nM concentrations are shown in Figure 3.
Figure 3.
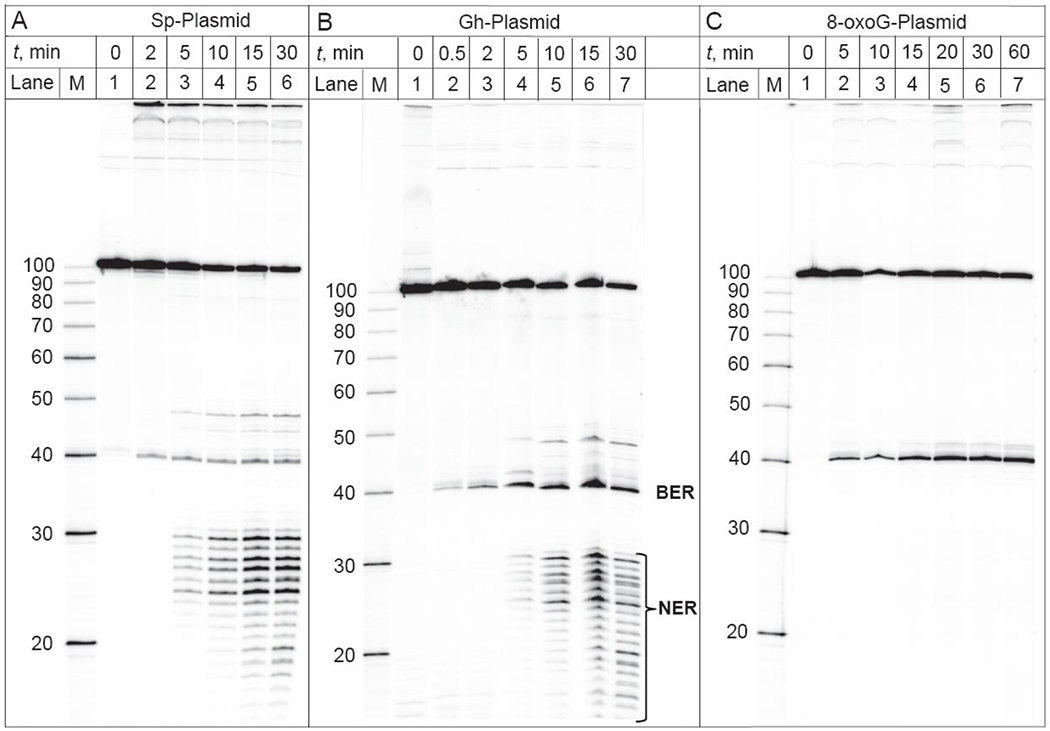
BER and NER incisions of the 32P-internally labeled covalently closed circular plasmids (0.2 nM) harboring S-Sp, Gh or 8-oxoG lesions as a function of incubation time in the same HeLa cell extract. Lanes M: oligonucleotide size markers.
The 40-mer and 101-mer oligonucleotide fragments are observable in each case, while the 40-mers are observed only if BER activity has occurred. The NER activity yields the usual ladders of dual incision products ~ 24 – 32 nt in lengths30, 31 in the case of Sp- and Gh-modified plasmids (Figures 3A and 3B). In the case of incubation times longer than ~ 15 min, the degradation of these dual incision products by nonspecific exonucleases32 becomes apparent since oligonucleotide sequences less than 24 nt in lengths become apparent (lanes 5, 6 and 6, 7 in Figures 3A and 3B, respectively). The 8-oxoG plasmid is exclusively repaired by the BER pathways and the NER dual incision products are absent in the case of this substrate (Figure 3C).
In addition to the well-characterized bands due to the major BER incision product (40 nt) and the NER dual incision products (the 24 – 32 nt ladder), additional weak bands between 40 and 50 nt are formed in a time-dependent manner (Figure 3). We found that the yields of these products are typically less than 10% of the BER and NER excision products. The bands that appear to migrate with mobilities close to those of the BER incision product (40 nt) are observable in all cases (Sp, Gh and 8-oxoG substrates). The BER bands can be attributed to the products of glycosylase/AP lyase activities that can result in three different 3’-ends: a 3′-phosphate group (β,δ-elimination), an α,β-unsaturated aldehyde (β-elimination), and a 3′-OH group derived from removal of 3′-phosphate/aldehyde groups.8 The mobilities of these BER products follow the following trend: 3’-phosphate > 3’-OH > 3’-α,β-unsaturated aldehyde.33 The slower migrating bands (near 48 nt) are clearly observed in the case of the NER substrates (Sp and Gh) only (Figure 3) and can be assigned to the fragments derived from the 3′-uncoupled NER incisions.34, 35 For instance, Evans et al. studied the repair of 1,3-intrastrand d(GpTpG)-cisplatin crosslinked lesions in closed circular plasmids;35 they observed 3′-uncoupled incisions at the 8th and 9th phosphodiester bonds 3′ to the lesion site, together with the usual NER dual incisions (the 24 – 32 nt ladder). Furthermore, the yields of NER products generated by the 3′-uncoupled incisions were much lower than the yields of the dual NER incisions, as in our experiments.
The BER and NER incision kinetics from the covalently closed circular plasmids harboring S-Sp, Gh and 8-oxoG lesions are depicted in Figure 4.
Figure 4.
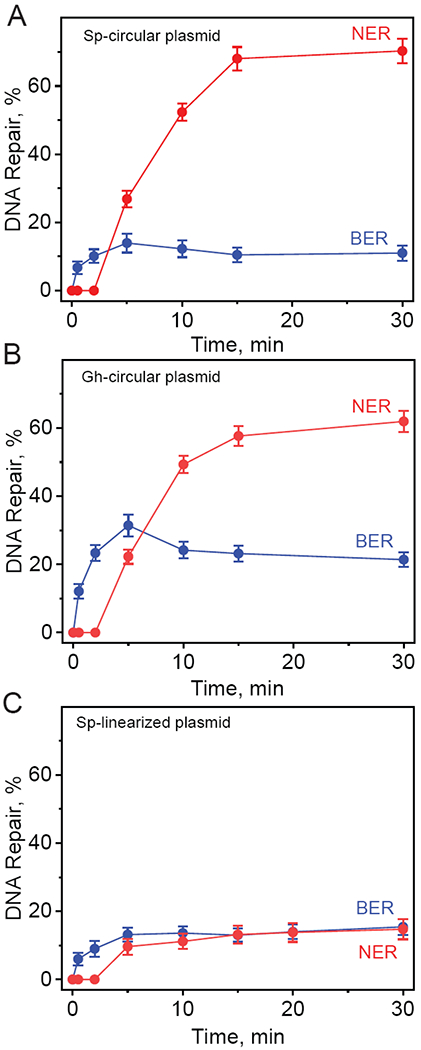
The kinetics of appearance of BER and NER incision products derived from the 32P-internally labeled covalently closed circular plasmids containing S-Sp (A), and Gh (B) lesions and linearized plasmid containing S-Sp (C) lesions as a function of incubation time in the same HeLa cell extract. The results of three independent experiments and their standard deviations are shown.
In the case of the Sp and Gh plasmid substrates the detectable levels of the NER dual incision products appear after the end of the incubation period (~2 min), and the reaction yields increase as a function of time and reach a maximum level of about 62 – 70% within 15 – 30 min after the start of the reaction (Figures 4A and 4B). By contrast, the BER incision products appear immediately after the start of the reaction and the BER yields approach maximum values of ~15% (Sp) and ~31% (Gh) ~ 5 min after mixing the reactants. The NER yields in covalently closed circular DNA are enhanced by a remarkable and astonishing factor of ~5 relative to the NER yields in the linearized form of the same plasmid (Figures 4A and 4C). By contrast, the BER yields of Sp remain practically unchanged.
A control experiment for BER activity with 8-oxoG, which is known to be efficiently excised by the glycosylase OGG1 (but is not known as a substrate of NER), is shown in Figure 5.
Figure 5.
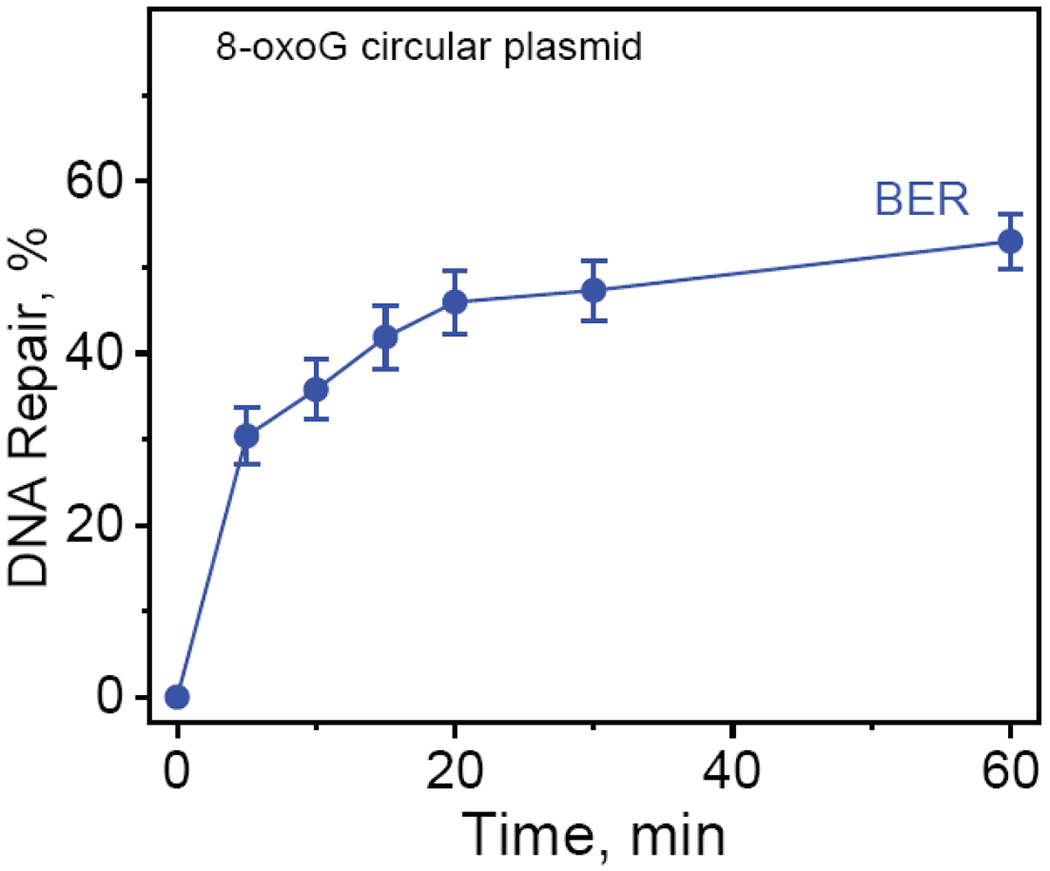
The kinetics of appearance of BER incision products derived from the 32P-internally labeled covalently closed circular plasmids containing 8-oxoG lesions as a function of incubation time in HeLa cell extract. The results of three independent experiments and their standard deviations are shown.
The yield of 8-oxoG BER products increases as a function of time and reaches a maximum level of ~53% after 30 – 60 min, showing that normal BER activity exhibited by a known BER substrate, is not inhibited in cccDNA. In the case of the Sp and Gh substrates, the combined yields of NER + BER excision products approaches ~ 80% (Figures 4A and 4B), and the low yield of BER products can be attributed to the efficient competition by NER proteins for binding to Sp or Gh.
NER/BER Incision Ratios.
In principle, the ratios of incision products generated at a given concentration of DNA substrates could be limited by the relative concentrations of one or the other kind of DNA repair protein. Such effects could manifest themselves at different DNA substrate concentrations at constant BER and NER protein concentrations. A five-fold rise of DNA concentrations from 0.2 nM to 1 nM induces a decrease in the NER/BER yield ratios from ~4.3 to ~2.7 and ~2.0 to ~1.2 in the case of Sp- and Gh-plasmids, respectively (Figure S1, Supporting Information). These results indicate that the observed small decreases in NER/BER ratios as a function of DNA substrate concentration, are due to limiting concentrations of NER proteins in cell extracts.
Impact of plasmid linearization on the interplay between BER and NER pathways.
In these experiments, we compared the relative BER and NER incision efficiencies of the site-specifically inserted hydantoin lesions in a covalently closed plasmid and the same, but linearized plasmid with blunt ends. The latter was prepared by the selective cleavage of the parent circular plasmid with a unique ScaI restriction enzyme that generates the linearized plasmid substrate with the lesion positioned at the 945th nucleotide counted from the 5′-end (Figure 6A).
Figure 6.
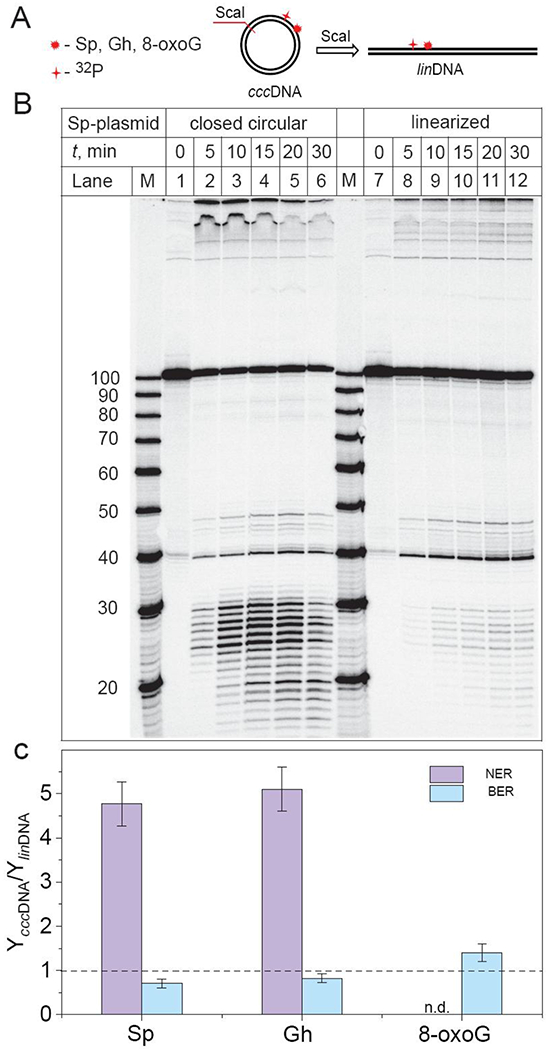
Impact of plasmid linearization on the ratios of the relative yields of BER and NER incision products (YcccDNA/YlinDNA) after incubation of 32P-internally labeled plasmids (0.2 nM) harboring S-Sp, Gh, and 8-oxoG lesions in HeLa cell extract. (A) Schematic representations of linearized plasmids (linDNA) obtained by the treatment of covalently closed circular plasmids (cccDNA) with ScaI restriction enzyme. (B) BER and NER incisions of the 32P-internally labelled covalently closed circular and linearized plasmids containing the same molar concentrations of DNA molecules bearing single S-Sp lesions (0.2 nM), as a function of incubation time in identical HeLa cell extracts (lanes labelled 0, 5, 10, 15, 20, 30 min). Lanes M: oligonucleotide size markers. (C) The ratios of the relative BER and NER yields were calculated by integration of the histograms (at 15 – 30 min) derived from the denaturing gel autoradiographs (Panel B and Figures S2 and S3 in Supporting Information). The designation n.d. above 8-oxoG, signifies that no detectable NER products were observable. The results of three independent experiments and their standard deviations (P < 0.05) are shown. The P values were calculated by using a paired t-test.
The DNA repair assays in HeLa cell extracts obtained with these substrates clearly demonstrate a significant suppression of NER dual incisions in the linearized plasmid in comparison with covalently closed circular plasmid (Figure 6B and Figure S2 in Supporting Information). The ratios of the relative yields of BER and NER incision products (YcccDNA/YlinDNA) in covalently closed circular plasmids and linearized plasmids are summarized in Figure 6C. This figure shows that the ratios of the relative yields of the NER dual incision products are 4.8±0.5 (Sp) and 5.1±0.5 (Gh) indicating that the NER activities of both hydantoin lesions in circular plasmids are greater than in linearized plasmids. By contrast, the BER activities of the hydantoin lesions in circular plasmids are slightly smaller than in linearized plasmids, because the ratios of the BER yields are in the range of 0.7 – 0.8 for both lesions. As expected, only BER activity is observed in the case of 8-oxoG lesions embedded in either cccDNA or linDNA (Figure S3, Supporting Information). A somewhat greater BER activity by a factor of up to 1.4±0.2 in cccDNA is observed (Figure 6C). By contrast, smaller BER yields of the Sp and Gh-lesions are observed in circular than in linear DNA (Figure 6C). This effect is attributed to the competition between XPC and BER enzyme for binding to the Sp or Gh lesions,14 thus leading to a smaller BER yield since the NER yield is highly favored in the circular DNA molecules. A likely BER protein that incises the Sp and Gh lesions is NEIL1 (molecular weight 43.7 kDa).36
The greater NER yields in circular than in linearized DNA suggests that the NER-initiating factor XPC-RAD23B has a greater affinity of recognizing NER substrates in circular plasmid DNA than in linear DNA of the same contour lengths (2686 base pairs). This observation can be understood if it is assumed that the termini of the linearized DNA molecules act as partial sinks of DNA-bound XPC molecules because of greater dissociation rates at the ends than within the interior of linearized DNA molecules.17 This hypothesis is supported by the observations of Mason et al.37 who demonstrated that NER excision efficiencies are significantly enhanced in human cell extracts when an NER substrate is embedded in a 149-mer linear duplex with the ends blocked by streptavidin-biotin complexes that prevented XPC molecules from sliding off the ends of DNA molecules.
Interplay of BER and NER pathways in supercoiled and relaxed plasmid substrates.
The sealing of a nicked strand in circular double-stranded plasmids by ligases, generates a relaxed form of plasmid DNA (rxDNA).23 Negative supercoiling of plasmids and chromosomal DNA is catalysed by gyrases in an ATP-dependent manner.38 In this work we compared the relative BER and NER incision efficiencies of site-specifically inserted guanine lesions in the relaxed and in the supercoiled form (scDNA) of covalently closed plasmids. The latter were prepared by supercoiling rxDNA with E. coli DNA gyrase.24 DNA repair assays in HeLa cell extracts clearly demonstrate that the yields of BER and NER incision yields in scDNA and rxDNA are similar (within ±10%) in scDNA and rxDNA (Figure S4, Supporting Information). These results indicate that the supercoiling does not significantly affect the competitive incisions between the NER and BER pathways, which determine the relative yields of NER and BER incision products.
How DNA repair proteins search and identify DNA lesions in a sea of unmodified DNA has long been a subject of significant interest. The current most often discussed model is facilitated diffusion,39, 40 a blend of mechanisms that include the random one-dimensional diffusional sliding of the protein along the double-stranded DNA molecule, that can include dissociation and re-association hopping events between nearby, or more distant sites, and intersegmental hopping between nearby looped segments of the same DNA molecule. Valuable experimental insights into the mechanisms have been obtained from single molecule experiments that examined the diffusion of DNA repair proteins hOGG1,41 XPC18 and the yeast analog of XPC, RaD4.42 The results are consistent with an overall constrained motion of the repair proteins along stretched linear DNA molecules. Cheon et al.18 have shown that the diffusion coefficient of XPC in constrained regions of double-stranded DNA containing cyclobutene pyrimidine dimer UV photolesions is DBP ≈ 0.010 ± 0.020 μm2>/s at a 150 mM NaCl concentration. The diffusion coefficient of the smaller glycosylase OGG1 is DOGG = 0.02 μm2/s.41 This difference is reasonable since the molar mass of XPC-RAD23B (151.1 kDa)43 is about 4 times greater than the mass of hOGG1 (38.8 kDa).44 While these differences suggest that the search mechanism might be somewhat faster in the case of OGG1, the efficiencies of XPC capture per collision with either lesion, nor the size of the constrained region,18 are known for either lesion studied here. Another mechanism that could explain the unusually large NER efficiencies of removal of the bulky and non-bulky NER substrates is the hopping of XPC molecules between different DNA segments in supercoiled cccDNA that will need to be investigated.
CONCLUSIONS
The efficiencies of excision of two hydantoin DNA lesions that are substrates of the two major repair pathways, NER and BER, have been compared in covalently closed circular DNA and in the linearized form of the same plasmid DNA. The high NER yields observed in the case of covalently closed circular Sp/Gh plasmids (62 – 70%) suggest an important role of the free DNA termini that partially suppress NER activity in linear DNA duplexes. In the case of covalently closed circular plasmids the NER dual incision are more efficient than in linearized plasmids by factor of ~5, a phenomenon that has also been observed using a bulky, benzo[a]pyrene-derived NER substrate that is not repaired by the BER pathway.17 By contrast, the BER incisions of the Sp and Gh lesions is less efficient by a factor of ~ 1.3 in the circular DNA plasmid molecules than in their linearized form. In contrast to the differences in NER yields, the BER yields of Sp, Gh, and 8-oxoG are similar in covalently closed circular and linearized DNA.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institute of Environmental Health Sciences grant R01 ES-027059 to V.S. and R21 ES-028546 to N. E. G.
ABBREVIATIONS
- BER
base excision repair
- NER
nucleotide excision repair
- 8-oxoG
8-oxo-7,8-dihydroguanine
- Gh
5-guanidinohydantoin
- Sp
spiroiminodihydantoin
- cccDNA
covalently closed circular plasmid
- linDNA
linearized plasmid
- scDNA
supercoiled plasmid
- rxDNA
relaxed plasmid
- nt
nucleotide
- bp
base pair
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/.
Ratios of the yields of BER and NER incision products from the closed circular plasmids harboring S-Sp, and Gh lesions as a function of the substrate concentration in the HeLa cell extract (Figure S1), BER and NER incisions of the closed circular and linearized plasmids containing 0.2 nM DNA molecules bearing single Gh (Figure S2), and 8-oxoG (Figure S3) lesions as a function of incubation time in the HeLa cell extracts, the yields of BER and NER incision products of single S-Sp, Gh and 8-oxoG lesions in supercoiled and relaxed closed circular plasmids incubated in HeLa cell extracts (Figure S4)(PDF).
The authors declare no competing financial interest.
REFERENCES
- (1).Loeb LA, and Harris CC (2008) Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 68, 6863–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lonkar P, and Dedon PC (2011) Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer 128, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Basu AK (2018) DNA damage, mutagenesis and cancer. Int. J. Mol. Sci 19, 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ferguson LR, Chen H, Collins AR, Connell M, Damia G, Dasgupta S, Malhotra M, Meeker AK, Amedei A, Amin A, Ashraf SS, Aquilano K, Azmi AS, Bhakta D, Bilsland A, Boosani CS, Chen S, Ciriolo MR, Fujii H, Guha G, Halicka D, Helferich WG, Keith WN, Mohammed SI, Niccolai E, Yang X, Honoki K, Parslow VR, Prakash S, Rezazadeh S, Shackelford RE, Sidransky D, Tran PT, Yang ES, and Maxwell CA (2015) Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin. Cancer Biol. 35 Suppl, S5–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tubbs A, and Nussenzweig A (2017) Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 168, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Scott TL, Rangaswamy S, Wicker CA, and Izumi T (2014) Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair. Antioxid Redox Signal 20, 708–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, and Bohr VA (2009) Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 30, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wallace SS, Murphy DL, and Sweasy JB (2012) Base excision repair and cancer. Cancer Lett. 327, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Marteijn JA, Lans H, Vermeulen W, and Hoeijmakers JH (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell. Biol. 15, 465–481. [DOI] [PubMed] [Google Scholar]
- (10).Kusakabe M, Onishi Y, Tada H, Kurihara F, Kusao K, Furukawa M, Iwai S, Yokoi M, Sakai W, and Sugasawa K (2019) Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 41, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tsutakawa SE, Tsai CL, C Y, Bralić A, Chazin WJ, Hamdan SM, Schärer OD, Ivanov I, and Tainer JA (2020) Envisioning how the prototypic molecular machine TFIIH functions in transcription initiation and DNA repair. DNA Repair (Amst) 96, 102972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, and David SS (2013) Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. Journal of the American Chemical Society 135, 13851–13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Shafirovich V, Kropachev K, Anderson T, Liu Z, Kolbanovskiy M, Martin BD, Sugden K, Shim Y, Chen X, Min JH, and Geacintov NE (2016) Base and nucleotide excision repair of oxidatively generated guanine lesions in DNA. J. Biol. Chem. 291, 5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kolbanovskiy M, Shim Y, Min JH, Geacintov NE, and Shafirovich V (2020) Inhibition of Excision of Oxidatively Generated Hydantoin DNA Lesions by NEIL1 by the Competitive Binding of the Nucleotide Excision Repair Factor XPC-RAD23B. Biochemistry 59, 1728–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shafirovich V, Kropachev K, Kolbanovskiy M, and Geacintov NE (2019) Excision of oxidatively generated guanine lesions by competing base and nucleotide excision repair mechanisms in human cells. Chem. Res. Tox. 32, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Markkanen E (2017) Not breathing is not an option: How to deal with oxidative DNA damage. DNA Repair (Amst) 59, 82–105. [DOI] [PubMed] [Google Scholar]
- (17).Kolbanovskiy M, Aharonoff A, Sales AH, Geacintov NE, and Shafirovich V (2020) Remarkable enhancement of nucleotide excision repair of a bulky guanine lesion in a covalently closed circular DNA plasmid relative to the same, but linearized plasmid Biochemistry 59, 2842−2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cheon NY, Kim HS, Yeo JE, Scharer OD, and Lee JY (2019) Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 47, 8337–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).McKinney K, Mattia M, Gottifredi V, and Prives C (2004) p53 linear diffusion along DNA requires its C terminus. Mol. Cell. 16, 413–424. [DOI] [PubMed] [Google Scholar]
- (20).Wang H, and Hays JB (2001) Simple and rapid preparation of gapped plasmid DNA for incorporation of oligomers containing specific DNA lesions. Molecular Biotechnology 19, 133–140. [DOI] [PubMed] [Google Scholar]
- (21).David SS, O’Shea VL, and Kundu S (2007) Base-excision repair of oxidative DNA damage. Nature 447, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shivji MK, Moggs JG, Kuraoka I, and Wood RD (2006) Assaying for the dual incisions of nucleotide excision repair using DNA with a lesion at a specific site. Method. Mol. Biol. 314, 435–456. [DOI] [PubMed] [Google Scholar]
- (23).Depew DE, and Wang JC (1975) Conformational fluctuations of DNA helix. Proc. Natl. Acad. Sc.i U. S. A. 72, 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gu M, Berrido A, Gonzalez WG, Miksovska J, Chambers JW, and Leng F (2016) Fluorescently labeled circular DNA molecules for DNA topology and topoisomerases. Sci. Rep. 6, 36006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Smeaton MB, Miller PS, Ketner G, and Hanakahi LA (2007) Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 35, e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kang TH, Reardon JT, Kemp M, and Sancar A (2009) Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. U. S. A. 106, 2864–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shafirovich V, Kolbanovskiy M, Kropachev K, Liu Z, Cai Y, Terzidis MA, Masi A, Chatgilialoglu C, Amin S, Dadali A, Broyde S, and Geacintov NE (2019) Nucleotide excision repair and impact of site-specific 5′,8-cyclopurine and bulky DNA lesions on the physical properties of nucleosomes. Biochemistry 58, 561–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Huang JC, Hsu DS, Kazantsev A, and Sancar A (1994) Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. U. S. A. 91, 12213–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gillet LC, and Scharer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106, 253–276. [DOI] [PubMed] [Google Scholar]
- (30).Huang JC, Svoboda DL, Reardon JT, and Sancar A (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5’ and the 6th phosphodiester bond 3’ to the photodimer. Proc. Natl. Acad. Sci. U. S. A. 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Huang JC, and Sancar A (1994) Determination of minimum substrate size for human excinuclease. J. Biol. Chem. 269, 19034–19040. [PubMed] [Google Scholar]
- (32).Kemp MG, Reardon JT, Lindsey-Boltz LA, and Sancar A (2012) Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J. Biol. Chem. 287, 22889–22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hatahet Z, Kow YW, Purmal AA, Cunningham RP, and Wallace SS (1994) New substrates for old enzymes. 5-Hydroxy-2’-deoxycytidine and 5-hydroxy-2’-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2’-deoxyuridine is a substrate for uracil DNA N-glycosylase. J. Biol. Chem. 269, 18814–18820. [PubMed] [Google Scholar]
- (34).Matsunaga T, Mu D, Park CH, Reardon JT, and Sancar A (1995) Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem. 270, 20862–20869. [DOI] [PubMed] [Google Scholar]
- (35).Evans E, Moggs JG, Hwang JR, Egly JM, and Wood RD (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. The EMBO journal 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, and Mitra S (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mason TM, Smeaton MB, Cheung JC, Hanakahi LA, and Miller PS (2008) End modification of a linear DNA duplex enhances NER-mediated excision of an internal Pt(II)-lesion. Bioconjug. Chem. 19, 1064–1070. [DOI] [PubMed] [Google Scholar]
- (38).Reece RJ, and Maxwell A (1991) DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26, 335–375. [DOI] [PubMed] [Google Scholar]
- (39).Lomholt MA, van den Broek B, Kalisch SM, Wuite GJ, and Metzler R (2009) Facilitated diffusion with DNA coiling. Proc. Natl. Acad. Sci. U. S. A. 106, 8204–8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gorman J, and Greene EC (2008) Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol. 15, 768–774. [DOI] [PubMed] [Google Scholar]
- (41).Blainey PC, van Oijen AM, Banerjee A, Verdine GL, and Xie XS (2006) A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U. S. A. 103, 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kong M, Liu L, Chen X, Driscoll KI, Mao P, Bohm S, Kad NM, Watkins SC, Bernstein KA, Wyrick JJ, Min JH, and Van Houten B (2016) Single-Molecule Imaging Reveals that Rad4 Employs a Dynamic DNA Damage Recognition Process. Mol. Cell. 64, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lee YC, Cai Y, Mu H, Broyde S, Amin S, Chen X, Min JH, and Geacintov NE (2014) The relationships between XPC binding to conformationally diverse DNA adducts and their excision by the human NER system: Is there a correlation? DNA Repair (Amst) 19, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Radicella JP, Dherin C, Desmaze C, Fox MS, and Boiteux S (1997) Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94, 8010–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


