Abstract
The angiotensin-converting enzyme 2 (ACE2) receptor has been proved for SARS-CoV-2 cell entry after auxiliary cellular protease priming by transmembrane protease serine 2 (TMPRSS2), but the co-effect of this molecular mechanism was unknown. Here, single-cell sequencing was performed with human conjunctiva and the results have shown that ACE2 and TMPRSS2 were highly co-expressed in the goblet cells with genes involved in immunity process. This identification of conjunctival cell types which are permissive to virus entry would help to understand the process by which SARS-CoV-2 infection was established. These finding might be suggestive for COVID-19 control and protection.
Keywords: SARS‐CoV‐2, Conjunctival goblet cell, ACE2, TMPRSS2
The emerging coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has pushed several countries into public health emergency all over the world (Al-Qahtani, 2020). COVID-19 patients demonstrate symptoms of fever, dry cough, dyspnea, and fatigue that can develop to pneumonia (Cascella et al., 2020; Pal et al., 2020). SARS-CoV-2 is infectious and it is transmitted mainly by inhaling aerosols or droplets released by the infected ones, appearing to be more transmissible than either severe acute respiratory syndrome (SARS) or middle east respiratory syndrome (MERS) (Hui, 2017; Koh et al., 2020). It was well known SARS was primarily transmitted through contact with mucous membranes in the eyes, mouth, or nose (Hui and Zumla, 2019). But the modes of transmission of SARS-CoV-2 have been key knowledge gaps (Luo et al., 2020). Since the outbreak of SARS-CoV-2, clinicians were speculating that the disease could also be transmitted by the conjunctiva, underlying the need for further investigations on its potential transmission pathways (“Beijing Daily of a doctor Guangfa Wang's SARS CoV-2 infection. http://www.bjnews.com.cn/news/2020/01/23/678189.html,” n.d.; Lu et al., 2020). SARS-CoV-2 was detected in the tears and conjunctival secretions of patients with SARS-CoV-2 infection(Xia et al., 2020).Further investigation found conjunctival congestion in 0.8% of patients was with laboratory-confirmed SARS-CoV-2 infection (Guan et al., 2020). Noticeably, Deng et al. inoculated rhesus macaques with SARS-CoV-2 conjunctivally and showed that infection via the conjunctival route is possible in primates(Deng et al., 2020).
SARS-CoV-2 is assumed to gain cell entry through the angiotensin-converting enzyme 2 (ACE2) receptor after auxiliary cellular protease priming by transmembrane protease serine 2 (TMPRSS2) (Hoffmann et al., 2020). RNA sequencing analysis have proved that ACE2 was expressed in human conjunctival tissue, especially in diseased conjunctiva (li et al., 2020). But the cell types and molecular mechanism for the process of cell entry was unknown. Single-cell sequencing has shown that ACE2 and TMPRSS2 were co-expressed in nasal epithelial cells with genes involved in innate immunity (Lange et al., 2020). Therefore, identifying the conjunctival cell types that are permissive to virus entry would help to understand the process by which SARS-CoV-2 infection was established.
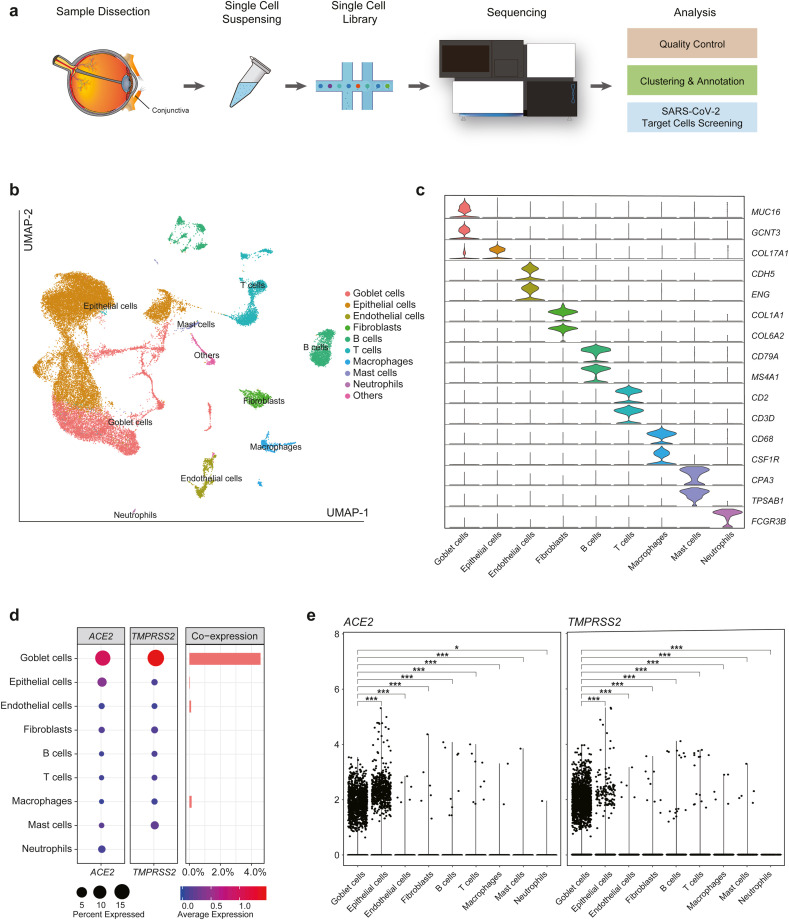
Here we assessed ACE2 and TMPRSS2 expression in human conjunctiva at single-cell level (Fig. 1 a). To perform the analysis, two samples were collected from conjunctival tissue. Two subjects (one male and one female) were included in this study. Detailed information of each subject was listed in Table S1. All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by Fudan Eye & ENT Hospital (2020044–1) following the Code of Ethics of the World Medical Association.
Fig. 1.
Single Cell Transcriptome Analysis of the Conjunctiva. (a) Schematic view of the experimental design. (b) Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plot showing the cellular composition of conjunctiva. Cells are colored according to the cell types. (c) Violin plots showing the expression level of cell type specific marker genes.(d) Dot plots showing the expression ratio and the average expression of ACE2 (left) and TMPRSS2 (middle). The expression ratio is indicated by dots size while average expression is indicated by color. Bar plot showing the co-expression ratio (right) of ACE2 and TMPRSS2 in different cell types. (e) Violin plots showing the expression pattern of ACE2 and TMPRSS2 in each cell type and the significance of the difference between goblet cells and each other cell type (*: 0.01 < p≤0.05; **: 0.001 < p≤0.01; ***: p≤0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Samples were collected during orbital exenteration surgery at normal sites of the donors’ conjunctiva. Following surgical resection, fresh tissue samples were immediately minced into small pieces, disintegrated with 0.25% trypsin (Gibco) at 37 °C for 10 min, treated with 10% FBS to terminate digestion, and filtered through sterile 70 μm cell strainers (BD Falcon) to obtain single-cell suspension. The single-cell suspension was further treated with Red Blood Cell Lysis Buffer (Solarbio) for 10 min, centrifuged at 350 g for 5 min, and resuspended in 10% FBS for cell counting and viability assessment.
Single-cell libraries from cell suspension were generated in the 10X Genomics V3 kit according to the manufacturer's instructions. After performing the library conversions using the MGIEasy Universal DNA Library Preparation Reagent Kit, the libraries were sequenced on BGISEQ-500 sequencing platform.
Raw sequencing data was processed by Cell Ranger 3.0.2 (10X Genomics). Then, Seurat 3.1.2(Stuart et al., 2019) was applied for downstream analysis. The gene expression matrices obtained from Cell Ranger were filtered based on the following criteria: (1) Genes detected in less than three cells were discarded; (2) Cells with genes number less than 200 were removed; (3) Cells with mitochondrial genes percentage greater than 10% were filtered out. After QC, a total of 34,382 high quality cells were acquired for analysis.
Seurat 3.1.2(Stuart et al., 2019) was applied to perform unsupervised clustering. First, LogNormalize method was used to perform normalization by a scale factor 100,000. Next we calculated the variance scores for each gene and defined the top 2000 genes as highly variable genes (HVGs). Then principal component analysis (PCA) was performed using HVGs and principal components (PCs) significance was calculated using JackStraw function. Following, clusters were identified using FindClusters function and visualized using RunUMAP. Besides, DoubletFinder R package(McGinnis et al., 2019) was employed to identified and removed the doublet cells. Finally, we applied FindIntegrationAnchors and IntegrateData functions to integrate these two sequencing libraries and performed the PCA dimensionality reduction and Uniform manifold approximation and projection (UMAP) visualization. The high-quality cells were grouped into nine cell types by UMAP (Fig. 1b, Table S2).(Li et al., 2020) (Luwei and Fen, 2020).
Differentially expressed genes (DEGs) were identified using FindAllMarkers function implemented in Seurat. Genes with adjusted P-value (Bonferroni method) less than 0.05 were defined as DEGs (Table S3). GO term enrichment were performed employing R package clusterProfiler(Yu et al., 2012)and BH method was used for multiple test correction (Table S4).
To annotate these clusters, we characterized several cell-type specific marker genes whose expression patterns and biological functions have been well characterized. For example, the representative cluster marker gene MUC16 and GCNT3 were highly and specifically enriched in the corresponding goblet cells, while CD2 and CD3D were enriched in the T cells (Fig. 1c). To allow users fully exploit our resources, the expression patterns of every single gene in each cell type is available in a freely accessible online platform https://db.cngb.org/VThunter/D2/.
We found that ACE2 and TMPRSS2 showed the highest expression in conjunctival goblet cells, while the two genes were highly co-expressed in this cell type as well (Fig. 1d, Table S5). This result is consistent with previous finding that ACE2 and TMPRSS2 were co-expressed in nasal epithelial cells (Sungnak et al., 2020), indicating both conjunctival and nasal mucosa might function as the entry sites of SARS-CoV-2. To further characterize the differential expression between various cell types, we compared ACE2 and TMPRSS2 in a detailed view individually. Notably, both ACE2 and TMPRSS2 showed significantly higher expression in goblet cells relative to other cell types (p≤0.001) (Fig. 1e, Figure S1a).(
Human ACE2 express together with innate immune genes in nasal goblet cells (Sungnak et al., 2020), to explore if this trend holds true in conjunctiva, we identified ACE2 positively correlated genes (adjusted p value≤0.05) in ACE2 expressing goblet cells. Genes with Pearson's correlation coefficient more than 0 and adjusted P-value (Bonferroni method) less than 0.05 were defined as ACE2 positively correlated genes. Then ACE2 positively correlated genes were used to perform GO term enrichment. Intriguingly, we noticed that those genes were associated with immunity process, covering a variety of biological pathways including “neutrophil degranulation” (C3/HSPA1A/SERPINA1/TCN1), “positive regulation of receptor-mediated endocytosis” (C3), “positive regulation of innate immune response” (HSPA1A/MUC15), “immune response-activating cell surface receptor signaling pathway” (EZR/MUC15), “positive regulation of inflammatory response” (PDCD4/C3) and “acute inflammatory response” (C3/SERPINA1) (Figures S1b-c, Tables S6-7).
In conclusion, we classified the human conjunctival into various clusters at single-cell resolution, and annotated these cell types by marker genes. The transcriptome analysis proved ACE2 and TMPRSS2 showed highest co-expression in certain cell types, especially goblet cells. These findings might be helpful to unravel SARS-CoV-2 transmission, as well as infection prevention and control for the medical workers and civilians.
Author contributions
Conceptualization, D.C., J.W. and J.Q; methodology, R.M, D.C., J.W. and J.Q; software, P.D.; formal analysis, P.D.; resources, R.M and L.G; writing—original draft preparation, R.M and S.J.; writing—review and editing, S.J., D.C. and P.D; visualization, P.D.; supervision, D.C., J.W., and J.Q.; project administration, D.C., J.W., and J.Q.; funding acquisition, J.W. and J.Q. All authors have read and agreed to the published version of the manuscript.
Funding
JQ was supported by the National Natural Science Foundation of China (grant number 81970835, 81800867) and the Natural Science Foundation of Shanghai (grant number 20ZR1409500). JHW was supported by Outstanding Academic Leaders in Shanghai (20XD1401100), Program for Outstanding Medical Academic Leader (2019LJ01), Aging and Women's and Children's Health Special Project of Shanghai Municipal Health Commission (2020YJZX0102), Xuhui District Health and Family Planning Commission Key Disease Joint Project (XHLHGG201807).
Declaration of competing interest
The authors have no conflict of interest.
Acknowledgements
We are thankful to the production team of China National GeneBank, Shenzhen, China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.exer.2021.108501.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Al-Qahtani A.A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): emergence, history, basic and clinical aspects. Saudi J. Biol. Sci. 2020 doi: 10.1016/j.sjbs.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. 2020. Features, Evaluation and Treatment Coronavirus (COVID-19), StatPearls. StatPearls Publishing. [PubMed] [Google Scholar]
- Deng W., Bao L., Gao H., Xiang Z., Qu Y., Song Z., Gong S., Liu Jiayi, Liu Jiangning, Yu P., Qi F., Xu Y., Li F., Xiao C., Lv Q., Xue J., Wei Q., Liu M., Wang G., Wang S., Yu H., Chen T., Liu X., Zhao W., Han Y., Qin C. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in rhesus macaques. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Yu, Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang Jin-lin, Liang Z., Peng Y., Wei L., Liu Y., Hu Ya-hua, Peng P., Wang Jian-ming, Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S. Epidemic and emerging coronaviruses (severe acute respiratory syndrome and Middle East respiratory syndrome) Clin. Chest Med. 2017 doi: 10.1016/j.ccm.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S.C., Zumla A. Severe acute respiratory syndrome: historical, epidemiologic, and clinical features. Infect. Dis. Clin. 2019 doi: 10.1016/j.idc.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh W.C., Naing L., Rosledzana M.A., Alikhan M.F., Chaw L., Griffith M., Pastore R., Wong J. What do we know about SARS-CoV-2 transmission? A systematic review and meta-analysis of the secondary attack rate, serial interval, and asymptomatic infection. medRxiv. 2020 doi: 10.1101/2020.05.21.20108746. 2020.05.21.20108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Wolf J., Auw‐Haedrich C., Schlecht A., Boneva S., Lapp T., Horres R., Agostini H., Martin G., Reinhard T., Schlunck G. Expression of the COVID‐19 receptor ACE2 in the human conjunctiva. J. Med. Virol. 2020;92:25981. doi: 10.1002/jmv.25981. jmv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- li s, li d, fang j, liu q, sun x, xu g, cao w. COVID-19 receptor ACE2 is expressed in human conjunctival tissue, expecially in diseased conjunctival tissue. medRxiv. 2020 doi: 10.1101/2020.05.21.20109652. 2020.05.21.20109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., wei Liu, fen X., fang Z. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020 doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Liu D., Liao X., Wu X., Jing Q., Zheng J., Liu F., Yang S., Bi B., Li Z., Liu J., Song W., Zhu W., Wang Z., Zhang X., Chen P., Liu H., Cheng X., Cai M., Huang Q., Yang P., Yang X., Huang Z., Tang J., Ma Y., Mao C. 2020. Modes of Contact and Risk of Transmission in COVID-19 Among Close Contacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337. doi: 10.1016/j.cels.2019.03.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12 doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Banovich N.E., Barbry P., Brazma A., Collin J., Desai T.J., Duong T.E., Eickelberg O., Falk C., Farzan M., Glass I., Gupta R.K., Haniffa M., Horvath P., Hubner N., Hung D., Kaminski N., Krasnow M., Kropski J.A., Kuhnemund M., Lako M., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K.B., Miao Z., Misharin A.V., Nawijn M.C., Nikolic M.Z., Noseda M., Ordovas-Montanes J., Oudit G.Y., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P.R., Rawlins E.L., Regev A., Reyfman P.A., Rozenblatt-Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H.B., Schultze J.L., Seibold M.A., Seidman C.E., Seidman J.G., Shalek A.K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S.A., Theis F.J., Tsankov A.M., Vallier L., van den Berge M., Whitsett J., Xavier R., Xu Y., Zaragosi L.E., Zerti D., Zhang H., Zhang K., Rojas M., Figueiredo F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Tong J., Liu M., Shen Y., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020;92:589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.