Abstract
SARS-CoV-2 has infected more than 30 million persons throughout the world. A subset of patients suffer serious consequences that require hospitalization and ventilator support. Current tests for SARS-CoV-2 generate qualitative results and are vital to make a diagnosis of the infection. However, they are not helpful to follow changes in viral loads after diagnosis. The ability to quantitatively assess viral levels is necessary to determine the effectiveness of therapy with anti-viral or immune agents. Viral load analysis is also necessary to determine the replicative potential of strains with different mutations, emergence of resistance to anti-viral agents and the stability of viral nucleic acid and degree of RT-PCR inhibition in different types of collection media. Quantitative viral load analysis in body fluids, plasma and tissue may be helpful to determine the effects of the infection in various organ systems. To address these needs, we developed two assays to quantitate SARS-CoV-2. The assays target either the S or E genes in the virus, produce comparable viral load results, are highly sensitive and specific and have a wide range of quantitation. We believe that these assays will be helpful to manage the clinical course of infected patients and may also help to better understand the biology of infection with SARS-CoV-2.
Keywords: SARS-CoV-2, Quantitative analysis, Viral load, Therapy response, Prognosis
1. Introduction
SARS-CoV-2 infection has spread extensively throughout the world and remains a formidable challenge for clinical testing. SARS-CoV-2 is a positive sense single stranded RNA virus that belongs to the β-coronavirus genus and is related to other coronaviruses such as SARS-CoV and MERS (Ren et al., 2020; Dhama et al., 2020). The virus is highly infectious and transmissible, but has a lower mortality rate than SARS-CoV or MERS (Wang et al., 2020). The RNA genome is 29,903 nt (Genbank NC_045512.2) and contains 14 open reading frames that include Orf1ab, S, 3a, 3b, E, M, 6, 7a, 7b, 8, 9b, 9c, N and 10 from the 5’ to 3’ end (Romano et al., 2020). Several molecular assays have been developed to detect SARS-CoV-2 in clinical specimens such as nasopharyngeal swabs, bronchoalveolar lavage and saliva (Lai et al., 2020; Russo et al., 2020; Corman et al., 2020). The assays predominantly target Orf1ab, N, S or E genes in the virus. Orf1ab (21,290 nt) spans almost 44 % of the genome and codes for a polyprotein that is cleaved into 15 peptides that include the 3C-like proteinase and RNA-dependent RNA polymerase (Yoshimoto, 2020). The S gene (3,822 nt) codes for the spike glycoprotein that binds ACE2 and TMPRSS2 receptors on the host cell surface (Ziegler et al., 2021). The N gene (1260 nt) codes for a nucleocapsid protein that is involved in viral synthesis and assembly (Cong et al., 2020). The E gene is quite short at 227 nt and codes for a protein that is associated with the viral envelop (Sarkar and Saha, 2020).
MAFFT alignment shows that SARS-CoV-2 (NC_045512.2) is 79.91 % identical to SARS-CoV Tor2 (NC_004718.3) and 55.47 % identical to MERS (NC_019843.3) (Lu et al., 2020). Sequence differences between SARS-CoV-2, SARS-CoV and MERS have allowed the development of specific assays to detect SARS-CoV-2. Most assays include more than one gene or site within a gene to avoid false negative results due to mutational drift in the virus. Molecular assays predominantly use one step reverse transcription and real time PCR (RT-qPCR) to detect the virus (Yan et al., 2020). Transcription mediated amplification and CRISPER based assays have also been developed (Gorzalsk et al., 2020; Patchsung et al., 2020). RT-qPCR assays have high analytical sensitivity depending on the starting amount of specimen used for nucleic acid extraction and the amount of extract used in the reaction. Even though the assays are performed by real time PCR, the results are reported as Detected or Not detected and are qualitative in nature. Qualitative assays are critical to make a diagnosis of SARS-CoV-2 infection but are not enough to follow the clinical course of infection in patients. Just like in HIV or HCV, after a diagnosis is made, it is necessary to determine if the viral load decreases upon treatment. The therapeutic benefit of potential anti-viral drugs and the replicative potential of different strains of the virus can only be assessed by viral load assays. Quantitative analysis is necessary to better understand the biology of the infection. For example, determining viral loads in organs such as the heart or kidneys may help to understand the pathogenic effects of the virus and the resultant morbidity that follows. Viral load assays may help to determine the infective dose in nasal and throat secretions and threshold levels that confer increased risk of transmission. The effect of storage and transportation conditions can be assessed objectively by quantitative viral assays and may lead to novel ways of collecting and transporting clinical specimens. Lastly, the effect of swabs made with new materials or different types of transport media upon viral stability and RT-qPCR inhibition can be assessed only by quantitative viral loads. To address these issues, we developed two viral load assays, one that targets the S gene and the other the E gene, that are highly sensitive and specific to quantitate SARS-CoV-2.
2. Materials and methods
This study was approved by the institutional review board at the University of Illinois at Chicago (Protocol no. 2020-0817)
2.1. Specimens
Left over nucleic acid extracts were used for the study after clinical testing was completed by the CDC assay protocol (CDC-006-00019, Revision: 02, 3/15/2020). The extracts were derived from nasopharyngeal swabs in M4RT transport media (VTM, Remel, Lenexa, KS, Cat no. R12505). 200 μl of VTM was added to an equal volume of Buffer AVL (Qiagen, Germantown, MD, Cat. No. 19073,) in a BSL2 hood, vortexed and extracted on an EZ1 Advanced XL instrument (Qiagen) using the EZ1 virus mini kit v2.0 (Qiagen, Cat no. 955134). The final elution volume was 60 μl. The extracts were stored at -80° until further analysis. 5 μl of the extract was used as template for quantitative RT-qPCR in duplicate reactions. Extracts that were either positive (n = 113) or negative (n = 50) by the CDC assay protocol (N1, N2) were used for the study. Frozen patient VTM specimens from 2017 and 2018 that were positive for influenza A, B, RSV, rhinovirus, enterovirus, parainfluenza, adenovirus and coronaviruses 229E, NL63, OC43, HKU1 (FilmArray respiratory panel, Biofire Diagnostics, Salt Lake City, UT) were re-extracted and analyzed to determine the specificity of the assays. In addition, 10 μl of whole inactivated SARS-CoV-2 virus (ATCC, Manassas, VA, Cat no. VR-1986HK) was spiked into 190 μl of a negative patient VTM, extracted on the EZ1 Advanced machine and eluted in 60 μl. Based on the certificate of analysis and assuming 100 % extraction efficiency, the estimated copies in the extract was 32,000 copies/μl. 2 μl of this extract was analyzed in the E and S assays.
2.2. Synthesis of in-vitro transcribed RNA
In-vitro transcribed RNA was used to generate standard curves for real time RT-qPCR because an internationally calibrated quantitative standard for SARS-CoV-2 is not yet available. gBlocks containing E and S gene sequences were obtained from Integrated DNA Technologies, Inc.
(IDT, Coralville, Iowa). nt26161-26520 and nt21394-21963 of the SARS-CoV-2 sequence were included in the E and S gBlocks respectively (Genbank accession no. NC_045512.2). A T7 RNA polymerase promoter (5’TAATACGACTCACTATAG3’) was attached to the 5’ end of each gBlock to enable in-vitro transcription. A short sequence (GAAAT) was added to the 5’ end of the promoter to stabilize T7 RNA polymerase binding. Upon receipt, gBlocks were dissolved in 10 mM Tris, pH7.5 to a concentration of 5 ng/μl and stored at -20° until further use. gBlocks were first amplified with SgeneF2-T7/Sgene-R2 gBLOCK or EgeneF1-T7/Egene-R1 primer pairs to generate enough template for in-vitro transcription (Table 1 ). PCR amplification was performed in 50 μl with 20pmols of each primer, 0.2 mM dNTP mix, 1X PCR buffer with 1.5 mM MgCl2 and 0.5 μl of HotStarTaq DNA polymerase (Qiagen, Cat no. 203203). Each PCR reaction contained 3 × 104 molecules of gBlock as template. PCR conditions were 95° for 15 min. followed by 32 cycles of 95° for 20 s, 52° for 30 s and 68° for 30 s. PCR products were run on a 2 % agarose gel to confirm amplification (S gene 541bp, E gene 380bp) and purified using the QIAquick PCR purification kit (Qiagen, Cat no. 28104). Next, in-vitro transcription was performed with the MEGAScript T7 Transcription kit (Thermofishe Scientific, Waltham, MA, Cat no. AM1334). Each 20 μl reaction contained 2ul of 10X buffer, ATP, CTP, GTP, UTP, T7 RNA polymerase and 1 pmol of T7-tagged amplicon. After incubating at 37° for 6 h, DNA was digested with 1 μl of TURBO-DNase at 37° for 15 min. In-vitro transcribed RNA was subsequently purified by the QIAamp RNA blood mini kit (Qiagen, cat no. 1067924) and eluted in 30 μl of dH2O. RNA concentration and purity were measured by the NanoDrop 2000 spectrophotometer. In addition, serial dilutions were also quantitated with the Qubit 3.0 fluorometer using the Qubit RNA HS assay kit (Thermofisher, Cat no. Q32852). The lengths of the S and E gene transcripts were 519 nt and 363 nt respectively. Copies/ng were calculated using the molecular weight of each transcript and Avogadro’s number (1 mol = 6.023 × 1023 molecules). In-vitro transcribed RNA was serially diluted in TTE buffer (1 mM Tris, pH 8.0, 0.1 mM EDTA, 0.1 % Trition X-100) and stored at -80°.
Table 1.
Primers used to amplify S and E gBlocks. The T7 RNA polymerase promoter sequence is highlighted in Bold.
| Gene | Primer | Sequence (5’>3’) | Amplicon |
|---|---|---|---|
| S | SgeneF2-T7 gBLOCK | GAAATTAATACGACTCACTATAGGGACATG | 541bp |
| Sgene-R2 gBLOCK | GGACTGGGTCTTCGAATCTAAAGTAGTA | ||
| E | EgeneF1-T7 | GAAATTAATACGACTCACTATAGGGAATCCAG | 380bp |
| Egene-R1 | TCGTTTAGACCAGAAGATCAGGAACT |
2.3. Primers and probes for RT-qPCR
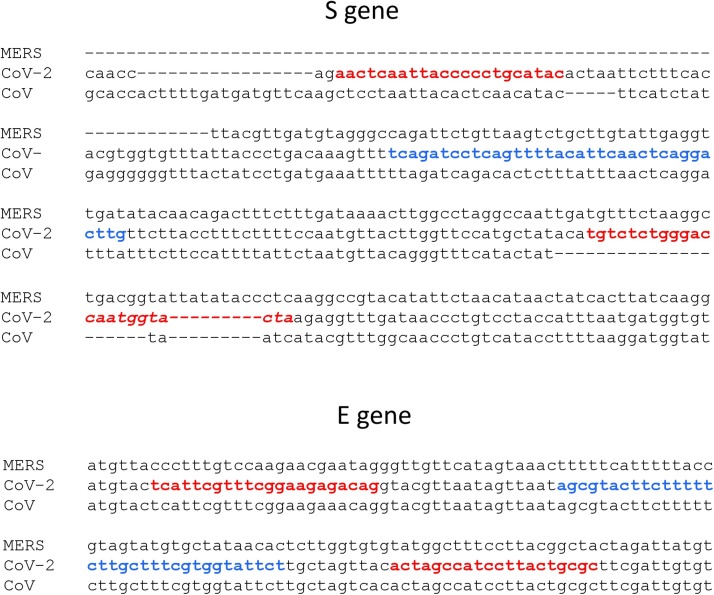
Primer and probe sequences were designed using Oligo v6.71 primer analysis software (Table 2 and Fig. 1 ). The probes were labeled with 6-FAM at the 5’ end and double quenched with Iowa Black at the 3’end and Zen internally (IDT). The specificity of primer and probe sequences was determined by BLASTn alignment with an expect threshold of 1000, word size of 7, reward/penalty ratio of 1/-1 and 20,000 target sequences (www.ncbi.gov). The S gene primer and probe sequences are 100 % identical to 15,600 SARS-CoV-2 sequences and based on the deletions and number of mismatches, are expected to amplify and detect only SARS-CoV-2 and not SARS-CoV or MERS viruses. The E gene primers and probe are 100 % identical to 15,864 SARS-CoV-2 sequences. With a single mismatch in the forward primer, the E gene primers are also expected to amplify SARS-CoV. Based on the number of mismatches, the E gene primers and probe are not expected to amplify MERS. After filtering out SARS-CoV-2 in the BLAST search, there was no significant identity across the entire length to any human, bacterial, fungal or other viral sequences indicating that the S gene primers and probes were specific to SARS-CoV-2 and the E gene primers and probe to SARS-CoV-2 and SARS-CoV.
Table 2.
S and E gene primers and probe sequences for one step RT-qPCR with sequence identity to SARS-CoV-2, SARS-CoV and MERS.
| Sequence identity (nt) |
||||||
|---|---|---|---|---|---|---|
| Gene | Primer | Sequence (5’>3’) | SARS-CoV-2 | SARS-CoV | MERS | Amplicon |
| S | Virion Sgene-F2 | AAC TCA ATT ACC CCC TGC ATA C | 22/22 | 14/22 | Deleted (0/23) | 167bp |
| Virion Sgene-R2 | TAG TAC CAT TGG TCC CAG AGA CA | 23/23 | 2/23, 18/23 deleted | 15/23 | ||
| SgeneProbe-2 | TCAGATCCTCAGTTTTACATTCAACTCAGGACTTG | 35/35 | 21/35 | 10/35 | ||
| E | EgeneF2 RQ-PCR | TCA TTC GTT TCG GAA GAG ACA G | 22/22 | 21/22 | 11/22 | 103bp |
| EgeneR2 RQ-PCR | GCG CAG TAA GGA TGG CTA GT | 20/20 | 20/20 | 12/20 | ||
| EgeneProbe-2 | AGCGTACTTCTTTTTCTTGCTTTCGTGGTATTCT | 34/34 | 34/34 | 10/34 | ||
Fig. 1.
CLUSTAL OMEGA ALIGNMENT (https://www.ebi.ac.uk/tools/msa/clustalo/) OF THE S AND E GENES OF SARS-COV-2 (NC_045512.2), SARS-COV (NC_004718.3) AND MERS (NC_019843.3): The S gene primers and probe are specific to SARS-CoV-2. The E gene primers and probe will amplify SARS-CoV-2 and SARS-CoV, but not MERS. Primers are in red, probes are in blue.
2.4. One step RT-qPCR
The TaqPath™ 1-Step RT-qPCR Master Mix, CG was used for reverse transcription and real time PCR (Thermo Fisher Scientific, Cat no. A15300). Viral loads were analyzed separately in S and E gene viral load assays. Each reaction contained 6.25 μl of mastermix, 10 pmols of forward and reverse primers, 5 pmols of labeled probe and 5 μl of nucleic acid extract in a volume of 25 μl. One-step RT-qPCR was performed on an ABI7500FastDx machine (Thermofisher, Cat no. 4406985) in Standard mode with the following conditions: 50° for 20 min. (RT), 95° for 3 min. and 45 cycles of 95° for 10 s and 56° for 30 s. Fluorescent signal was collected in the annealing/extension stage at 56°. Real time PCR curves were analyzed using ABI software v1.4.1 with the auto baseline option. The threshold was set at 0.05 for S and 0.09 for E gene curves respectively. Each patient specimen was analyzed in duplicate.
2.5. Standard curves
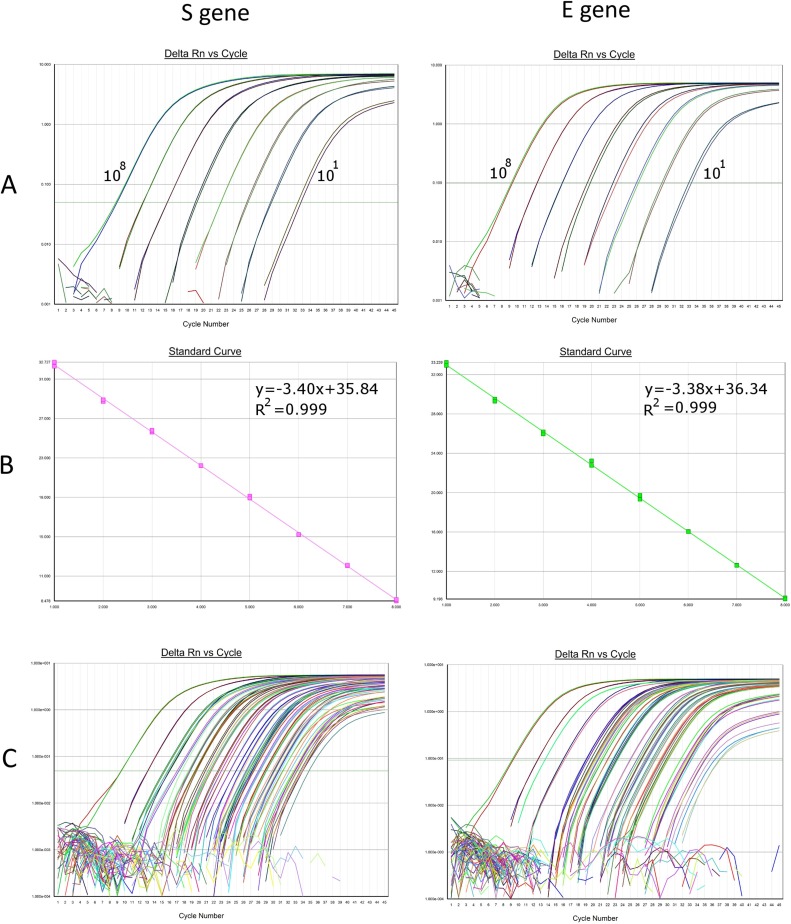
A 10-fold serial dilution of in-vitro transcribed RNA was used to generate standard curves (Fig. 2 ). The series contained 108 to 10 copies/reaction with each level run in duplicate. A standard curve was included in every run. S and E gene viral load/reaction was calculated by interpolation to the respective standard curve. Because 200 μl of patient VTM was extracted and eluted in 60 μl, of which 5 μl was analyzed in each reaction, the mean viral load/reaction was multiplied by 60 to obtain the mean viral load/ml in the sample. The range of quantitation was 6 × 102 to 6 × 109 copies/ml for both assays.
Fig. 2.
STANDARD CURVES FOR THE S AND E GENE ASSAYS: A. S and E amplification curves in duplicate with a ten-fold dilution series of in-vitro transcribed RNA (101 to 108 copies/reaction). B. Linear regression with Log10 standards on X-axis and Ct on the Y-axis. Slopes and correlation coefficients are shown. C. Representative amplification curves with standards and patient specimens included.
2.6. Limit of detection
At first, a “pure LOD” was calculated by directly spiking known amounts of S or E in-vitro transcribed RNA into RT-qPCR reactions. The “pure LOD” was intended to exclude confounding factors of extraction loss and PCR inhibition. Different amounts of in-vitro transcribed RNA were spiked and analyzed in a several replicate reactions (total of 224 reactions for the S and 176 for the E transcript). The amounts spiked and the number of replicate reactions analyzed are shown in Table 5. The amount that yielded a 95 % detection probability by Probit regression was determined to be the “pure LOD” of the transcript. Next, contrived samples were created by spiking 100 copies each (2X pure LOD) of S and E in-vitro transcribed RNA into 200 μl of negative VTM from twenty patient samples to determine the “extracted LOD”. Another batch of twenty contrived specimens was created by spiking 100 copies of a SARS-CoV-2 Standard obtained from Exact Diagnostics (Fort Worth, TX, Cat no. SKU COV019). This standard contains synthetic RNA transcripts from E, N, Orf1ab, RdRp and S genes of SARS-CoV-2, each at 200,000 copies/ml. Next, to determine the “extracted LOD” of the whole virus, a positive patient sample with an average viral load of 1.71 × 108 copies/ml (mean of S and E assays) was diluted serially in VTM to a concentration of 50 copies/μl. Twenty contrived specimens were created by spiking 1.5 μl (75 copies) into 200 μl of negative patient VTM. The contrived specimens were then extracted and analyzed.
Table 5.
Estimation of the “pure LOD” by probit analysis. Copies/reaction of in-vitro transcribed RNA, replicates analyzed and number detected are shown.
| S in-vitro transcribed RNA |
E in-vitro transcribed RNA |
||||
|---|---|---|---|---|---|
| Copies/reaction | Replicates | Detected | Copies/reaction | Replicates | Detected |
| 1 | 42 | 29 | 1 | 45 | 24 |
| 3 | 42 | 40 | 3 | 45 | 40 |
| 5 | 56 | 54 | 5 | 50 | 49 |
| 10 | 48 | 48 | 10 | 12 | 12 |
| 16 | 12 | 12 | 15 | 12 | 12 |
| 32 | 12 | 12 | 25 | 12 | 12 |
| 63 | 12 | 12 | |||
2.7. Statistical analysis
Statistical analysis was performed with MedCalc® Statistical Software version 19.5 (MedCalc Software Ltd, Ostend, Belgium; https://www.medcalc.org; 2020). Passing-Bablok regression was used to compare S and E gene viral loads and Ct values in patient specimens. Bland-Altman analysis was used to determine the bias between the two assays. The Wilcoxon paired t-test was used to compare median S and E viral loads and S and E Ct values. Probit regression analysis was used to calculate the 95 % detection probability (“pure LOD”) of E and S transcripts.
2.8. Clinical course
The clinical course of the patients was accessed from the electronic medical record. Clinical course was classified as mild if the patient was not hospitalized, moderate if hospitalized, but not on ventilator support and severe if on ventilator support or the outcome was death.
3. Results
3.1. Patient specimens
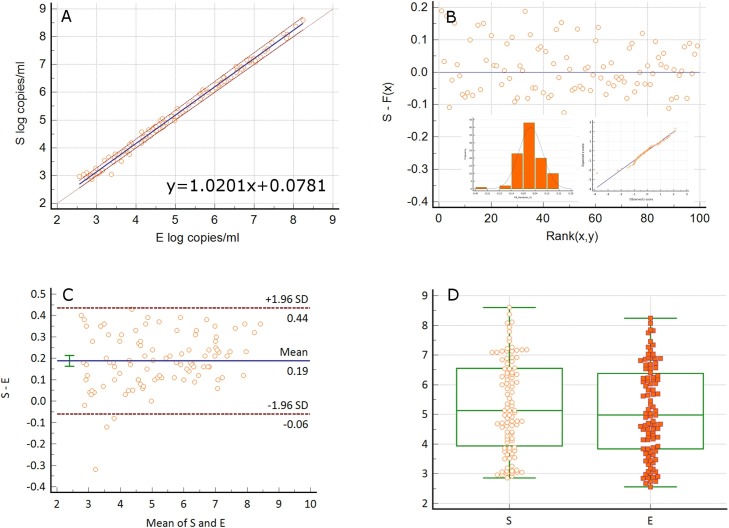
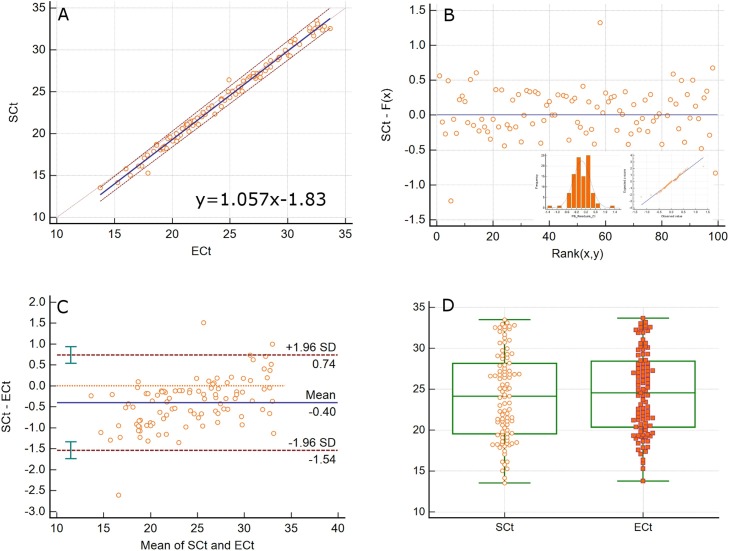
We developed two viral load assays to quantitate the S and E genes of SARS-CoV-2 in extracted nucleic acids from nasopharyngeal swabs in VTM. The specimens were previously determined to be positive for SARS-CoV-2 by the validated CDC N1 and N2 protocol. RNaseP amplified in all specimens with Ct values of 25–31 indicating adequate specimen quality. All patient samples that were positive by the CDC assay (n = 113) were detected for SARS-CoV-2 by E and S assays with 100 % positive percent agreement. 99/113 of these specimens had viral loads in the quantitation range of both assays (600 to 6 × 109 copies/ml). 14/113 specimens were detected for SARS-CoV-2 but were below the quantifiable range of both assays. Of the 14 samples, 13 amplified in both S and E gene assays with Ct values of 33.8–38.5. One sample amplified for the E gene with a Ct value of 38.5, but did not amplify for the Sgene. PCR amplification was robust with low background and unequivocal amplification curves (Fig. 2C). S and E log10 viral load values were highly correlated (Spearman rank correlation 0.995, 95 % CI 0.993-0.997, p < 0.0001) with a slope of 1.0201 (Passing-Bablok regression, 95 % CI 1.0038–1.0374, y = 1.0201 + 0.0781). S gene viral loads were higher by 0.19 logs/ml (Bland-Altman, 95 % CI -0.06 to 0.44). Although close, the median S gene viral load of 5.13 logs/ml (95 % CI 4.74–5.91) was significantly different from the E gene viral load of 4.98 logs/ml (95 % CI 4.54–5.66) (Wilcoxon test, p < 0.0001). The viral loads ranged from 2.86 to 8.6 logs/ml for the S gene and 2.56–8.24 logs/ml for the E gene. These results are summarized in Fig. 3 . The E and S Ct values were also highly correlated (Spearman rank correlation 0.995, 95 % CI 0.992-0.996) with a slope of 1.057 (Passing-Bablok regression, 95 % CI 1.036–1.077, y = 1.057x-1.83). The S gene values were lower by 0.4 Ct units when compared to the E values (Bland-Altman, 95 % CI -1.54 to 0.74). The median Ct values of 24.14 (95 % CI 21.90–26.37) and 24.55 (95 % CI 22.33–26.13) were significantly different with a range of 13.53–33.5 for the S and 13.76–33.67 for the E assay (Wilcoxon test, p < 0.0001). These results are summarized in Fig. 4 . The mean inter-run and intra-run CV of log10 viral load values from repeat analysis of three patient specimens were 1.37 % and 0.4 % for the S and 1.92 % and 0.61 % for the E genes respectively. The corresponding mean inter-run and intra-run CV of untransformed viral loads were 12.52 % and 4.8 % for the S and 17.15 % and 4.56 % for the E gene respectively. In addition, 10 μl of heat inactivated SARS-CoV-2 whole virus from ATCC was spiked into 200 μl of negative patient VTM, extracted and analyzed by both assays. The viral load of the stock was calculated to be 4.38 × 105 copies/μl with the S gene and 1.35 × 105 copies/μl with the E gene assay. According to the certificate of analysis, the viral load for the lot number received was 1.92 × 105 copies/μl by ddPCR. Using this reference value, the viral load was 0.36 logs higher by the S gene and 0.15 logs lower by the E gene assay. Nonetheless, when compared to changes that are clinically significant such as with HIV (± 0.5 logs), the differences between the ATCC value and the viral loads obtained with the S and E gene assays seem clinically insignificant.
Fig. 3.
COMPARISON OF S AND E VIRAL LOADS IN PATIENT SPECIMENS: A. Passing-Bablok regression of S and E log10 viral loads/ml from patient specimens. B. Plot of residuals with histogram and Q-Q plot showing a non-gaussian distribution. C. Bland-Altman analysis of the difference between S and E log10 viral load/ml values. The mean of S and E viral loads is plotted against their difference. D. Wilcoxon test (paired samples) analysis of median log10 viral load/ml, p < 0.0001.
Fig. 4.
COMPARISON OF S AND E Ct VALUES IN PATIENT SPECIMENS: A. Passing-Bablok regression of S and E Ct values from patient specimens. B. Plot of residuals with histogram and Q-Q plot showing a non-gaussian distribution. C. Bland-Altman analysis of the difference between S and E Ct values. The mean of S and E Ct values is plotted against their difference. D. Wilcoxon test (paired samples) analysis of the Ct values, p < 0.0001.
Fifty extracts that were negative for SARS-CoV-2 by the CDC assay were also negative by S and E assays. RNaseP (CDC protocol) amplified in all negative specimens indicating that the specimen quality and extraction were adequate and that there was no PCR inhibition. Nucleic acid extracts from VTM specimens collected in previous years that were positive for influenza A, B, RSV, hMPV, parainfluenza, adenovirus, rhino/enterovirus and coronaviruses 229E, NL63, OC43 and HKU1 were negative by both S and E gene assays indicating that the primers and probes used were specific to SARS-CoV-2. Because the forward primer sequence is deleted in MERS and the reverse primer sequence is deleted in SARS-CoV, it is impossible to amplify SARS-CoV or MERS in the S gene assay. The E assay will however amplify SARS-CoV in addition to SARS-CoV-2. There was 100 % negative percent agreement between the CDC protocol results (N1 and N2) and E and S assays. These results indicate that the E and S gene assays are capable of quantitating SARS-CoV-2 effectively and precisely in patient specimens with high specificity.
3.2. Standard curves
Both assays had a wide range of quantitation from 101 to 108 copies/reaction or 6 × 102 to 6 × 109 copies/ml (Fig. 3, Fig. 4). At < 10 copies/reaction, the detection frequency was < 100 % (Table 5) with Ct values that were imprecise (CV > 5 %). Based on the 100 % detection frequency and high precision, 10 copies/reaction was chosen as the lowest standard (Table 3, Table 4 ). Because an internationally calibrated standard is not available, we used in-vitro transcribed S and E gene RNA to generate the standard curves. Even though quantified whole viral RNA is available from few commercial sources, the stated copies/unit volume lacks verification by a standardized assay or agency. Hence we used gBlocks containing E and S gene sequences tagged with T7 RNA polymerase promoter at the 5’end to synthesize in-vitro transcribed RNA. The procedure generated an inexhaustible quantity of transcripts that could be used ad libitum. Because the in-house generated transcripts contain a defined sequence length, it was possible to accurately calculate copies/ng transcript based on the molecular weight of the transcript. As such, we were able to add accurate amounts of in-vitro transcribed RNA in the RT-qPCR reactions to generate standard curves. The performance characteristics of the standard curves are shown in Table 3, Table 4 and Fig. 2. The results show that the Ct values, correlation co-efficient and slope of the E and S gene assays were similar and precise.
Table 3.
Performance characteristics of standard curves – S gene assay. Mean, standard deviation, 95 % CI and CV are shown for the Ct value of each standard (columns 2–9), slope and correlation co-efficient, R2 (columns 10–11) from 6 runs performed on different days. S gene (A), E gene (B).
| 108 | 107 | 106 | 105 | 104 | 103 | 102 | 101 | Slope | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | 9.4 | 12.96 | 16.05 | 19.53 | 23.12 | 26.64 | 29.93 | 33.33 | −3.425 | 0.9995 |
| SD | 0.04 | 0.14 | 0.09 | 0.16 | 0.19 | 0.32 | 0.24 | 0.36 | 0.044 | 0.0006 |
| 95 % CI | 9.36, 9.45 | 12.81, 13.10 | 15.96, 16.14 | 19.36, 19.69 | 22.92, 23.32 | 26.31, 26.98 | 29.68, 30.18 | 32.95, 33.72 | −3.47, -3.38 | 0.998, 1.000 |
| CV | 0.45 % | 1.09 % | 0.54 % | 0.8 % | 0.83 % | 1.2 % | 0.8 % | 1.09 % | 1.28 % | 0.06 % |
Table 4.
Performance characteristics of standard curves – E gene assay. Mean, standard deviation, 95 % CI and CV are shown for the Ct value of each standard (columns 2–9), slope and correlation co-efficient, R2 (columns 10–11) from 6 runs performed on different days.
| 108 | 107 | 106 | 105 | 104 | 103 | 102 | 101 | Slope | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | 9.41 | 12.8 | 16.13 | 19.61 | 22.97 | 26.28 | 29.64 | 33.29 | −3.394 | 0.9988 |
| SD | 0.019 | 0.033 | 0.057 | 0.13 | 0.15 | 0.114 | 0.17 | 0.47 | 0.05 | 0.0005 |
| 95 % CI | 9.38, 9.44 | 12.74,12.85 | 16.04, 16.22 | 19.4, 19.81 | 22.72, 23.22 | 26.09, 26.46 | 29.36, 29.92 | 32.54, 34.03 | −3.47, -3.31 | 0.998, 0.9995 |
| CV | 0.2 % | 0.26 % | 0.35 % | 0.66 % | 0.67 % | 0.44 % | 0.59 % | 1.41 % | 1.48 % | 0.05 % |
3.3. Limit of detection
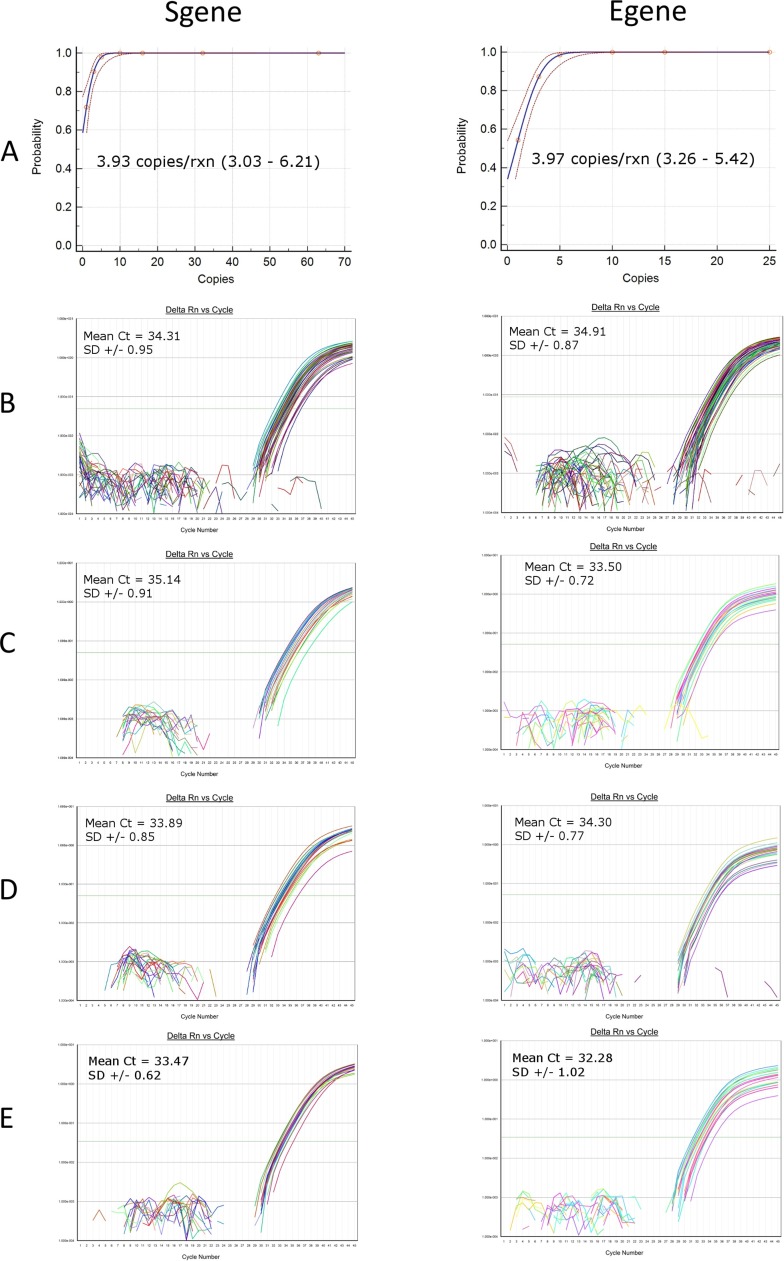
At first, the “pure LOD” of the S and E quantitative assays was determined by spiking different amounts of in-vitro transcribed RNA into replicate RT-qPCR reactions. Copies/reaction, replicates analyzed, and number detected are shown in Table 5. The probability of detection was calculated by Probit regression analysis (Fig. 5 ). The 95 % probability of detection (pure LOD) was 3.93 copies/reaction (95 % CI 3.03–6.21) and 3.97 copies/reaction (95 % CI 3.26–5.42) for the S and E transcripts respectively (Fig. 5A). At around the LOD value, the mean Ct was 34.31 (± 0.95 SD, CV 2.8 %, range of 32.43–36.71) for the S and 34.91 (± 0.87 SD, CV 2.51 %, range of 33.14–37.03) for the E gene (Fig. 5B). The “pure LOD” represents the best case scenario free from the effects of extraction and PCR inhibition. Next, 100 copies of S and E in-vitro transcribed RNA were spiked into 200 μl of VTM from twenty negative VTM specimens and extracted. An amount higher than the “pure LOD” (2X) was spiked to account for any loss during extraction. 20/20 (100 %) specimens were detected for the S and E genes with mean Ct values of 35.14 (SD ± 0.91) and 33.5 (SD ± 0.72) respectively (Fig. 5C). Further, 100 copies of a SARS-CoV-2 standard obtained from Exact Diagnostics was spiked into 200 μl of negative patient VTM and extracted. 20/20 (100 %) specimens were detected for the S gene and 19/20 (95 %) for the E gene with mean Ct values of 33.89 (SD ± 0.85) and 34.30 (SD ± 0.77) respectively (Fig. 5D). These values are similar to the contrived specimens that contained 100 copies of S and E in-vitro transcribed RNA. These results show that the extracted LOD was 8.3 copies/reaction or 500 copies/ml for both the S and E gene assays. In addition, 75 copies of whole virus from a positive patient specimen were spiked into 200 μl of negative patient VTM, extracted and analyzed. 20/20 (100 %) contrived specimens were detected with mean Ct values of 33.47 (SD ± 0.62) and 32.28 (SD ± 1.02) for the S and E genes respectively (Fig. 5E). The extracted LOD is thus 375 copies/ml for patient derived whole intact virus for both assays.
Fig. 5.
PROBIT REGRESSION AND LOD AMPLIFICATION CURVES. A. Probit regression curves of the “pure LOD”. The dashed lines represent the upper and lower 95 % confidence intervals. B. “Pure LOD” for in-vitro transcribed RNA. C. “Extracted LOD” for in-vitro transcribed RNA, 500 copies/ml. D. “Extracted LOD” for the SARS-CoV-2 standard (Exact Diagnostics), 500 copies/ml. E. “Extracted LOD” for whole virus derived from a patient specimen, 375 copies/ml.
3.4. Viral load and clinical course
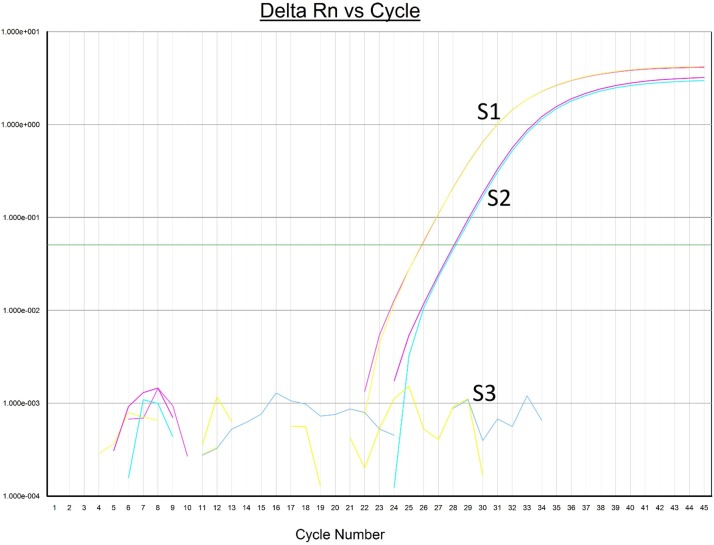
Medical records were available for review in 112/113 patients. 76/112 (68 %) of patients in the study were not hospitalized, whereas 36/112 (32 %) were hospitalized. All hospitalized patients had one or more co-morbidities such as diabetes, hypertension, chronic kidney disease, obesity, congestive heart failure, asthma, autoimmune disease, sickle cell disease, COPD, HIV, HCV and organ transplant. About half (39/76) of the non-hospitalized patients had diabetes and or hypertension. 13 hospitalized patients required ventilator support, out of which 8 ultimately died with an overall death rate of 7.1 %. Patients that required ventilator support and or died had multiple and severe co-morbidities. In the eight patients that died, the viral loads ranged from less than the lower limit of quantitation to over 6 logs/ml at the time the specimens were tested. The viral loads did not show any obvious trend with clinical severity i.e. hospitalization, ventilator support or death. Three specimens from one patient taken at weekly intervals showed decreasing viral loads by the S gene assay: 117,114 copies/ml, 25,781 copies/ml, and then undetected (Fig. 6 ). This patient was not hospitalized and had a mild clinical course.
Fig. 6.
SERIAL VIRAL LOAD ANALYSIS IN ONE PATIENT. Amplification curves from three serial specimens taken at weekly intervals in one patient showing reduction in viral load by the S gene assay.
4. Discussion
In this study we developed and validated two viral load assays to quantitate SARS-CoV-2. Because an internationally calibrated quantitative standard is not yet available, two assays were developed so that their performance could be compared against each other and to have confidence in the results. The two assays thus served to validate each other’s performance. More-over, in the unlikely occurrence of mutations in the primer/probe regions of one assay, the alternate assay could be used to quantify the viral load. Both assays were highly sensitive, specific and had a wide range of quantitation (7 logs). The viral loads and Ct values from the E and S gene assays were highly correlated with the S gene assay producing slightly higher values by about 0.19 logs/ml. Differences in reverse transcription efficiency, Ct values and slope between the two assays might have contributed to the ultimate difference in viral load values. Both assays were precise with low inter and intra-run variations. We used in-vitro transcribed RNA as standards for quantitation because these transcripts are of defined length and could be quantitated accurately based on their molecular weight. A significant advantage of the procedure was that it allowed us to rapidly synthesize an abundant quantity of transcripts without having to clone the viral sequences into an expression vector. A heat inactivated whole virus specimen (ATCC VR-1986HK) that was spiked into negative patient VTM, extracted and analyzed by both assays produced results that were quite close to the given value. The ATCC specimen was quantified with ddPCR, whereas our study used RT-qPCR. The minor differences observed may be related to the different methods used for viral load analysis, but in general the results were comparable indicating that the in-vitro transcribed RNA transcripts were suitable as standards to quantify SARS-CoV-2 in clinical specimens. Nonetheless, it would be helpful to confirm the accuracy of quantitation by prospective studies with another method such as ddPCR and by an internationally accepted calibrator when it becomes available.
The S and E assays were highly specific for SARS-CoV-2 and did not detect other common respiratory viruses. The primer/probe regions in both assays are relatively conserved in SARS-CoV-2 by BLASTn analysis (NCBI Genbank, >15,500 complete genomes) with no significant identity to bacterial, fungal, human or other viral sequences over their entire length. Further, the virus was not detected in fifty specimens that were clearly negative by the CDC N1 and N2 assays.
By probit regression, the 95 % detection probability of E and S in-vitro transcribed RNA was about 4 copies/reaction. This value represents the “pure LOD” after spiking the transcripts directly into the RT-qPCR reactions and required 400 replicate reactions. This value is close to the 95 % detection probability of E transcripts reported by Corman et al. (2020). The “pure LOD” also represents the best sensitivity for the transcripts free from the extraction procedure or PCR inhibition and serves as a baseline to compare LOD after extraction. After spiking negative VTM, the “extracted LOD” was 500 copies/ml for S and E in-vitro transcribed RNA. The same LOD was obtained with a SARS-CoV-2 standard from Exact Diagnostics, confirming the result from in-vitro transcribed RNA. Whole virus from a positive patient specimen spiked into VTM and extracted yielded a slightly better LOD of 375 copies/ml.
In one patient from whom we had serial specimens, the viral load (S gene assay) decreased in subsequent specimens and became negative after 3 weeks (Fig. 6). This patient had a mild clinical course and recovered fully by the end of 3 weeks suggesting that viral load analysis may be useful to follow-up patients after diagnosis. In other patients analyzed at a single time point, SARS-CoV-2 viral loads did not correlate with overt clinical severity. Further studies with serial specimens are necessary to assess the utility of quantitative analysis to determine prognosis. All hospitalized patients had co-morbidities that may have contributed to clinical severity. Our study shows that patients with co-morbidities may suffer severe clinical manifestations even with low viral loads in nasopharyngeal specimens. Preliminary reports suggest that dexamethasone may reduce mortality in hypoxic patients on ventilator support (Johnson and Vinetz, 2020; Solinas et al., 2020). However, steroids are generally contra-indicated if viral loads are high. Determining the viral load by the S or E gene assays may help decide whether steroids can be given safely. The assays can also be used to monitor the efficacy of anti-viral drugs such as remdesivir and antibody based therapies because of their high precision, sensitivity and wide range of quantitation. Rising viral loads may indicate emergence of resistance mutations and guide therapy with novel agents. However, variability in sample collection, assay design, PCR efficiency and analytical sensitivity of the assays used must all be considered when interpreting alterations in viral load. Strain changes in the virus over time may result in different replicative efficiencies and such changes can be monitored effectively with the S and E gene viral load assays. A strain with glycine at position 614 in the Spike protein (D614 G) has been reported to have a higher replication potential when compared to strains that contain a glutamic acid at that position (Korber et al., 2020). Determining the viral load may help to decide whether a person poses a high risk of spreading the infection. It is conceivable that patients who recover, but with very low viral loads below the quantitative range of the assays might not pose a serious risk of transmission and thus can safely be discharged from the hospital and quarantined at home. Qualitative results are not helpful in this regard. The extent of SARS-CoV-2 replication in various tissues and the mechanisms of organ damage are poorly understood at the present time. Quantitative analysis can help to determine replication levels in tissues and correlate pathology in organ systems with viral loads. The infectious potential of specimens can be assessed in a much more objective manner with viral load analysis, rather than with a qualitative result. For example, the S or E gene viral load assays could be used to determine threshold levels of the virus in saliva that confer an elevated risk of transmission because cough and sneeze aerosols contain mostly saliva. Detection by a molecular assay however does not necessarily mean infectivity, especially with low viral loads. Because molecular assays also detect fragmented viral RNA that is not infectious, it is important to determine infectivity and transmissibility by viral cultures or by visualization of intact virions by electron microscopy. Quantitative analysis of SARS-CoV-2 may help to determine the half-life of the virus deposited on inanimate surfaces and shed light on which of them pose the greatest hazard. Due to their wide range of quantitation, the S or E gene assays may be helpful not only to assess the efficiency, but also the magnitude of PCR inhibition with swabs and collection fluids that are manufactured with newer materials. Because nucleic acid extraction is a bottle neck in high throughput analysis, the effect of eliminating this step can be better assessed with the quantitative assays developed in this study. Because the S and E viral load assays are precise, sensitive and specific, they may be helpful to develop an internationally calibrated standard for quantitation of SARS-CoV-2.
5. Conclusions
In summary, we report the development and validation of two sensitive and specific viral load assays to quantitate SARS-CoV-2 viral load. These assays may help to better unravel the pathogenesis of the infection, its effect on various organ systems and be useful to guide therapy.
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. This work was supported by the Department of Pathology at the University of Illinois at Chicago.
Declaration of Competing Interest
The authors report no declarations of interest.
CRediT authorship contribution statement
Joshua Bland: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing - review & editing. Ashley Kavanaugh: Data curation, Investigation, Methodology, Project administration, Resources, Validation, Visualization. Lenny K. Hong: Data curation, Investigation, Methodology, Project administration, Resources, Validation, Visualization. Shrihari S. Kadkol: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Acknowledgements
We are grateful to Dr. Fred Behm, Head of the Department of Pathology, Dr. Sally Campbell-Lee, Director of Clinical Pathology and Peter A. Gashkoff, Administrative Director for their support and encouragement. This work would not have been possible without the unflinching efforts of Carol Dodge (Manager, Molecular Pathology laboratory), Mei Shen, Lynn Terrile (Supervisors), Adrian Tira, Jyothirmayi Ryali, Ermias Wedeyohannes, Meihong Zhang, Sadia Khan and Kristen Bradford (Medical Technologists).
References
- Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94 doi: 10.1128/JVI.01925-19. e01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., KW Chu D., Bleicker T., Brünink S., Schneider J., Schmidt M.L., GJC Mulders D., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken R., PG Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. pii=2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Khan S., Tiwari R., Sircar S., Bhat S., Malik Y.S., Singh K.P., Chaicumpa W., Bonilla-Aldana D.K., Rodriguez-Moralesg A.J. Coronavirus disease 2019 –COVID-19. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00028-20. e00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzalsk A.J., Tian H., Laverdure C., Morzunov S., Verma S.C., VanHooser S., Pandori M.W. High-throughput transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.M., Vinetz J.M. Dexamethasone in the management of covid -19. BMJ. 2020;370:m2648. doi: 10.1136/bmj.m2648. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group, McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Wang C.-Y., Ko W.-C., Hsueh P.-R. In vitro diagnostics of coronavirus disease 2019: technologies and application. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.05.016. S1684-1182(20)30140-30147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020 doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Ren Y.R., Golding A., Sorbello A., Ji P., Chen J., Saluja B., Witzmann K., Arya V., Reynolds K.S., Choi S.Y., Nikolov N.P., Sahajwalla C. A comprehensive updated review on SARS-CoV-2 and COVID-19. J. Clin. Pharmacol. 2020;60:954–975. doi: 10.1002/jcph.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A., Minichini C., Starace M., Astorri R., Calò F., Coppola N. Current status of laboratory diagnosis for COVID-19: a narrative review. Infect. Drug Resist. 2020;13:2657–2665. doi: 10.2147/IDR.S264020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M., Saha S. Structural insight into the role of novel SARSCoV-2 E protein: a potential target for vaccine development and other therapeutic strategies. PLoS One. 2020;15(8) doi: 10.1371/journal.pone.0237300. e0237300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas C., Perra L., Aiello M., Migliori E., Petrosillo N. A critical evaluation of glucocorticoids in the management of severe COVID-19. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.06.012. S1359-6101(20)30161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. Agents. 2020;55(6) doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30(3):e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus‑2 (SARS CoV‑2 or n‑COV19), the cause of COVID‑19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., et al. (more than 30 authors).2020. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2021;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]