Abstract
Background
We aimed to determine whether plasma cell-free DNA (cfDNA) concentration is associated with survival in patients with isocitrate dehydrogenase (IDH) wild-type glioblastoma (GBM).
Methods
Pre-operative and post-chemoradiotherapy blood samples were prospectively collected from patients with newly diagnosed IDH wild-type GBM. Patients underwent surgical resection or biopsy and received adjuvant radiotherapy with concomitant temozolomide. Cell-free DNA (cfDNA) was isolated from plasma and quantified using SYBR Green-based q polymerase chain reaction (qPCR).
Results
Sixty-two patients were enrolled and categorized into high vs. low cfDNA groups relative to the pre-operative median value (25.2 ng/mL, range 5.7–153.0 ng/mL). High pre-operative cfDNA concentration was associated with inferior PFS (median progression-free survival (PFS), 3.4 vs. 7.7 months; log-rank P = .004; hazard ratio [HR], 2.19; 95% CI, 1.26–3.81) and overall survival (OS) (median OS, 8.0 vs. 13.9 months; log-rank P = .01; HR, 2.43; 95% CI, 1.19–4.95). After adjusting for risk factors, including O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status, pre-operative cfDNA remained independently associated with PFS (HR, 2.70; 95% CI, 1.50–4.83; P = .001) and OS (HR, 2.65; 95% CI, 1.25–5.59; P = .01). Post-hoc analysis of change in cfDNA post-chemoradiotherapy compared to pre-surgery (n = 24) showed increasing cfDNA concentration was associated with worse PFS (median, 2.7 vs. 6.0 months; log-rank P = .003; HR, 4.92; 95% CI, 1.53–15.84) and OS (median, 3.9 vs. 19.4 months; log-rank P < .001; HR, 7.77; 95% CI, 2.17–27.76).
Conclusions
cfDNA concentration is a promising prognostic biomarker for patients with IDH wild-type GBM. Plasma cfDNA can be obtained noninvasively and may enable more accurate estimates of survival and effective clinical trial stratification.
Keywords: cell-free DNA, glioblastoma, liquid biopsy, overall survival, progression-free survival
Key Points.
High pre-operative plasma cfDNA concentration was independently associated with survival in newly diagnosed, IDH wild-type GBM.
An increase in cfDNA concentration following chemoradiotherapy compared to pre-operative baseline levels also provided accurate survival projections.
This noninvasive biomarker may allow for more tailored treatment planning and clinical trial stratification in IDH wild-type GBM.
Importance of the Study.
Although GBM remains incurable, disease course and survival in patients vary widely. Methylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter can predict prognosis but is limited by the need for tumor tissue and significant variation in detection methods. In this prospective cohort study of 62 patients with IDH wild-type GBM, high pre-operative plasma cfDNA concentration was associated with inferior PFS and OS even after adjusting for known prognostic factors, including MGMT promoter methylation and extent of surgical resection. At the 1 month post-chemoradiotherapy time point, increased cfDNA concentration compared to pre-operative baseline was also associated with inferior PFS and OS. Plasma cfDNA can be obtained noninvasively and may add significant value for prognostication and response assessment in IDH wild-type GBM.
The standard treatment paradigm for glioblastoma (GBM) has remained largely unchanged since 2005,1 consisting of maximal safe resection, adjuvant radiotherapy with concomitant temozolomide chemotherapy, and maintenance temozolomide cycles.2 Although the vast majority of patients with GBM ultimately succumb to the disease,3 progression-free survival (PFS) and overall survival (OS) outcomes vary widely due to demographic, clinical, and tumor molecular differences across patients.4,5 It is well-established that mutations in the isocitrate dehydrogenase genes (IDH1 or IDH2) are associated with distinct underlying biology and substantially better OS compared to IDH wild-type GBM.6,7 However, these mutations are only present in approximately 10% of primary GBMs,8 and outcomes are far more difficult to predict within the much larger population of patients with IDH wild-type tumors.5 Despite widespread recognition of this interpatient heterogeneity,9–11 clinical trials in IDH wild-type GBM continue to suffer from a dearth of accurate prognostic biomarkers for risk stratification.12–14 Until such biomarkers are routinely used for prognostic enrichment and stratification, efforts to detect a signal of efficacy in this highly heterogeneous patient population may remain elusive, even with exceptionally promising therapies in clinical development.15,16 In addition, improved prognostication in patients with GBM would lead to more personalized clinical decision-making in routine practice.17,18
At present, O6-methylguanine-DNA methyl-transferase (MGMT) promoter methylation is the most extensively studied prognostic biomarker for patients with IDH wild-type GBM, with MGMT methylation in the tumor being associated with better prognosis and increased sensitivity to temozolomide.19 However, routine implementation of this biomarker in clinical practice and trials has been challenging due to controversy around detection methods and optimal cutoff definitions.12,13 In addition, MGMT promoter methylation assays currently require tumor tissue acquired by invasive brain biopsies or surgical resections. A less technically challenging, noninvasive prognostic biomarker for GBM may allow for easier and potentially more accurate risk stratification for GBM patients, leading to improvements in both clinical trial design and standard clinical care.
Blood-based liquid biopsy has become part of routine clinical care for detection of therapeutically targetable mutations in nonsmall cell lung cancer,20 breast cancer,21 and other solid tumors. While the detection rate of somatic mutations in the plasma of patients with GBM has been relatively low,22,23 we and others have demonstrated the prognostic value of total cell-free DNA (cfDNA) concentration for GBM and other solid tumors, irrespective of the proportion of cfDNA that is tumor-derived.22,24–28 In addition, recent studies suggest that changes in variant allele fractions of detected mutations from longitudinal cfDNA samples can provide further prognostic value.29–32 To our knowledge, the dynamics of cfDNA concentration, and possible prognostic implications, have not yet been studied for patients with GBM.
We previously published a pilot study which suggested that cfDNA concentration may be associated with survival outcomes in patients with newly diagnosed GBM.22 Here we aimed to corroborate this finding in a larger, independent cohort of patients. In addition, we explored the prognostic value of on-therapy cfDNA dynamics.
Materials and Methods
Patients and Treatment
Patients with radiographically suspected high-grade glioma planned for initial surgical resection or biopsy at the University of Pennsylvania between February 2018 and March 2020 were enrolled in this prospective cohort study (IRB#828164, expiration date ofJanuary 9, 2021). Following surgery, patients remained in the study if (1) histopathology confirmed GBM, (2) the tumor was found to be IDH1/2 wild-type by next-generation sequencing, (3) and the patient initiated standard adjuvant radiotherapy and concomitant temozolomide chemotherapy following surgery. Pre-operative blood samples were collected from all patients, and 1 month post-chemoradiotherapy samples were collected from the subset of patients that elected to participate in the study longitudinally. Patients receiving experimental therapies and/or tumor-treating fields with first-line therapy were excluded from the current analysis to avoid the confounding impact of these therapies on survival outcomes. As of the July 28, 2020 data cutoff, pre-operative blood samples had been collected from 96 consecutive patients, all of whom had tumors determined to be IDH wild-type; 9 were taken off study because histopathology did not confirm GBM, and 25 were excluded because they received experimental therapies and/or tumor-treating fields following surgery. Baseline demographic and clinical variables collected included age, sex, MGMT promoter methylation status, Karnofsky performance status (KPS), and extent of surgical resection (biopsy or partial resection vs. near or gross total resection). Written informed consent was obtained from all patients under approval by the University of Pennsylvania Institutional Review Board.
IDH1/2 Mutational and MGMT Promoter Methylation Status Determination
IDH1/2 mutational and MGMT promoter methylation status was determined by analysis of DNA extracted from tumor tissue collected at the time of initial resection. Targeted NGS for IDH1/2 mutation determination was performed at the University of Pennsylvania’s Clinical Laboratory Improvement Amendments (CLIA)-certified Center for Personalized Diagnostics using their Solid Tumor Sequencing Panel consisting of 152 genes (Comprehensive Solid Tumor HaloPlexHS, version 2.0; Agilent Technology, Inc.) as previously described.22 MGMT promoter methylation analysis was performed in the CLIA-certified Molecular Pathology Laboratory at the University of Pennsylvania. Bisulfite-converted DNA was amplified with primers targeting differentially methylated region 2 (DMR2) of the MGMT promoter, and percent methylation was determined by pyrosequencing of the amplified product (PyroMark Q24, Qiagen).
Specimen Collection and Plasma Isolation
Whole blood samples were collected in either Streck® Cell-Free DNA BCT (Streck) or K2EDTA (Becton-Dickinson) blood collection tubes. Streck samples were stored and processed at room temperature; samples were banked within 24 h of collection. Streck whole blood was centrifuged at 1,600 ×g for 10 min; the plasma supernatant was then collected and centrifuged twice at 4,122 ×g for 15 min (swinging bucket rotor, brake-off). K2EDTA samples were stored and processed at 4°C; samples were banked within 1 h of collection. Whole blood was centrifuged at 1,900 ×g for 10 min; the plasma supernatant was isolated and centrifuged at 3,000 ×g for 15 min (swinging bucket rotor, brake-on). All plasma samples were aliquoted at 1 mL and stored at −80°C for future use.
cfDNA Analysis
Plasma was isolated from whole blood and cfDNA extraction performed using the QIAamp MinElute ccfDNA Mini Kit (Qiagen) according to the manufacturer’s instructions, and elution was performed in 2 consecutive steps (30 µL each). cfDNA was eluted in Ultra-clean Water (Qiagen) and stored at 4°C prior to quantification.
Quantification was performed using an SYBR Green-based qPCR assay for a 115 bp amplicon of the human ALU repeat. Amplification was performed using forward primer 5′-CCT GAG GTC AGG AGT TCG AG-3′ and reverse primer 5′-CCC GAG TAG CTG GGA TTA CA-3′ (Integrated DNA Technologies). DNA standard (Promega) curve dilutions and cfDNA samples were diluted 1:10 in nuclease-free water. Power SYBR Green PCR Master Mix (Applied Biosystems) was prepared according to the manufacturer’s instructions; qPCR was performed on 1 µL of sample and run in quadruplicate on a ViiA 7 Real-Time PCR System (Applied Biosystems). Results were analyzed using the QuantStudio Real-Time PCR Software (Applied Biosystems).
Patients were classified as high vs. low pre-operative cfDNA according to whether pre-operative cfDNA concentration was above vs. below the median value of 25.2 ng/mL for the cohort of 62 patients enrolled in this study. This value is slightly higher than the cut-off value of 13.4 ng/mL calculated for our previous study of 42 patients.22 In a post-hoc analysis of the prognostic impact of post-chemoradiotherapy cfDNA, patients were dichotomized by whether the cfDNA concentration at the time of first post-radiation MRI was higher vs. lower than the pre-operative cfDNA concentration.
Survival Analysis and Statistical Methods
Differences in baseline characteristics between patients with high vs. low cfDNA were evaluated with chi-square tests. The date of tumor progression was determined retrospectively using all available data from the electronic health record, including serial MRI scans and clinical notes (including clinical decline, outcomes of multidisciplinary tumor board discussions, and the start of new antineoplastic therapies) and pathology reports from biopsies/resections when patients underwent a repeat surgery. For the primary analysis of pre-operative cfDNA, PFS was defined as the time from initial surgery until the date of tumor progression or death from any cause, and OS was defined as the time from initial surgical resection until death from any cause. The reverse Kaplan–Meier (KM) method was used to determine the median follow-up time.
For the analysis of the prognostic impact of cfDNA dynamics, PFS was defined as the time from the patient’s first post-radiation MRI until the date of tumor progression or death from any cause. Three patients progressed prior to their first post-radiation MRI and were therefore excluded from this PFS analysis. OS was defined as the time from the patient’s first post-radiation MRI until the date of death from any cause.
The KM method was used to estimate median PFS and OS. Log-rank tests were used to assess crude differences in survival according to plasma cfDNA concentration. Cox regression was used to adjust for relevant prognostic variables including age (<65 vs. ≥65), MGMT promoter methylation status (methylated vs. unmethylated), the extent of surgical resection (gross/near total resection vs. subtotal resection or biopsy), and performance status (KPS ≥60 vs. <60). All statistical analyses were performed using Stata software, version 16 (StataCorp). There was no missing data in this study.
Results
Patient Characteristics
Pre-operative blood samples for 62 patients, all with ≥4 months of follow-up, comprised the cohort for the current analysis, with baseline demographic and clinical characteristics shown in Table 1. All patients underwent a collection of pre-operative blood samples with extraction and quantification of plasma cfDNA successfully completed for 62 patients (100%). The patients were then dichotomized into high- and low-cfDNA groups by the median pre-operative cfDNA concentration of 25.2 ng/mL (Supplementary Figure 1; range, 5.7–153.0; interquartile range [IQR], 13.4–40.4). There was no significant difference for any baseline demographic or clinical characteristics between the high- and low-cfDNA groups (Table 1).
Table 1.
Baseline Characteristics of the Low- and High-cfDNA Groups
| Variable | All (n = 62) |
Low (<Median) Plasma cfDNA (n = 31) |
High (>Median) Plasma cfDNA (n = 31) |
P value |
|---|---|---|---|---|
| Age | ||||
| Median (range) | 67 (39–85) | 65 (39–85) | 69 (49–84) | .68 |
| <65, n (%) | 26 (42%) | 13 (42%) | 13 (42%) | >.99 |
| ≥65, n (%) | 36 (58%) | 18 (58%) | 18 (58%) | |
| Sex, n (%) | ||||
| Male | 39 (63%) | 19 (61%) | 20 (65%) | .79 |
| Female | 23 (37%) | 12 (39%) | 11 (35%) | |
| Karnofsky Performance Status, n (%) | ||||
| <60 | 11 (18%) | 3 (5%) | 8 (13%) | .12 |
| ≥60 | 51 (82%) | 28 (45%) | 23 (37%) | |
| Surgical resection, n (%) | ||||
| Near Total/Gross Total | 23 (37%) | 12 (39%) | 11 (35%) | .79 |
| Biopsy Only/Partial | 39 (63%) | 19 (61%) | 20 (65%) | |
| MGMT promoter methylation status, n (%) | ||||
| Methylated | 25 (40%) | 12 (39%) | 13 (42%) | .80 |
| Unmethylated | 37 (60%) | 19 (61%) | 18 (58%) |
MGMT, O6-methylguanine-DNA methyltransferase.
Association of Pre-operative cfDNA Concentration with PFS and OS
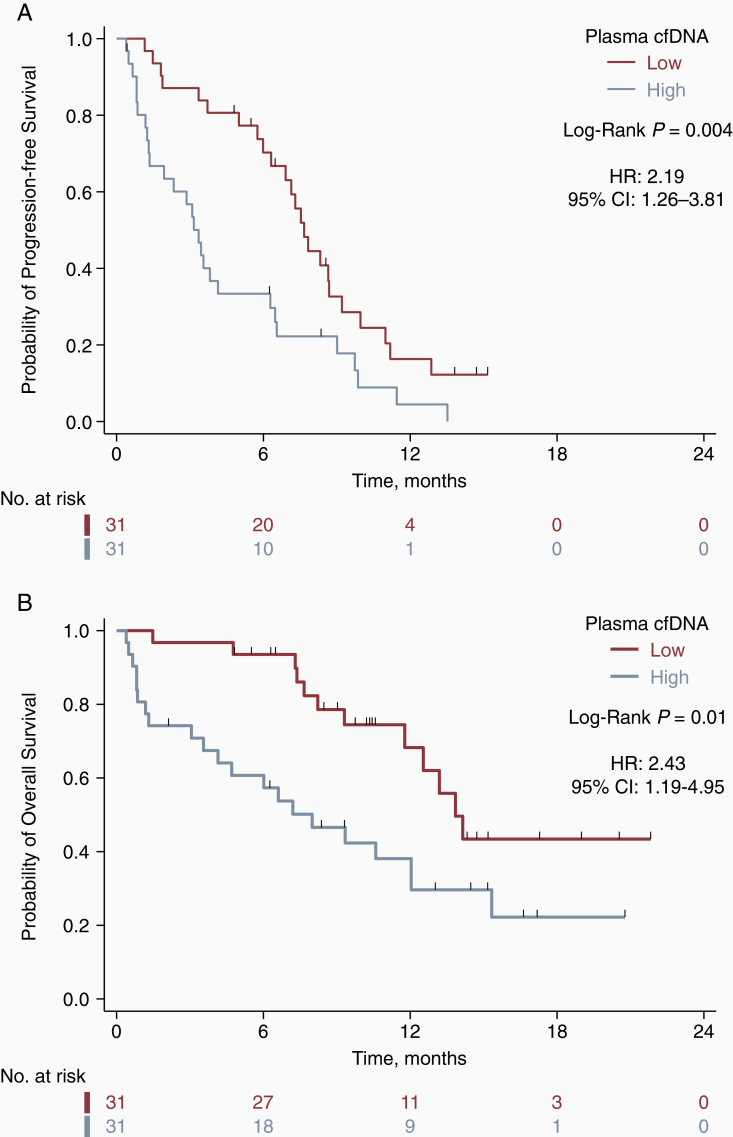
The median duration of follow-up was 14.5 months (95% CI, 10.3–16.6). Kaplan-Meier curves for PFS and OS, according to pre-operative cfDNA concentration are displayed in Figure 1. High cfDNA was associated with inferior PFS (median PFS, 3.4 vs. 7.7 months; P = .004 by the log-rank test; hazard ratio [HR], 2.19; 95% CI, 1.26–3.81) and OS (median OS, 8.0 vs. 13.9 months; P = .01 by the log-rank test; HR, 2.43; 95% CI, 1.19–4.95). Multivariable Cox regression analysis demonstrated the independent prognostic value of cfDNA concentration when adjusting for other established prognostic factors, including MGMT promoter methylation status, extent of surgical resection, age, and performance status (Table 2). These findings were consistent for both PFS (HR, 2.70; 95% CI, 1.50–4.83; P = .001) and OS (HR, 2.65; 95% CI, 1.25–5.59; P =.01).
Figure 1.
Pre-operative cfDNA concentration is associated with progression-free and overall survival in GBM. Kaplan–Meier survival curves for progression-free survival (A) and overall survival (B) by pre-operative cfDNA concentration. Low vs. high plasma cfDNA designates patients below or above the median pre-operative cfDNA concentration of 25.2 ng/mL, respectively.
Table 2.
Multivariable Analysis for Progression-free and Overall Survival
| Progression-free Survival | Overall Survival | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P value | HR (95% CI) | P value |
| Plasma cfDNA Concentration | ||||
| Low (<Median) | 1 [reference] | .001 | 1 [reference] | .01 |
| High (>Median) | 2.70 (1.50–4.83) | 2.65 (1.25–5.59) | ||
| MGMT Promoter Methylation | ||||
| Methylated | 1 [reference] | .02 | 1 [reference] | .08 |
| Unmethylated | 2.31 (1.17–4.56) | 2.20 (0.91–5.33) | ||
| Extent of Resection | ||||
| Near Total or Gross Total | 1 [reference] | .03 | 1 [reference] | .26 |
| Biopsy Only or Partial | 2.01 (1.06–3.81) | 1.59 (0.72–3.54) | ||
| Age | ||||
| <65 years old | 1 [reference] | .003 | 1 [reference] | .05 |
| ≥65 years old | 2.53 (1.37–4.7) | 2.14 (0.99–4.62) | ||
| Karnofsky Performance Status | ||||
| ≥60 | 1 [reference] | .02 | 1 [reference] | .009 |
| <60 | 2.56 (1.16–5.61) | 3.22 (1.34–7.73) | ||
CI, confidence interval; HR, hazard ratio; MGMT, O6-methylguanine-methyltransferase.
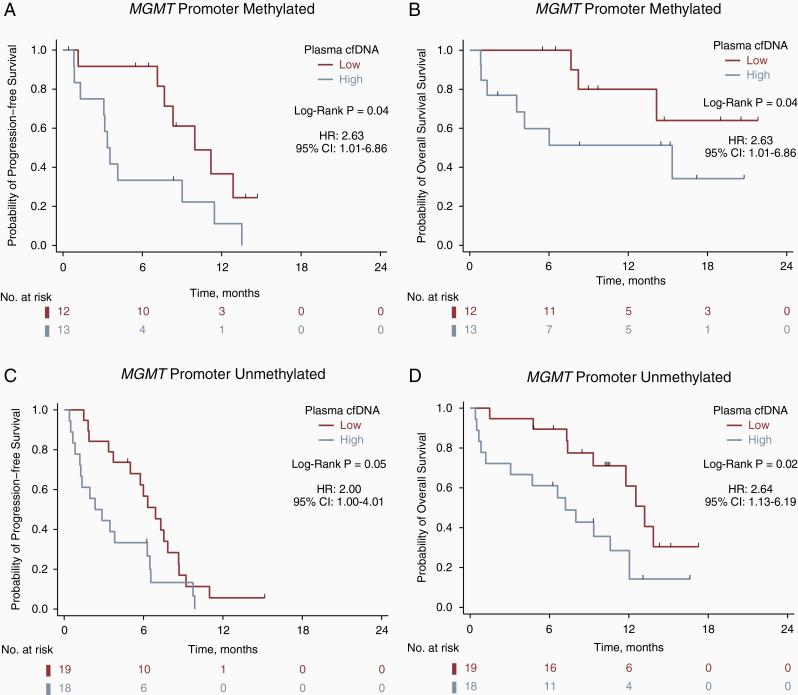
Given that MGMT promoter methylation status is the strongest predictive factor for outcome in temozolomide-treated patients with GBM,19 we assessed the prognostic impact of pre-operative cfDNA concentration separately within each MGMT subgroup. In patients with tumors with MGMT promoter methylation (n = 25), high cfDNA was associated with inferior PFS (median PFS, 3.4 vs. 10.0 months; P =.04 by the log-rank test; hazard ratio [HR], 2.63; 95% CI, 1.01–6.86) and OS (median OS, 15.33 vs. NR months; P =.11 by the log-rank test; HR, 2.90; 95% CI, 0.74–11.27), although the association with OS did not reach statistical significance (Figure 2A and B). In patients with tumors that lacked MGMT promoter methylation (n = 37), high cfDNA was associated with inferior PFS (median PFS, 2.3 vs. 6.9 months; P = .05 by the log-rank test; HR, 2.00; 95% CI, 1.00–4.01) and OS (median OS, 7.2 vs. 13.2 months; P = .02 by the log-rank test; HR, 2.64; 95% CI, 1.13–6.19) (Figure 2C and D).
Figure 2.
Pre-operative cfDNA concentration is associated with survival irrespective of MGMT promoter status. Kaplan–Meier survival curves for progression-free survival (A) and overall survival (B) for 25 MGMT promoter methylation positive patients, and for progression-free survival (C) and overall survival (D) for 37 MGMT negative patients. Low vs. high plasma cfDNA designates patients below or above the median pre-operative cfDNA concentration of 25.2 ng/mL, respectively.
Association of On-therapy Change in cfDNA Concentration with PFS and OS
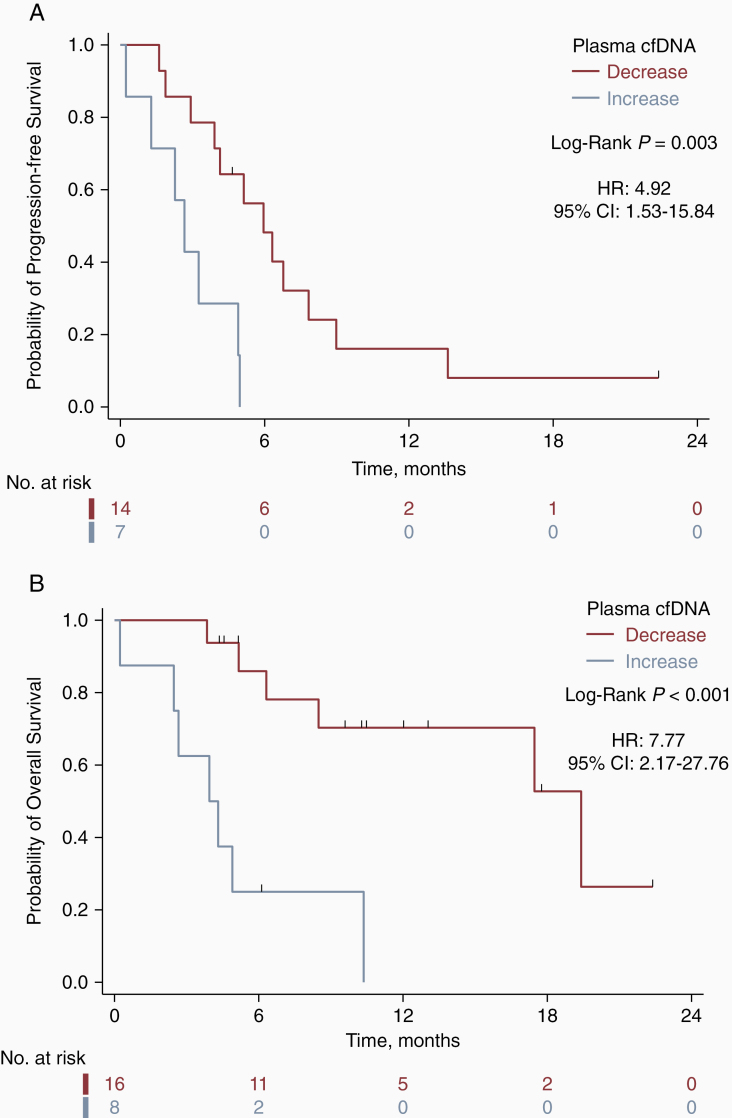
In a post-hoc analysis, we next examined whether the change in cfDNA concentration from pre-operative to 1 month post-chemoradiotherapy time points was associated with subsequent PFS and OS. This analysis included the 24 of 62 patients (39%) who had undergone post-chemoradiotherapy cfDNA collection and accrued ≥4 months of follow-up from the post-chemoradiotherapy collection time point. Eight of 24 patients (33%) experienced an increase in cfDNA, which was associated with worse PFS (median, 2.7 vs. 6.0 months; P = .003 by the log-rank test; HR, 4.92; 95% CI, 1.53–15.84) and OS (median, 3.9 vs. 19.4 months; P < .001 by the log-rank test; HR, 7.77; 95% CI, 2.17–27.76) (Figure 3).
Figure 3.
Change in cfDNA concentration between the pre-surgery and post-chemoradiotherapy time points is associated with both progression-free and overall survival. Kaplan-Meier survival analyses for a post-hoc cohort of 24 patients demonstrates higher progression-free survival (A) and overall survival (B) for patients who experience decreases in their cfDNA concentration from baseline to the post-chemoradiotherapy time point. Progression-free survival and overall survival are defined from the time of the patient’s first post-chemoradiotherapy MRI.
Discussion
These results build on our prior discovery22 of cfDNA concentration as a novel prognostic biomarker in GBM, confirming this finding in a larger, independent patient cohort. Importantly, the prognostic impact of pre-operative cfDNA concentration remains highly significant after consideration of multiple clinical variables, including MGMT promoter methylation status, the extent of surgical resection, age, and Karnofsky performance status (KPS). We also determined, for the first time to our knowledge, that an increase in cfDNA concentration from pre-operative to post-chemoradiotherapy is associated with worse subsequent PFS and OS, suggesting that on-therapy cfDNA dynamics may have a role for assessing therapeutic response in patients with GBM.
The ability to more accurately predict prognosis in patients with GBM is of great value in neuro-oncology, as the inter-individual heterogeneity of GBM has been well known for years.9,10,33,34 This represents a major challenge when attempting to accurately stratify patients with newly diagnosed GBM in clinical trials, as well as when discussing goals of care with patients and families. cfDNA may offer a noninvasive approach for improved prognostication in newly diagnosed GBM, with relevance for both neuro-oncology research and clinical practice. Of note, since plasma cfDNA can provide important prognostic information even before surgical resection of a new GBM, cfDNA may be useful in the design of neoadjuvant or window-of-opportunity studies in the newly diagnosed setting. Furthermore, in contrast to MGMT methylation, plasma cfDNA quantification can be performed noninvasively and represents a rapid turn-around, low-tech, and relatively inexpensive assay that can be completed using commercially available reagents. While future studies are needed to determine the optimal integration of cfDNA and MGMT methylation for prognostication in GBM, our results suggest that use of the 2 biomarkers together may better refine prognosis in an individual patient than either alone.
Our data also suggest that serial cfDNA measurements may offer an opportunity for assessing therapeutic response to chemoradiotherapy at the time of 1 month post-radiation imaging. This is a pivotal timepoint in the care of patients with GBM, as this first post-radiation MRI scan is used to assess initial response to front-line therapy and serves as the patient’s new baseline imaging moving forward.35 However, the first post-radiation MRI is notoriously difficult to interpret, as chemoradiotherapy can lead to breakdown of the blood-brain barrier and resultant contrast extravasation resembling tumor progression.36 When these radiographic changes occur but subsequently stabilize or regress in the absence of new therapeutic intervention, the phenomenon is referred to as pseudoprogression.37 Although pseudoprogression accompanies the initial chemoradiotherapy phase of treatment in at least 25% of patients with newly diagnosed GBM,38,39 currently available neuroimaging techniques are unable to reliably distinguish between true tumor progression and pseudoprogression.40 As a result, neuro-oncologists are often uncertain about whether chemoradiotherapy has been effective at this early juncture and if a change in therapy is warranted. In many cases, this uncertainty is only resolved once the patient has undergone multiple repeat MRI scans to monitor for resolution or progression of the radiographic changes, leading to wasted time on ineffective therapy for patients experiencing tumor progression and unnecessarily frequent imaging and anxiety for patients with pseudoprogression. If our results are validated in larger prospective cohorts, pre- vs. post-chemoradiotherapy plasma cfDNA dynamics may eventually be used to predict post-chemoradiotherapy outcomes irrespective of ambiguous post-treatment MRI findings.
Importantly, the plasma cfDNA biomarker used in this study was derived by quantifying all cell-free DNA in the plasma compartment, regardless of how much was tumor-derived (ie, circulating tumor DNA [ctDNA]). Although both cfDNA and ctDNA have previously demonstrated prognostic utility across multiple solid tumors,24–28,41–43 the biology of nontumor derived cfDNA and its connection with prognosis have been relatively understudied.44,45 In GBM, it is well established that the vast majority of plasma cfDNA is not tumor-derived.22,23,46 Nonetheless, we have shown that plasma cfDNA levels are markedly elevated in a subset of patients with GBM, and that these higher levels are strongly and independently associated with worse prognosis. Further studies are needed to determine the mechanism of cfDNA release into the circulation of patients with GBM, as well as to elucidate the tissue and cell(s) of origin for plasma cfDNA in these patients. Methylation profiling of cfDNA samples may be useful in this regard.47–49 Ultimately, improved understanding of the biology of plasma cfDNA in GBM may lead to even more accurate prognostication and, more importantly, potential therapeutic opportunities.
This single-center study has several limitations, including a relatively modest sample size. However, the pre-surgical results for our cohort of 62 patients do confirm those in our previous pilot study,22 which was conducted in a separate cohort of 42 patients. While multicenter validation is needed, the reproducibility of the association of cfDNA with prognosis in 2independent cohorts of patients with GBM strengthens our findings. In addition, the cfDNA dynamics analysis was conducted post-hoc on a subset of 24 patients for whom data was available and was underpowered to perform multivariate adjustment for other clinical variables. Nonetheless, such striking prognostic data certainly bear further investigation. Finally, another potential limitation is that we excluded patients who received experimental front-line therapy and/or tumor-treating fields. While this was necessary to avoid the confounding effects of additional therapies received by a minority of patients, these exclusions have 2important implications. First, our results may not be generalizable to patients receiving therapies other than standard radiation plus temozolomide. Second, the uniform treatment of all patients in our cohort means that we cannot determine whether plasma cfDNA is a prognostic biomarker in IDH wild-type GBM (ie, retains prognostic value regardless of what treatment is administered), a predictive biomarker of benefit from radiation plus temozolomide, or both. Future studies of cfDNA involving patients receiving treatments other than radiation and temozolomide are needed to address these issues.
Conclusions
cfDNA has the potential to be an early and noninvasive prognostic tool in patients with GBM. Our data suggest it may also have value in monitoring response to chemoradiotherapy. The strength of our findings supports further investigation of cfDNA as a prognostic biomarker in the setting of a multicenter validation study, which is currently being planned. cfDNA may ultimately be used for more accurate prognostication in clinical practice, as well as more refined stratification of patients in GBM clinical trials. Additional studies are needed to understand the mechanism through which elevated cfDNA is associated with poor prognosis in GBM.
Supplementary Material
Acknowledgments
We thank the patients and their families for participating in the study. We are grateful to all study investigators, nurses, and supporting staff for providing care to the patients and data management.
Funding
This study was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR001878 to S.J.B. and E.L.C.). Paul Calabresi Career Development Award for Clinical Oncology, National Institutes of Health K12 grant (K12 CA076931 to S.J.B.). Glioblastoma Translational Center of Excellence from the Abramson Cancer Center (S.J.B., Z.A.B., S.B., A.S.D., D.O., S.A.N., E.L.C.). Abramson Cancer Center Core grant (P30-CA016520 to Q.L. and E.L.C.).
Meeting presentation: This research was presented as an oral presentation at the 2020 American Society of Clinical Oncology Annual Meeting: May 30, 2020: Virtual Meeting.
Conflict of interest statement. S.J.B. reported receiving research funding to his institution from Eli Lilly and Company, Incyte Corp, GlaxoSmithKline, and Novocure; performing consultative services for Bayer and Novocure outside the submitted work. Q.L. reported receiving research funding to his institution from Bayer and Pfizer. E.L.C. reported receiving research funding to her institution from Merck, Becton-Dickinson, and Janssen Pharmaceuticals. S.A.N. reported receiving research funding to his institution from Blue Earth Diagnostics.
Authorship Statement. Concept and design: S.J.B., Till, S.A.N., and E.L.C. Acquisition, analysis, or interpretation of data: S.J.B., J.T., A.A., H.K.S., T.A.B., J.H., S.A.N., and E.L.C. Drafting of the manuscript: S.J.B., J.T., A.A., J.F., E.L.C. Critical revision of the manuscript for important intellectual content: Z.A.B., S.B., A.S.D., D.M.O., and E.L.C. Statistical analysis: S.J.B., J.T., and Q.L. Obtained funding: S.J.B., D.M.O., Q.L., and E.L.C. Administrative, technical, or material support: A.A., H.K.S., Yee, T.A.B., J.H., Z.A.B., and S.A.N.. Supervision: S.J.B., J.T., A.S.D., and E.L.C.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Wen PY, Weller M, Lee EQ, et al. Glioblastoma in adults: a society for neuro-oncology (SNO) and European society of neuro-oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22(8):1073–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burgenske DM, Yang J, Decker PA, et al. Molecular profiling of long-term IDH-wildtype glioblastoma survivors. Neuro Oncol. 2019;21(11):1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson RM, Phillips HS, Bais C, et al. Development of a gene expression-based prognostic signature for IDH wild-type glioblastoma [published online ahead of print, 2020 Sep 8]. Neuro Oncol. 2020;noaa157. doi: 10.1093/neuonc/noaa157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Louis DN, Perry A, Reifenberger G, et al. The 2016 World health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 8. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oh S, Yeom J, Cho HJ, et al. Integrated pharmaco-proteogenomics defines two subgroups in isocitrate dehydrogenase wild-type glioblastoma with prognostic and therapeutic opportunities. Nat Commun. 2020;11(1):3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mansouri A, Hachem LD, Mansouri S, et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019;21(2):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malmström A, Łysiak M, Kristensen BW, Hovey E, Henriksson R, Söderkvist P. Do we really know who has an MGMT methylated glioma? Results of an international survey regarding use of MGMT analyses for glioma. Neurooncol Pract. 2020;7(1):68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanguturi SK, Trippa L, Ramkissoon SH, et al. Leveraging molecular datasets for biomarker-based clinical trial design in glioblastoma. Neuro Oncol. 2017;19(7):908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prados MD, Byron SA, Tran NL, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro Oncol. 2015;17(8):1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skaga E, Kulesskiy E, Fayzullin A, et al. Intertumoral heterogeneity in patient-specific drug sensitivities in treatment-naïve glioblastoma. BMC Cancer. 2019;19(1):628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pollom EL, Sborov KD, Soltys SG, Asch SM, Sudore RL, Aslakson RA. Advance care planning needs in patients with Glioblastoma undergoing radiotherapy. J Pain Symptom Manage. 2018;56(6):e6–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fritz L, Zwinkels H, Koekkoek JAF, et al. Advance care planning in glioblastoma patients: development of a disease-specific ACP program. Support Care Cancer. 2020;28(3):1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 20. Aggarwal C, Thompson JC, Black TA, et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2019;5(2):173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turner NC, Kingston B, Kilburn LS, et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020;21(10):1296–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bagley SJ, Nabavizadeh SA, Mays JJ, et al. Clinical utility of plasma cell-free DNA in adult patients with newly diagnosed Glioblastoma: a pilot prospective study. Clin Cancer Res. 2020;26(2):397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller Bark J, Kulasinghe A, Chua B, Day BW, Punyadeera C. Circulating biomarkers in patients with glioblastoma. Br J Cancer. 2020;122(3):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res. 2016;22(23):5772–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nygaard AD, Holdgaard PC, Spindler KL, Pallisgaard N, Jakobsen A. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer. 2014;110(2):363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7(9):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Valpione S, Gremel G, Mundra P, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer. 2018;88:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Phallen J, Leal A, Woodward BD, et al. Early noninvasive detection of response to targeted therapy in non-small cell lung cancer. Cancer Res. 2019;79(6):1204–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raja R, Kuziora M, Brohawn PZ, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with Durvalumab. Clin Cancer Res. 2018;24(24):6212–6222. [DOI] [PubMed] [Google Scholar]
- 31. Goldberg SB, Narayan A, Kole AJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res. 2018;24(8):1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anagnostou V, Forde PM, White JR, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res. 2019;79(6):1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37(6):E11. [DOI] [PubMed] [Google Scholar]
- 34. Burger PC, Kleihues P. Cytologic composition of the untreated glioblastoma with implications for evaluation of needle biopsies. Cancer. 1989;63(10):2014–2023. [DOI] [PubMed] [Google Scholar]
- 35. Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiographic response assessment in Glioblastoma clinical trials. Neurotherapeutics. 2017;14(2):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ellingson BM, Wen PY, van den Bent MJ, Cloughesy TF. Pros and cons of current brain tumor imaging. Neuro Oncol. 2014;16(Suppl 7:vii2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Mieghem E, Wozniak A, Geussens Y, et al. Defining pseudoprogression in glioblastoma multiforme. Eur J Neurol. 2013;20(10):1335–1341. [DOI] [PubMed] [Google Scholar]
- 38. Brandes AA, Franceschi E, Tosoni A, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26(13):2192–2197. [DOI] [PubMed] [Google Scholar]
- 39. Taal W, Brandsma D, de Bruin HG, et al. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer. 2008;113(2):405–410. [DOI] [PubMed] [Google Scholar]
- 40. Akbari H, Rathore S, Bakas S, et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. 2020;126(11):2625–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamfjord J, Guren TK, Dajani O, et al. Total circulating cell-free DNA as a prognostic biomarker in metastatic colorectal cancer before first-line oxaliplatin-based chemotherapy. Ann Oncol. 2019;30(7):1088–1095. [DOI] [PubMed] [Google Scholar]
- 42. Viller Tuxen I, Barlebo Ahlborn L, Mau-Soerensen M, et al. Plasma total cell-free DNA is a prognostic biomarker of overall survival in metastatic solid tumour patients. Br J Cancer. 2019;121(2):125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fernandez-Garcia D, Hills A, Page K, et al. Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res. 2019;21(1):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Batth IS, Mitra A, Manier S, et al. Circulating tumor markers: harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann Oncol. 2017;28(3):468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li BT, Drilon A, Johnson ML, et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol. 2016;27(1):154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu X, Ren J, Luo N, et al. Comprehensive DNA methylation analysis of tissue of origin of plasma cell-free DNA by methylated CpG tandem amplification and sequencing (MCTA-Seq). Clin Epigenetics. 2019;11(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moss J, Magenheim J, Neiman D, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.