Dear Editor,
Spatial transcriptomics (ST) makes it possible to perform RNA-seq at hundreds of precisely located spots on the surface of a histological slice (Ståhl et al., 2016). Since mRNA diffusion is minimal during tissues permeabilization and mRNA capture, the transcriptome of each spot is thought to aggregate the transcriptomes of the cells it contains. The number of cells within a spot and their transcriptional output depend on their type, state, and overall local morphology. ST shares some limitations with single-cell RNA-seq, including high dropout rate. So far, ST studies have relied on preprocessing pipelines inspired by single-cell RNA-seq studies (Ståhl et al., 2016; Asp et al., 2017; Giacomello et al., 2017; Berglund et al., 2018; Lundmark et al., 2018). These include normalization of gene-wise read counts in a cell/spot by the total number of reads collected from that cell/spot. But the number of reads obtained from a spot could reflect its cellular content or technical variation in RNA capture and amplification. Thus, whether read count normalization is warranted in the context of ST remains an open question. We addressed it by quantifying the cellular content of individual spots from image analysis and by comparing it with read counts.
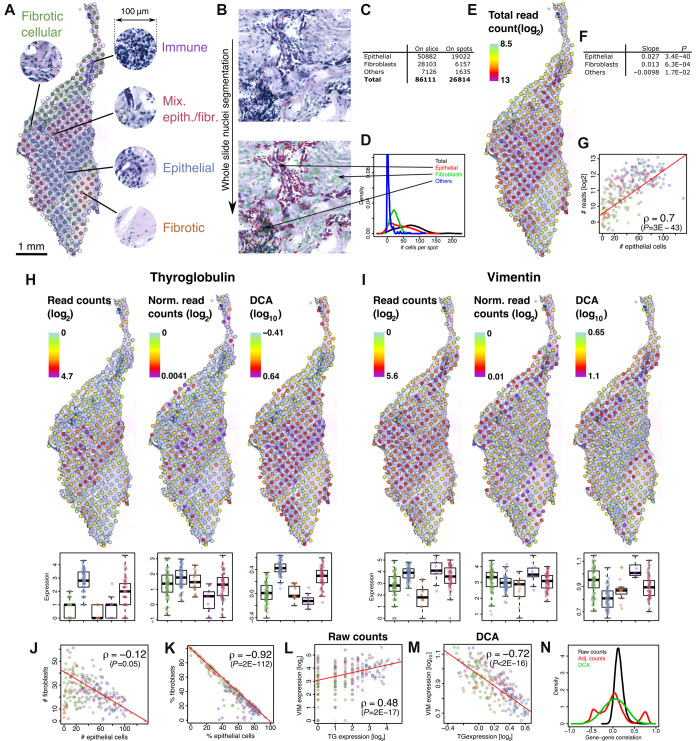
A BRAF V600E-mutated papillary thyroid cancer was profiled with ST (Supplementary material). Pathology review of the H&E image revealed five major types of morphologies (Figure 1A). This qualitative approach was complemented by whole-slide machine learning-based localization of nuclei, and their classification within three categories (Figure 1B): epithelial cells, fibroblasts, and ‘other cells’, which mostly contain immune cells. Among 86111 detected nuclei (Figure 1C), 31% were located within ST spots. The mean number of cells per spots varied from 0 to 197 (median 67).
Figure 1.
Variation of total read counts is related to morphology and number of cells of different types. (A) Five types of morphological regions were determined from pathology. The transcriptome was measured in each spot with ST. (B) The nuclei on the H&E image were segmented and classified with a machine learning-based algorithm (Supplementary material). (C) Nuclei counts. (D) Distributions of the number of cells per ST spot. (E) Total read count per spot. (F) Multivariate analysis of association between cell numbers and the log2 of total read count per spot. (G) Each point represents a ST spot with same color code as in A. ρ denotes the Spearman’s correlation. (H and I) Expression of TG (H) and VIM (I) without normalization (left), with adjustment for total read counts (center), and with DCA normalization (right). Boxplots represent the expression of spots in the regions shown in A (same color code). (J and K) Points represent spots (same color code as before) with the absolute number (J) and cell-type proportion (K) of epithelial cells (x-axis) and fibroblasts (y-axis). The large negative correlation stems from the low number of ‘others’ in C. (L and M) Expressions of TG and VIM are compared using raw counts or auto-encoder-based normalization. (N) Distribution of gene–gene correlation across spots for raw counts and normalized data.
Spot-wise read coverage varied from 356 to 8749 across the slide (Figure 1E), a 25-fold variation. It was associated with the number of cells of all types in a multivariate analysis (Figure 1G), particularly epithelial cells (Figure 1F). As expected, total read count per spots was highest in dense epithelial areas and lowest in low cell density fibrotic zones (Figure 1E and G). We concluded that total read counts per spot reflects relevant quantitative and qualitative features of tissue morphology.
To assess the effect of normalization, we compared raw read counts with raw counts normalized for total counts and with scale-normalized expression estimates generated by the Deep Count Autoencoder (DCA; Eraslan et al., 2019), a neural network-based algorithm developed in the context of single-cell RNA-seq. Figure 1H and I show expression of thyroglobulin (TG, a thyroid differentiation marker) and vimentin (VIM, a mesenchymal intermediate filament), respectively. Raw counts and normalized expression of TG all closely followed epithelial density and total counts (compare Figure 1H to Figure 1A and E, and see also Supplementary Figure S1). VIM raw counts were substantial in the epithelial areas but also in the cellular fibrosis and immune foci. Normalization, however, revealed a dramatically different picture, particularly for DCA: while remaining high in cellular fibrosis and immune foci, VIM expression was lower in epithelium (Figure 1I; Supplementary Figure S1).
Thus, normalization affected the spatial expression pattern of VIM, but not TG. The absolute numbers of epithelial cells and fibroblasts per spot were weakly associated (Figure 1J), while their relative proportions, i.e. their number divided by the total number of cells within a spot, were massively anti-correlated (Figure 1K). The raw counts of TG and VIM are positively correlated (Figure 1L), while their normalized values are negatively correlated (Figure 1M). The positive correlation between TG and VIM raw counts (Figure 1L) suggests that the tumoral epithelium expresses VIM and could undergo an epithelial–mesenchymal transition. This transition has been reported in BRAF V600E-mutated tumors (Knauf et al., 2011) such as this one. VIM is also expressed in primary cultures of normal thyrocytes treated with epidermal growth factor, which inhibits differentiation, but not of thyrocytes treated with thyroid-stimulating hormone, which promotes thyroid differentiation—while both are mitogenic (Coclet et al., 1991). Alternatively, VIM expression by fibroblasts could be promoted by nearby epithelial cells. Overall, raw counts seemed more related to the number of cells of a given cell type, while normalized expression captured cell types’ relative proportions.
To rule out possible artifacts related to TG and VIM in this particular tissue slice, we reproduced the above analysis (i) to the thyroid-stimulating hormone receptor (TSHR) and collagen III α1 (COL3A1) in another slice of the same thyroid cancer (Supplementary Figure S2) and (ii) to the estrogen receptor (ESR1) and VIM in a publicly available breast cancer slice profiled on the recent Visium platform (10x Genomics; Supplementary Figure S3). Taken together, these controls support the generality of the effect of normalization on the relation between epithelial differentiation and mesenchymal markers and their relevance to Visium slides, which have a 4-fold higher resolution than the first generation of ST slides shown in Figure 1.
To gain insights on the global effect of normalization, we calculated the distribution of gene−gene correlations across spots for all three expression metrics (Figure 1N). Raw counts were positively correlated for most pairs of genes. By contrast, normalized expression correlations were centered on 0. This implies that genes tend to show a similar expression pattern that reflects total transcriptional output when raw counts are considered, while normalized expression highlights contrasts between genes.
We showed that the variation of total read counts is largely determined by local cell density in ST data. Thus, total counts per spot are biologically informative and do not necessarily need to be normalized out. Importantly, normalization and denoising are technically independent operations. For example, DCA estimates read count scale factors, but users are free to use its scale-adjusted or un-adjusted outputs (Eraslan et al., 2019). Our study shows that both options are valid but address different purposes.
Raw read counts inform about the absolute density of cell types, while normalized expression informs about their relative proportions. It is remarkable that normalized expression better detects specific morphologies such as pure epithelium and cellular fibrosis (see boxplots in Figure 1H and I, and compare the DCA panels in Figure 1H and I with the pathology annotation in Figure 1A), while raw counts do reflect actual cell-type local densities and may highlight atypical expression patterns, as exemplified here for VIM in regions of high epithelial density.
The resolution of commercially available ST will eventually reach sub-cellular resolution (Vickovic et al., 2019). Given that some cells, e.g. cancer cells, produce more RNA than others (Lovén et al., 2012), it begs the question of to what extent our argument also applies at single-cell level. Cell-level phenotypic information measured independently of transcription must be available together with matched cell transcriptomes in order to unambiguously address this question.
[Supplementary material is available at Journal of Molecular Cell Biology online. Code and data are available at https://github.com/vdet/st-normalization. We thank Annelie Mollbrink and Jose Navarro for help with the ST protocol and bioinformatics. This work was supported by the ‘Les Amis de l’Institut Bordet’, Fondation Naets (J1813300), the Fondation Belge Contre le Cancer (2016-093), and Fond National de la Recherche Scientifique (FNRS; U.N019.19 and J006120F). M.S. is supported by FNRS and J.R.-V. is supported by the Fonds National de la Recherche, Luxembourg (11587122). M.S. performed experiments with support of J.L. and A.T. J.R.-V. and V.D. performed computational analysis. L.C., A.S., and D.L. handled sample banking and pathology review. G.A. resected the tumor. C.M. and V.D. supervised the research and wrote the manuscript.]
Supplementary Material
References
- Asp M., Salmén F., Ståhl P.L., et al. (2017). Spatial detection of fetal marker genes expressed at low level in adult human heart tissue. Sci. Rep. 7, 12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund E., Maaskola J., Schultz N., et al. (2018). Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 9, 2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coclet J., Lamy F., Rickaert F., et al. (1991). Intermediate filaments in normal thyrocytes: modulation of vimentin expression in primary cultures. Mol. Cell. Endocrinol. 76, 135–148. [DOI] [PubMed] [Google Scholar]
- Eraslan G., Simon L.M., Mircea M., et al. (2019). Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun. 10, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello S., Salmén F., Terebieniec B.K., et al. (2017). Spatially resolved transcriptome profiling in model plant species. Nat. Plants 3, 17061. [DOI] [PubMed] [Google Scholar]
- Knauf J.A., Sartor M.A., Medvedovic M., et al. (2011). Progression of BRAF-induced thyroid cancer is associated with epithelial–mesenchymal transition requiring concomitant MAP kinase and TGFβ signaling. Oncogene 30, 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Orlando D.A., Sigova A.A., et al. (2012). Revisiting global gene expression analysis. Cell 151, 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark A., Gerasimcik N., Båge T., et al. (2018). Gene expression profiling of periodontitis-affected gingival tissue by spatial transcriptomics. Sci. Rep. 8, 9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl P.L., Salmén F., Vickovic S., et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82. [DOI] [PubMed] [Google Scholar]
- Vickovic S., Eraslan G., Salmén F., et al. (2019). High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.