Abstract
Post-transcriptional methylation of N6-adenine and N1-adenine can affect transcriptome turnover and translation. Furthermore, the regulatory function of N6-methyladenine (m6A) during heat shock has been uncovered, including the enhancement of the phase separation potential of RNAs. In response to acute stress, e.g. heat shock, the orderly sequestration of mRNAs in stress granules (SGs) is considered important to protect transcripts from the irreversible aggregation. Until recently, the role of N1-methyladenine (m1A) on mRNAs during acute stress response remains largely unknown. Here we show that the methyltransferase complex TRMT6/61A, which generates the m1A tag, is involved in transcriptome protection during heat shock. Our bioinformatics analysis indicates that occurrence of the m1A motif is increased in mRNAs known to be enriched in SGs. Accordingly, the m1A-generating methyltransferase TRMT6/61A accumulated in SGs and mass spectrometry confirmed enrichment of m1A in the SG RNAs. The insertion of a single methylation motif in the untranslated region of a reporter RNA leads to more efficient recovery of protein synthesis from that transcript after the return to normal temperature. Our results demonstrate far-reaching functional consequences of a minimal RNA modification on N1-adenine during acute proteostasis stress.
Keywords: stress response, stress granules, N1-methyladenine
Introduction
Polypeptide-coding mRNAs are coated by specialized proteins at any stage of their lifecycle (Singh et al., 2015). Although mRNAs lose the associated proteins during ribosomal translation, a particular arrangement of ribosomes protects naked RNA from interactions with other molecules (Brandt et al., 2010). One of few circumstances when free mRNA appears in the cytosol is the disassembly of polysomes during stress-induced shutdown of protein synthesis (Bounedjah et al., 2014). Under these conditions, mRNAs are able to associate with phase-separating proteins, an initial step of membrane-less RNA–protein granule formation (Alriquet et al., 2019). Stress granules (SGs), induced by heat shock and other misfolding-promoting stressors, are a prime example of such membraneless assemblies that grow, collapse, and fuse to serve a number of purposes (Protter and Parker, 2016).
Through engagement in interactions with proteins, RNA is able to affect the formation and composition of phase-separated multi-molecular assemblies (Shevtsov and Dundr, 2011; Zhang et al., 2015; Garcia-Jove Navarro et al., 2019; Sanchez de Groot et al., 2019). Recent in vitro and in vivo models provided evidence for RNA-dependent granulation (Han et al., 2012; Maharana et al., 2018). The effect on protein sequestration is not restricted to coding transcripts and, indeed, intergenic noncoding RNAs have been shown to drive the assembly of subnuclear A-bodies (Wang et al., 2018). In addition to RNA–protein associations, also RNA–RNA interactions are emerging as an important determinant of cellular phase separations. It was recently found that RNAs alone, without the involvement of proteins, can phase-separate in case they contain diseases-associated extensions of CAG, CUG, and GGGGCC motifs (Jain and Vale, 2017). Furthermore, purified cellular RNAs can form assemblies in vitro analogous to SGs (Van Treeck et al., 2018). The secondary structure of involved mRNAs was shown to determine the molecular composition of Whi3 droplets in Ashbya cells (Langdon et al., 2018) by driving RNA self-association.
Next to the role of protein-coding by mRNAs, different classes of RNAs have additional cellular functions arising from their structural features (Ganser et al., 2019). This additional functionality is significantly extended by the post-transcriptional modifications of the transcripts (Harcourt et al., 2017; Nachtergaele and He, 2018). While the first modifications on mRNAs were identified long ago, their involvement in cellular processes has just recently begun to be revealed (Davalos et al., 2018). N6-adenine methylation (m6A) is the most prevalent mRNA modification and is involved in the regulation of the stability of its containing transcripts (Wang et al., 2014; Huang et al., 2018) and their translation (Meyer et al., 2015; Zhou et al., 2015). A complex epitranscriptome machinery of m6A writers and erasers has evolved to sustain the dynamic character of mRNA tagging. The machinery involves the core methyltrasferase complex METTL3/METTL14 and the two demethylating dioxygenases FTO and ALKBH5 (Wu et al., 2017). The methylation effects are mediated through a number of m6A tag readers from the families of YTH domain-containing proteins (YTHDC1 and YTHDF1–YTHDF3) and insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1–IGF2BP3) (Nachtergaele and He, 2018).
Although methylation of N1-adenine (m1A) is less prevalent, it produces a strong effect on RNA structure. By introducing a positive charge, the methyl group on N1-adenine disturbs Watson–Crick base pairing and leads to local duplex melting. The changes of folding equilibria through the m1A have been well characterized in short RNAs (Voigts-Hoffmann et al., 2007; Zhou et al., 2016). However, the structural and functional role of m1A in protein-coding RNAs is yet poorly understood. In regard to m1A on mRNAs, TRMT6/61A, TRMT61B, and TRMT10C were implicated as writers (Li et al., 2017; Safra et al., 2017) and ALKBH3 was shown to be the respective demethylase (Dominissini et al., 2016; Li et al., 2016). YTHDF1–YTHDF3 and YTHDC1 have been suggested as m1A readers (Dai et al., 2018; Seo and Kleiner, 2020). As expected, m1A in coding sequences of mRNAs affected the translation of methylated transcripts (Li et al., 2017; Safra et al., 2017). In addition, m1A is a key modification of several other classes of RNAs, including the large ribosomal subunit 28S rRNA (Sloan et al., 2017). From ~90 modifications present on tRNA molecules, m1A was found on five different positions including A58, which in humans is methylated by the heterodimer TRMT6/61A RNA methyltransferase (Oerum et al., 2017). Many human tRNA species are hypomodified at A58 with exception of the initiator tRNA (Saikia et al., 2010). The eukaryotic initiator tRNA forms a hydrogen-bonded cluster involving N1-methylated A58, which is critical for the stability of the molecule. Interestingly, the deletion of the ALKBH1 demethylase which removes the methyl group from A58 leads to the increased cellular level of initiator tRNA (Liu et al., 2016) thus indicating at the dynamic character of this modification.
Dynamic nature of RNA modifications and their impact on RNA structure and on RNA–protein interactivity make them well suited to regulate RNA granulation into ribonucleoprotein assemblies following environmental and developmental cues. Specifically, m6A readers contain low-complexity amino acid sequences that can drive phase separation of proteins. For example, YTHDF1 was shown to localize with SGs upon arsenite treatment in an RNA-dependent manner (Wang et al., 2015). Similarly, arsenite was shown to induce m6A in the 5ʹ vicinity of the transcripts, which is then utilized by YTHDF3 to direct modified RNAs to SGs (Anders et al., 2018). YTHDF1, YTHDF2, and YTHDF3 can phase-separate in vitro and in vivo and this separation is enhanced by mRNAs with multiple m6As (Ries et al., 2019).
Conformational stress, such as acute heat shock or aggregation of mutant proteins, can disturb proteome homeostasis, the proteostasis (Vabulas et al., 2010; Labbadia and Morimoto, 2015; Hipp et al., 2019). Under these conditions, aggregation of proteins into reversible assemblies and phase-separated membrane-less compartments are thought to prevent proteome damage and promote cellular fitness (Wallace et al., 2015; Audas et al., 2016; Rabouille and Alberti, 2017; Franzmann et al., 2018). On the other side, faulty aggregation of proteins was shown to contribute to cytotoxicity (Bolognesi et al., 2016; Mateju et al., 2017). RNA is the key component of phase-separated cellular compartments, yet how it contributes to the proteostasis is less clear. Notably, significant amounts of protein-free mRNA can be released from polysomes into cytosol during stress. In stressed lysates, N1-adenine methyltransferase TRMT6/61A was found to be the top interactor with free mRNA (Alriquet et al., 2019). In this work, we establish the molecular link between RNA and protein homeostasis, which involves the effect of TRMT6/61A and its product m1A on stress-induced granulation.
Results
The m1A motif is enriched in SG mRNAs
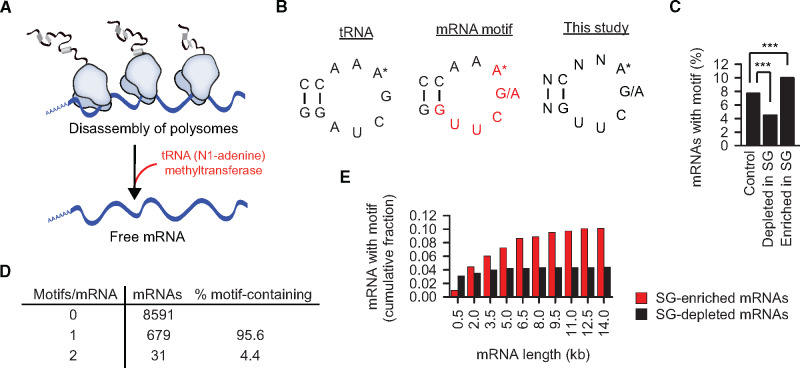
In the previous work, we analyzed interactors of free RNA in heat-shocked HeLa lysates (Alriquet et al., 2019). The TRMT6 and TRMT61A subunits of the methyltrasferase complex were found enriched over the background on average 42- to 88-fold, respectively, and in vitro analysis proved the capacity of the complex to bind mRNAs directly (Figure 1A). We set out to investigate the role of this interaction in detail. Two recent studies suggested TRMT6/61A as an m1A writer targeting mRNAs (Li et al., 2017; Safra et al., 2017), although initially the heterodimer was described as the N1-adenine methyltrasferase modifying adenine 58 in the T-loop of tRNAs (Anderson et al., 1998; Ozanick et al., 2005). Thus not surprisingly, a fraction of the m1A-containing mRNAs displays a sequence motif resembling that used by TRMT6/61A in tRNAs (Li et al., 2017; Safra et al., 2017; Figure 1B, left and middle). We took advantage of a comprehensive list of mRNAs enriched in mammalian SGs to analyze the occurrence of the m1A motif in this dataset (Khong et al., 2017). The stability of oligonucleotide loops is known to depend on the size of the stem formed by the complementary strands. We chose stem-length of two base pairs in our analysis (Figure 1B, right). The analysis revealed that the TRMT6/61A-targeted transcripts are indeed significantly enriched in the SG-sequestered mRNAs (150 out of 1434) (Figure 1C). The presence of the methylation motif is an indirect, yet strong evidence of the role of m1A in granulation, especially, in combination with the TRMT6/61A recruitment and the m1A enrichment in SG as reported below. In 150 transcripts containing the m1A motif, two GO categories were significantly increased: ‘Regulation of RNA metabolic processes’ (1.7×, 59 proteins) and ‘Axonogenesis’ (4.6×, 13 proteins). The latter set (Supplementary Figure S1A) represents an intriguing hit, because the build-up of long neuronal axons requires packing and transport of RNA granules to the sites of local translation (Kiebler and Bassell, 2006) The enrichment of axonogenesis proteins suggests that the m1A-linked granulation might be a more general mechanism of the mRNA metabolism not restricted only to stress conditions. Structurally, the m1A-motif-containing transcripts are longer than control mRNAs (Supplementary Figure S1B) and the 3ʹ-untranlated regions (UTRs) are longer than those of the control RNAs. Differently from the m6A-driven granulation where multiple m6A tags are needed for sequestration of the modified mRNAs into SGs (Ries et al., 2019), 96% of the m1A motif-containing mRNAs have only one motif (Figure 1D). Interestingly, the difference in the fraction of motif-containing mRNAs between SG-enriched and -depleted sets increased with the increasing length of transcripts (Figure 1E).
Figure 1.
The m1A motif is enriched in SG mRNAs. (A) TRMT6/61A methyltrasferase binds free RNAs in heat-shocked cytosol of HeLa cells (Alriquet et al., 2019). (B) tRNA, TΨC arm of tRNA where adenine 58 (A*) is N1-methylated by TRMT6/61A. mRNA, mRNA motif (red) targeted by TRMT6/61A for adenine N1-methylation (Li et al., 2017). This study, motif used here to predict mRNA targets of TRMT6/61A-mediated adenine N1-methylation. (C) Fraction of mRNAs enriched or depleted in SGs (Khong et al., 2017) and containing the m1A motif: 150 out of 1434 and 60 out of 1353, respectively. Control, mRNA set neither enriched nor depleted in SG; ***P < 0.001, chi-square analysis. (D) The number of m1A motifs in the mRNAs analyzed in Figure 1C. (E) Cumulative fraction of mRNAs at different transcript size cutoff.
Partial knockout of TRMT6/61A sensitizes cells to heat shock and arsenite stress
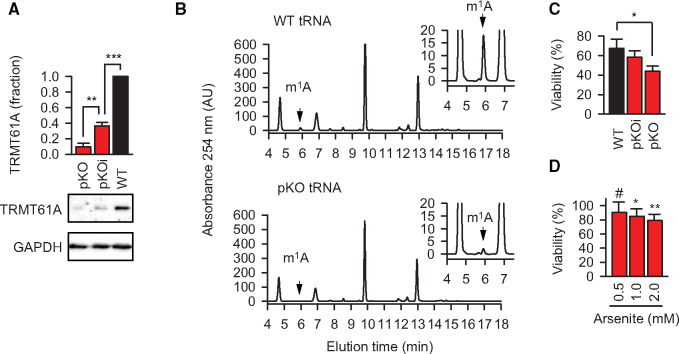
TRMT6 and TRMT61A belong to the ‘core’ essential genes in human cell lines, so that their deficiency is lethal (Blomen et al., 2015). To reduce the levels of the enzymatic subunit TRMT61A of the methyltrasferase, we used the CRISPR/Cas9 system in the aneuploidic HeLa cell line known to harbor up to six sets of chromosomes (Landry et al., 2013; Figure 2A). As intended, in gene-edited cells the amount of m1A in tRNAs was strongly, but not completely reduced; thus, they have been called partial knockout cells (Figure 2B; Supplementary Figure S2A). The residual methylation of tRNAs was obviously sufficient for translation, because protein synthesis and cellular viability in partial knockout cells were not significantly changed and the proliferation was only slightly slower (Supplementary Figure S2B–D). In contrast, cellular viability upon heat shock for 2 h was reduced and correlated with the levels of TRMT61A (Figure 2C). Likewise, TRMT61A activity was needed to protect cells from another acute proteostasis stressor, arsenite, an oxidative agent broadly used to induce SGs (Figure 2D). Noteworthy, SG formation during arsenite treatment was affected in the partial knockout cells (Figure 3A). SGs effect biological reactions by shifting the equilibria of interacting molecules toward associated states and by limiting the interactions of sequestered components with the bulk cytosol (Protter and Parker, 2016). Thus, impaired granulation in TRMT61A-deficient cells could be partially responsible for the reduced viability of the cells during acute stress.
Figure 2.
Partial knockout of TRMT6/61A sensitizes cells to heat shock and arsenite stress. (A) TRMT61A amount determined by western blotting. GAPDH was used as loading control. WT, wild-type HeLa cells; pKOi, partial knockout with intermediate level of TRMT61A; pKO, partial knockout with strong reduction of TRMT61A level. **P < 0.01, ***P < 0.001, two-tailed t-test; N = 3 independent experiments (mean + SD). (B) High-pressure liquid chromatography (HPLC) analysis of N1-methylated adenosine (m1A) from cellular tRNAs in wild-type HeLa cells (WT) and TRMT61A partial knockout cells (pKO). One representative out of three independent experiments is shown. (C) Reduced amounts of the methyltransferase correlate with the increased sensitivity to heat shock. WT, wild-type cells with normal amount of TRMT61A; pKO and pKOi, partial knockout cells with strongly reduced and intermediate amounts of TRMT61A, respectively. *P < 0.05, two-tailed t-test; N = 3 independent experiments (mean + SD). (D) Viability of TRMT61A knockout cells after arsenite treatment for 1 h in comparison (%) to wild-type cells at the same concentrations of arsenite. *P < 0.05, **P < 0.01, two-tailed t-test; #, not significant difference. N = 3 independent experiments (mean + SD).
Figure 3.
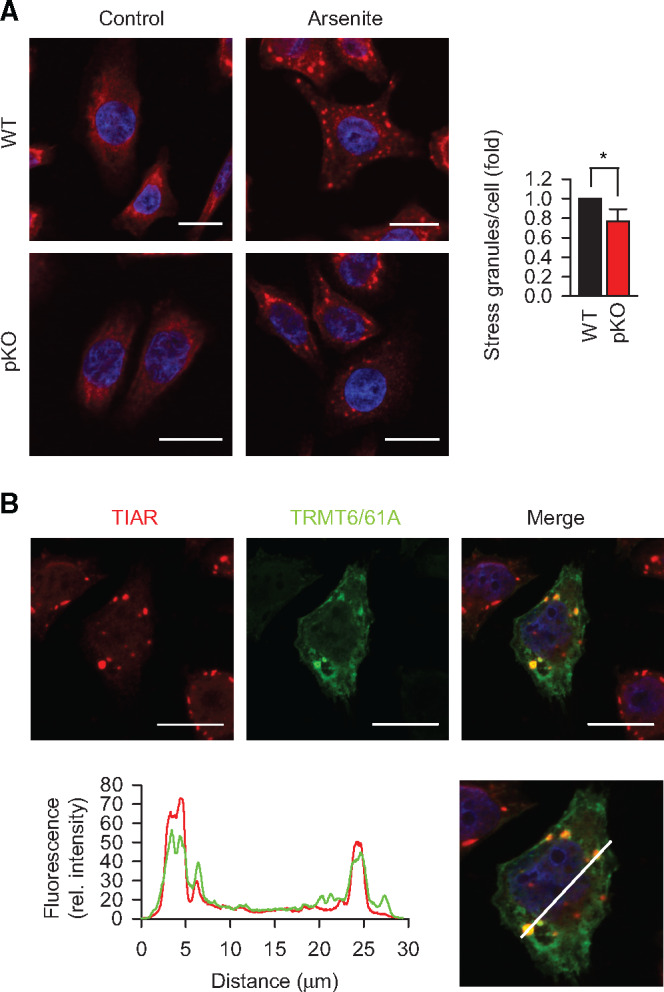
TRMT6/61A is involved in stressed-induced granulation. (A) Immunofluorescence staining of TIAR (red) to detect SG formation upon arsenite treatment for 30 min in serum-free medium. DAPI staining (blue), nuclei. Scale bar, 20 μm. WT, wild-type HeLa cells; pKO, partial knockout of TRMT61A. *P < 0.05, two-tailed t-test; N = 3 independent experiments (mean + SD). (B) Localization of TRMT6/61A in arsenite-induced SGs that are detected with anti-TIAR antibody. A representative image from three independent experiments. Scale bar, 20 μm. The white bar in the lower picture indicates the line where the fluorescence rel. intensity extracted. Red, TIAR; green, TRMT6/61A.
Molecular chaperones are key in sustaining cellular functions during proteostasis stress (Vabulas et al., 2010). Specifically, HSP70 was shown to be a part of the machinery which regulates SG composition and dynamics (Ganassi et al., 2016). We compared cellular levels of the constitutive HSC70 and the inducible HSP70 in wild-type and partial knockout cells and did not detect significant difference (Supplementary Figure S3A). The inducibility of HSP70 during heat shock was not affected either, arguing against the classical cellular damage due to defect in the molecular chaperone system.
m1A accumulates in SGs
In addition to its tRNA-related functions, TRMT6/61A has been proposed to act also as mRNA N1-adenine methyltransferase (Li et al., 2017; Safra et al., 2017), and m1A in mRNAs was shown to increase upon heat shock (Dominissini et al., 2016). These findings suggest that TRMT6/61A might be involved in adaptive RNA granulation during stress (Figure 3A). Indeed, we found that TRMT6/61A methyltransferase localizes to SG under stress indirectly pointing to the m1A involvement in granulation (Figure 3B; Supplementary Figure S3B). For visualization of SGs, we used antibodies against the SG marker TIAR, a protein involved in the assembly of mammalian granules (Kedersha et al., 1999). To quantify the accumulation of methyladenine in SGs by mass spectrometry (MS), we combined SG isolation (Khong et al., 2018) and targeted selected ion monitoring (tSIM) procedures. The original protocol from Roy Parker’s lab represents a procedure combining differential centrifugation and antibodies to purify SG cores. We used a simplified version omitting the affinity purification step, which yielded sufficient amount of TIAR-positive RNA–protein fraction for quantitative MS (Supplementary Figure S4A). Robust chromatography conditions together with high-resolution, accurate-mass performance of an Orbitrap analyzer allowed unambiguous identification and quantification of m1A and m6A (Figure 4A; Supplementary Figure S4B). Actually, the amount of m1A was underestimated under these conditions because of the partial conversion of m1A to m6A under alkaline conditions, which is known as Dimroth rearrangement (Macon and Wolfenden, 1968; Figure 4A, lower panel). A significant enrichment of m1A (calculated as m1A/A fraction) in granules was detected as compared to cytosolic mRNAs from 0.01% m1A in cytosolic mRNAs to 0.088% in SG RNA (Figure 4B and C). At the same time, the ratio m6A/A in SG RNA increased less compared to that in cytosolic mRNAs. The m1A signal from ribosomal RNA in SG can be ruled out, because the only ribosomal m1A in human ribosome is on the 60S subunit (Piekna-Przybylska et al., 2008; Sloan et al., 2017) which is not present in SGs (Kedersha et al., 2002). As there are 1218 unmodified adenines in human rRNA, the expected m1A fraction in ribosomes is 0.082%. This means that even in the presence of contaminating rRNA in SGs, mRNAs would contain higher than 0.082% m1A to reach the measured 0.088%. Furthermore, the insignificant contamination with tRNA can be inferred from the low variations of methyladenine amounts in our measurements (low standard deviations in Figure 4B): the ratio m1A/A in tRNA is very high and thus would readily manifest as large variation due to fluctuating levels of contaminating tRNA from prep to prep.
Figure 4.
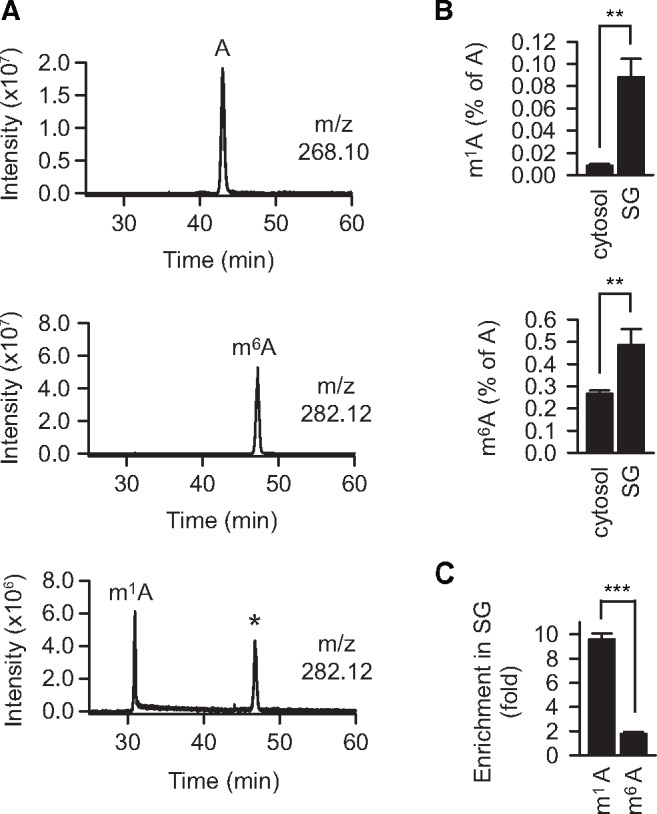
m1A accumulates in SGs. (A) Extracted ion chromatograms of indicated m/z with a range of ±0.002 Da. Top, a mixture of four standard ribonucleosides plus N1-methyladenosine and N6-methyladenosine; middle, N6-methyadenosine only; bottom, N1-methyladenosine only. A, adenosine. *, Dimroth rearrangement of m1A to m6A. (B) The fraction of methylated adenine in SG RNAs (SG) is increased compared to that in the cytosolic mRNA pool (cytosol) as measured by selective ion monitoring MS. **P < 0.01, two-tailed t-test. N = 3 independent experiments (mean + SD). (C) m1A enriched in SG RNA is stronger than m6A. m1A, fraction of N1-methyladenosine over unmodified adenosine; m6A, fraction of N6-methyladenosine over unmodified adenosine. The fractions of m1A and m6A in cytosolic mRNAs were set as 1. ***P < 0.001, two-tailed t-test; N = 3 independent experiments (mean + SD).
m1A safeguards mRNAs during heat shock
Both modifications of mRNA, m1A and m6A, have been shown to increase during heat shock (Zhou et al., 2015; Dominissini et al., 2016). We used tSIM MS to confirm this effect in our experimental system. The fraction m1A/A in cytosolic mRNAs increased from 0.009% to 0.012%, while the fraction of the more prevalent m6A increased from 0.27% to 0.38% (Supplementary Figure S5A). Although small, these changes were significant, which supports the notion of stress-related roles of both modifications.
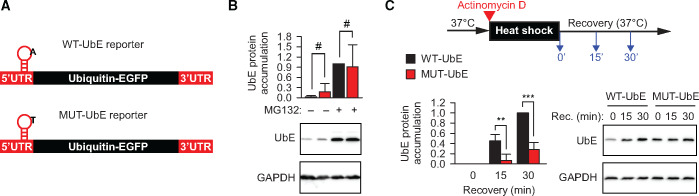
To test functionally whether the m1A-installing TRMT6/61A can protect mRNAs during proteostasis stress, m1A motif-containing reporters were prepared. The m1A motif from the 5ʹ-UTR of the PRUNE1 transcript was found to be N1-adenine methylated in two recent studies (Li et al., 2017; Safra et al., 2017) and to be significantly enriched in SGs 1.5-fold (Khong et al., 2017). We inserted the m1A motif into the 5ʹ-UTR of an Ubiquitin-EGFP (UbE) construct (Figure 5A; Supplementary Figure S5B). The UbE protein is highly unstable and its accumulation can be detected with the precision of several minutes upon inhibition of proteasomal degradation in HeLa cells (Vabulas and Hartl, 2005). Consequently, the accumulation of the UbE protein would indicate the functional amount of its coding mRNA in the cytosol from the time point when new mRNA synthesis and protein degradation are stopped. In addition to the wild-type reporter WT-UbE, a control reporter MUT-UbE was generated (Supplementary Figure S5B, lower sequence). MUT-UbE contains thymine (uracile in the transcribed RNA) instead of adenine in the motif loop such that methylation cannot take place. Under normal conditions, the accumulation of UbE protein encoded by the wild-type and mutant reporters did not differ significantly (Figure 5B).
Figure 5.
m1A safeguards mRNAs during heat shock. (A) Schematic depiction of the m1A motif-containing reporter. UTR, untranslated region. The position of the motif on the transcript is indicated. The nucleotide sequence of the motif is shown in Supplementary Figure S5B. (B) Accumulation of UbE from WT-UbE and MUT-UbE transcripts during 3 h-long inhibition of proteasomal degradation with 20 μM MG132 (+MG132). One representative anti-EGFP western blot from three independent experiments is shown. GAPDH was used as loading control. #, not significant difference; two-tailed t-test; N = 4 independent experiments (mean + SD). (C) Accumulation of UbE protein from WT-UbE and MUT-UbE transcripts during recovery after heat shock in the presence of actinomycin D to block synthesis of new mRNA molecules. UbE amount at time point 0 was subtracted from all values during recovery. One representative anti-EGFP western blot is shown. GAPDH was used as loading control. ***P < 0.001, **P < 0.01, two-tailed t-test; N = 4 independent experiments (mean + SD).
To test the m1A reporters during heat shock and recovery, transcription of new mRNA was inhibited with actinomycin D, which resulted in comparable amounts of the respective transcripts during the experimental time scale (Supplementary Figure S5C). The heat-misfolded proteome overloaded the degradation capacity of the proteasome leading to small accumulation of proteins from both reporters as seen by the residual amounts of UbE protein at the recovery time point 0 (Figure 5C, lanes 1 and 4). Notably, the amount of UbE protein from MUT-UbE at the recovery time point 0 was consistently higher, which suggests that the heat-induced shutdown of the MUT-UbE mRNA translation was less efficient and took longer time. By contrast, upon returning to 37°C, the protein from the WT-UbE reporter accumulated significantly faster (Figure 5C). These data indicate that the m1A motif-containing mRNAs (i) were removed from the translatable pool more efficiently upon onset of heat shock and (ii) during recovery became again functional (translatable) faster.
Discussion
The main effect of m1A is the block of base pairing, which induces local melting in RNA molecules (Zhou et al., 2016). Considering that m1A increases on mRNAs during stress (Dominissini et al., 2016) and that the secondary RNA structure is able to determine the specificity of stress-related associations (Langdon et al., 2018; Sanchez de Groot et al., 2019), we hypothesized that the impairment of TRMT6/61A activity might lead to aberrant interactions. Thereby, the following mechanistic scenario can be assumed: (i) less of the orderly sequestration of mRNAs during proteostasis stress, (ii) increased coaggregation of unsequestered mRNAs with stress-misfolded proteins, (iii) increased recruitment of mRNA-binding proteins to the coaggregated mRNAs, (iv) co-aggregation of the recruited mRNA-binding proteins. RNA repeat expansion disorders are known cases of clinical consequences following the entrapment of RNA-binding proteins (La Spada and Taylor, 2010; Cid-Samper et al., 2018). Our results indicate that also normal RNAs can get involved in similar pathogenic loops, if those RNAs are not processed and sequestered properly during stress.
There are several arguments in support to the requirement of TRMT6/61A for mRNAs during proteostasis stress. (i) In stressed HeLa cytosol, TRMT6/61A was found to be the top interactor with free mRNA (Alriquet et al., 2019). TRMT6 and TRMT61A were enriched on mRNA bait 42-fold and 88-fold, respectively, while the median of all 79 significantly enriched proteins was as low as 7.3. (ii) The crystal structure of the human TRMT6/61A in complex with RNA revealed that the binding of RNA molecule is determined not by the motif loop alone, but instead is very extensive and involves the T-stem and the acceptor stem as well (Finer-Moore et al., 2015). This association is sequence-unspecific, because it relies on the nucleic acid backbone. (iii) The mRNA reporters differ by the methylation-targeted nucleotide only (T instead of A), yet this difference is sufficient to determine different fate of otherwise identical mRNAs during proteostasis stress (Figure 5C).
Our data uncovered a link between m1A and cellular reaction to proteostasis stress. One strategy to cope with severe variations of external and internal milieu is the attenuation of protein synthesis (Holcik and Sonenberg, 2005; Liu and Qian, 2014). As a consequence, significant amounts of free RNA can be released from disassembled polysomes, which then engage into non-native or stress-specific associations (Bounedjah et al., 2014; Alriquet et al., 2019). The large size and loose compaction of freed RNA make them susceptible to aberrant interactions with misfolded proteins that also accumulate under these conditions. The aberrant associations might lead to irreversible RNA–protein co-aggregates (Figure 6). An orderly and reversible sequestration of freed RNAs into RNA–protein granules has evolved to counteract the RNA–protein aggregation. Our data show that TRMT6/61A methyltransferase is involved in granulation and safeguarding of mRNAs during stress. How m1A contributes to RNA sequestration remains to be determined. The small size of the methyl tag on adenine as opposed to substantial cellular consequences suggests existence of specialized m1A reader proteins responsible for the effect.
Figure 6.
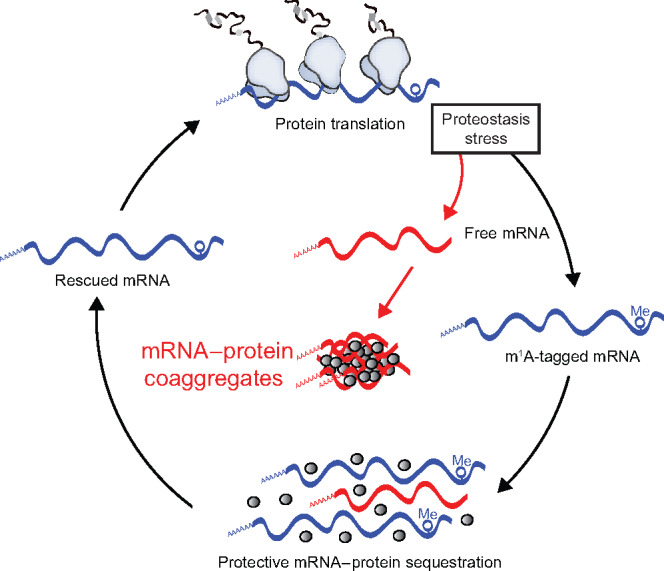
Different fates of free RNA during proteostasis stress. Polysome disassembly during stress releases free RNA into cytosol. The aberrant RNA–RNA and RNA–protein associations might lead to irreversible co-aggregates. We hypothesize that an orderly and reversible sequestration of free RNAs into RNA–protein granules has evolved to counteract the irreversible aggregation.
The diversity and importance of the epitranscriptome phenomena have become compellingly evident (Harcourt et al., 2017; Peer et al., 2017; Nachtergaele and He, 2018; Xiong et al., 2018), which created the pressing demand of new and reliable methods to quantify modified nucleotides with high sensitivity and resolution (Dominissini and Rechavi, 2017). Yet the heterogeneity of the epitranscriptome system is obvious even now. For example, m1A tagging of mRNA was shown to fall into three categories: cytosolic TRMT6/61A-dependent, TRMT6/61-independent and mitochondrial TRMT6/61B-dependent (Li et al., 2017). Under normal conditions, only ~10% of all m1A-containing mRNAs depend on TRMT6/61-driven methylation. It remains to be clarified whether the TRMT6/61-dependent subset could increase under stress conditions. The significantly stronger direct association of TRMT6/61A with RNAs in heated lysates supports this scenario (Alriquet et al., 2019). Likewise, the sequence-specificity of TRMT6/61A methylation might be modulated during stress, further increasing the complexity of mRNA modification in the cellular environment.
Materials and methods
Reagents, plasmids, and antibodies
2,3-Bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt (XTT), phenazine methosulfate (PMS), and cycloheximide were from Sigma-Aldrich, and TRIzol and RiboGreen were from Invitrogen. All other chemicals were from Sigma-Aldrich unless otherwise indicated.
Flag-tagged TRMT6 and TRMT61A mammalian expression vectors were purchased from Genscript. Two additional Flag tags where inserted into the vector coding TRMT61A. An m1A motif from human PRUNE1 5ʹ-UTR was introduced into the UbE construct (Vabulas and Hartl, 2005) to prepare WT-UbE reporter. The motif was mutated such that the to-be methylated adenine was exchanged into uracil to prepare MUT-UbE. Expression constructs generated for this study were prepared by standard molecular biology techniques and coding sequences were verified.
The following antibodies were used: anti-TRMT6 (A303-008A-M) from Bethyl; anti-TRMT61A (sc-107105) from Santa Cruz Biotechnology; anti-GAPDH (2118), anti-TIAR (8509), HRP-conjugated anti-rabbit-IgG (7074), and Alexa Fluor 647-conjugated anti-rabbit IgG (4414) from Cell Signaling; anti-Flag (F1804), anti-GFP (11814460001), and HRP-conjugated anti-mouse IgG (A9044) from Sigma-Aldrich.
Cell lines
The wild-type HeLa and TRMT61A partial knockout cell lines were cultured in DMEM supplemented with 10% (v/v) FBS, 2 mM L-glutamine, 100 IU/ml penicillin G, 100 μg/ml streptomycin sulfate, and nonessential amino acids. HeLa cells with low levels of TRMT61A (pKO) were prepared from wild-type HeLa cells by transfection with 1 μg TRM61A Double Nickase Plasmids (h) mix (SantaCruz Biotechnology) and selection with puromycin.
The B16-F10 cell line was cultured in DMEM supplemented with 10% (v/v) FBS, 2 mM L-glutamine, 100 IU/ml penicillin G, 100 μg/ml streptomycin sulfate, and nonessential amino acids.
Western blotting
Protein concentration of lysates used for western blotting was normalized and reducing SDS sample buffer was added. Samples were resolved using 10% SDS–PAGE and transferred onto nitrocellulose membranes. Membranes were blocked with 5% skim milk or 5% bovine serum albumin (BSA) in Tris-buffered saline/0.1% Tween 20, probed with the indicated antibodies and developed using the SuperSignal West Pico PLUS. Chemiluminescence images were acquired with the Chemidoc MP imaging system and bands quantified using the Image Lab 5.0 software.
Cellular viability assay
HeLa cells were seeded at 1.25 × 104 cells/well in duplicates in a 48-well plate in 200 μl DMEM supplemented with 25 mM Hepes–NaOH, pH7.5. The plates were immediately placed in an incubator at 37°C or 45°C for 2 h and then back to 37°C. After 14 h, medium in each well was discarded and replaced by 200 μl DMEM with 0.33 mg/ml XTT + 12.5 μg/ml PMS . After 2 h of incubation at 37°C, A475 and A600 were measured. Duplicates were averaged and specific absorbance calculated. To compare sensitivity to arsenite, wild-type HeLa and TRMT61A partial knockout cells were seeded in two 48-well plates at 1.25 × 104 cells/well in triplicates. Next day, the cells were treated with arsenite for 1 h at 37°C. The cells were washed with phosphate-buffered saline (PBS), and then 200 μl 0.33 mg/ml XTT + 12.5 μg/ml PMS in DMEM was added per well. After 4 h of incubation at 37°C, A475 and A600 were measured. Triplicates were averaged and specific absorbance was calculated.
WT-UbE and MUT-UbE reporter assay
HeLa cells were seeded in a 12-well plate at 1 × 105 cells per well. Next day, they were transfected with 300 ng reporter constructs using polyethylenimine (PEI; the ratio DNA:PEI was 1:6 using a 1 mg/ml PEI solution). After 24 h, the cells were treated with 20 μM MG-132 for 3 h, collected and analyzed by western blotting using anti-GFP and anti-GAPDH antibodies.
For the heat shock recovery assay, 12 × 106 HeLa cells were electroporated with 20 to 30 µg WT-UbE or MUT-UbE and seeded into two 10-cm dishes per transfection. After 5 h, cells were collected, washed, and resuspended in serum-free DMEM supplemented with 25 mM Hepes–NaOH, pH7.5 and 10 µM Actinomycin D. The cells were heated at 45°C for 30 min and then transferred to 37°C for recovery (time point 0). During recovery at time points 0, 15, and 30 min, aliquots were collected directly into the preheated SDS sample buffer and then analyzed by western blotting using anti-GFP and anti-GAPDH antibodies. To quantify the accumulation of newly synthesized reporter protein, the amount of UbE at time point 0 was subtracted.
Fluorescence microscopy
For TRMT6/61A and SG colocalization analysis, HeLa cells were electroporated with 15 µg of each Flag-tagged TRMT6 and TRMT61A and seeded at 0.5 × 106 cells/well in a 12-well plate on polylysine-coated slides. After 24 h, medium was replaced by serum-free DMEM for additional 2 h. The cells were treated with 0.25 mM arsenite for 30 min, washed with PBS and fixed with 3.7% paraformaldehyde/PBS for 9 min at room temperature (RT). They were then permeabilized with acetone at −20°C for 5 min, blocked with 1% BSA/PBS for 1 h at RT and incubated with anti-TIAR in 1% BSA/PBS at RT for 1 h. Incubation with anti-Rabbit IgG-AlexaFluor647 and anti-Flag M2-Cy3 was done in 1% BSA/PBS at RT for 1 h, followed by three washing steps with PBS and DAPI staining. Slides were mounted in PBS and imaged. ImageJ (Schindelin et al., 2012) was used to compare fluorescence profiles along a set line.
To compare SG formation upon arsenite treatment, wild-type HeLa and TRMT61A partial knockout cells were seeded in a 12-well plate with polylysine-coated cover slides at 1.5 × 105 cells/well. Next day, medium was replaced by serum-free medium and the cells were kept at 37°C for 2 h. Arsenite was then added at 0.0625 mM for 30 min at 37°C. Cells were washed with PBS, fixed with 3.7% paraformaldehyde at RT for 10 min, permeabilized with acetone at −20°C for 5 min, blocked with 1% BSA/PBS for 1 h at RT, and incubated with anti-TIAR for 1 h, followed by incubation with anti-Rabbit-IgG Alexa Fluor647 conjugate for 1 h, three washes with PBS, and staining with DAPI . Slides were imaged using a Zeiss LSM-780 inverted confocal microscope with a 63× oil immersion objective. The experiment was performed in triplicates and at least 200 cells were analyzed per condition and per repetition. Quantification was performed using CellProfiler by detecting nuclei and identifying by propagation from the nuclei. SGs were identified and counted within each cell.
Quantitative PCR
Total RNA was extracted with TRIzol. cDNAs were prepared using RevertAid kit and diluted 10×. Reporter and GAPDH sequences were amplified using respective primers and KiCqStart SYBR Green qPCR ReadyMix. The cycling conditions used were the following: 3 min at 95°C, 39× (15 sec 95°C, 30 sec 58°C, 15 sec 72°C). Each reaction was performed in triplicates and results were analyzed as described (Livak and Schmittgen, 2001) with GAPDH as a reference. Analysis was performed using the CFX Manager Software (Bio-Rad).
tRNA isolation for HPLC analysis
RNAs <200 nucleotides were extracted using mirVana miRNA Isolation kit. The extracted RNAs were separated in an 8% Polyacrylamide-TBE-urea gel, the tRNA band was visualized by UV shadowing and cut out of the gel. The gel pieces were incubated overnight with 0.5 M ammonium acetate at 25°C and 600 rpm. Next day, the gel pieces were discarded and 1 ml 100% ethanol was added per 400 μl of extract. The mixture was incubated at −80°C for 1 h and tRNAs were pelleted at 17000× g for 30 min at 4°C. Pellets were washed with 70% ethanol, air-dried, and frozen until further processing.
HPLC analyses
tRNA pellets were resuspended with 16 µl of MS grade water and digested to nucleosides by incubation with 0.1 U nuclease P1 and 0.1 U bacterial alkaline phosphatase BAP C75 in P1 buffer (20 mM NH4OAc, pH5.3) at 37°C for 1 h. Chromatographic separation was performed on an UltiMate 3000 HPLC (Thermo Fisher Scientific) equipped with a diode array detector (DAD). Five microliters of the nucleoside mixture were loaded on a Synergy Fusion RP column (4 mm particle size, 80 Å pore size, 250 mm length, 2 mm inner diameter) from Phenomenex with 100% buffer A (5 mM NH4OAc, pH5.3) and separated at 35°C with a flow rate of 0.35 ml/min using a linear gradient of buffer B (100% acetonitril) to 20% in 20 min, then to 40% in 2 min. At the end of the gradient the column was washed with 95% buffer B for 7 min and re-equilibrated with buffer A for 4 min. The nucleosides were monitored by recording the UV chromatogram at 254 nm.
Quantification of modified nucleotides by MS
SGs were prepared as described (Khong et al., 2018) omitting the affinity purification step. HeLa cells were treated with 0.5 mM Arsenite at 37°C for 60 min, washed, pelleted, and flash-frozen in liquid nitrogen. The pellets were thawed, resuspended in 500 µl lysis buffer (50 mM Tris–HCl, pH7.4, 100 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM DTT, 50 µg/ml Heparin, 0.5% NP40, protease inhibitor, and RNasin), passed seven times through a 26 G needle and centrifuged at 1000× g for 5 min at 4°C. The lysate was transferred to a new tube and centrifuged at 17000× g for 20 min at 4°C. The resulting supernatant was called ‘Soluble 1’. The pellet was resuspended in 500 µl lysis buffer and centrifuged again at 17000× g for 20 min at 4°C. The resulting supernatant was called ‘Soluble 2’. The pellet was resuspended in 150 µl lysis buffer, spinned at 850× g for 2 min at 4°C. The supernatant was called ‘SG pellet’ and constituted the SG core enriched fraction. Total RNA was extracted from the SG pellet by Trizol extraction and RNA precipitation and stored at −80°C until MS analysis.
PolyA mRNA for comparison was prepared from HeLa cells cultivated at 37°C. The cells were lysed in 500 µl lysis buffer (20 mM Tris–HCl, pH7.5, 500 mM LiCl, 0,5% LiDS, 1 mM EDTA, 5 mM DTT), incubated at RT for 5 min and passed 5× through a 20 G needle. The lysate was added to 100 μl of washed oligo-d(T)25 magnetic beads and incubated at RT for 10 min. The beads were washed twice with WB-I (20 mM Tris–HCl, pH7.5, 500 mM LiCl, 0.1% LiDS, 1 mM EDTA, 5 mM DTT), twice with WB-II (20 mM Tris–HCl, pH7.5, 500 mM LiCl, 1 mM EDTA), and once with low salt buffer (20 mM Tris–HCl, pH7.5, 200 mM LiCl, 1 mM EDTA). The beads were resuspended in 100 µl elution buffer (20 mM Tris–HCl, pH7.5, 1 mM EDTA), mRNAs were eluted by heating at 50°C for 2 min, precipitated and stored at −80°C until MS analysis.
RNA from SGs or cytosolic mRNAs from heated HeLa cells were resuspended in 10 μl of MS water and digested with 0.05 units of nuclease P1 (Wako) and 0.05 units of bacterial alkaline phosphatase (BAP C75, TaKaRa) in P1 buffer (20 mM ammonium acetate, pH5.3) for 1 h at 37°C. Samples were store at −20°C before MS.
Chromatographic separation of the nucleosides was performed with an Easy-nLC1000 on an in-house packed column (100 µm inner diameter, 50 cm length, 4 μm Synergi Fusion 80 Å pore size from Phenomenex using a gradient from mobile phase A (5 mM ammonium formate, pH5.3) to 32% mobile phase B (100% acetonitrile) for 50 min followed by a second step to 40% B for 8 min, with a flow rate of 200 nl/min.
Retention times were monitored using 10 nM solutions of nucleoside standards purchased from Sigma-Aldrich (main nucleosides) and Carbosynh (N1-methyladenosine and N6-methyladenosine).
Nucleosides were injected into a Q Exactive Plus mass spectrometer equipped with a Nanospray Flex Ion-Source and analyzed in the positive mode with a method consisting of full MS and targeted SIM scans. Full scans were acquired with AGC target value of 106, resolution of 70.000, and maximum injection time of 100 ms. Adenosine, N1-methyladenosine and N6-methyladenosine were monitored at the respective retention time with a 4-amu isolation window, AGC target value of 2 × 105, 140.000 resolution, and maximum injection time of 500 ms.
The correct identification of N1-methyladeosine and N6-methyladenosine was proved by fragmentation of the parent ion using a normalized collision energy of 30 and recording MS/MS scans at a resolution of 70000, 4.0 m/z isolation window, 2 × 105 ACG target, and maximum fill time of 120 ms.
Manual identification and quantification of nucleosides was performed using Thermo Xcalibur Qual Browser. Extracted ion chromatograms were generated with a deviation of 0.002 Da from the theoretical m/z value and the peaks were integrated using the manual peak annotation function. Absolute amounts of nucleosides were calculated generating an external calibration curve in the range 0.2–3000 fmol by serial dilutions of the nucleoside standards. N1-methyladenosine (m1A) and N6-methyladenosine (m6A) amounts were reported as percentage of adenosine (A).
m1A motif
The reference set of human genes (11195 genes) was taken from Khong et al. (2017). Ensemble Biomart (version 90) was used to select all transcript variants (69090 transcripts) and to retrieve the cDNA sequences. The unique transcripts were filtered to select those which match the exact length reported in Khong et al. (2017) (final set of 9301 transcripts). The motifs centered on TTCAA (AGTTCAANNCT, CGTTCAANNCG, GGTTCAANNCC, TGTTCAANNCA) and centered on TTCGA (AGTTCGANNCT, CGTTCGANNCG, GGTTCGANNCC, TGTTCGANNCA) were searched separately in all the sequences. A search algorithm was written in Python for this specific task.
Statistical analyses
All repetitions in this study were independent biological repetitions performed at least three times if not specified differently. Two-tailed t-test was used for statistical significance testing. In case of categorical data, Mann–Whitney test was used.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
We thank Prof. H. Schwalbe (Goethe University Frankfurt) for critical discussion.
Funding
This work was funded by the European Research Council grants MetaMeta_311522 (R.M.V.), RIBOMYLOME_309545 (G.G.T.), and ASTRA_855923 (G.G.T). G.G.T. acknowledges support of the H2020 projects IASIS_727658 and INFORE_825080, the Spanish Ministry of Economy and Competitiveness BFU2017-86970-P, as well as the collaboration with Peter St. George-Hyslop financed by the Wellcome Trust. R.M.V. acknowledges support by the Deutsche Forschungsgemeinschaft (DFG) grant EXC115. M.H. is funded by DFG CRC902 ‘Molecular Principles of RNA-based Regulation’.
Conflict of interest: none declared.
Author contributions: R.M.V. conceived and supervised the project with the help of G.G.T. R.M.V., M.H., and G.G.T. conceived the experiments. M.A., A.M.-L., and G.H. designed and performed the experiments and analyzed the data. G.C. designed and performed the mass spectrometry experiments and analyzed the data. R.D.P., G.G.T., and G.C. performed bioinformatics analyses. R.M.V. and G.G.T. wrote the manuscript with contribution from all authors.
References
- Alriquet M., Martínez-Limón A., Hanspach G., et al. (2019). Assembly of proteins by free RNA during the early phase of proteostasis stress. J. Proteome Res. 18, 2835–284. [DOI] [PubMed] [Google Scholar]
- Anders M., Chelysheva I., Goebel I., et al. (2018). Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J., Phan L., Cuesta R., et al. (1998). The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas T.E., Audas D.E., Jacob M.D., et al. (2016). Adaptation to stressors by systemic protein amyloidogenesis. Dev. Cell 39, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V.A., Májek P., Jae L.T., et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. [DOI] [PubMed] [Google Scholar]
- Bolognesi B., Lorenzo Gotor N., Dhar R., et al. (2016). A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 16, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O., Desforges B., Wu T.-D., et al. (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42, 8678–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F., Carlson L.-A., Hartl F.U., et al. (2010). The three-dimensional organization of polyribosomes in intact human cells. Mol. Cell 39, 560–569. [DOI] [PubMed] [Google Scholar]
- Cid-Samper F., Gelabert-Baldrich M., Lang B., et al. (2018). An integrative study of protein–RNA condensates identifies scaffolding RNAs and reveals players in fragile X-associated tremor/ataxia syndrome. Cell Rep. 25, 3422–3434.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Wang T., Gonzalez G., Wang Y. (2018). Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal. Chem. 90, 6380–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V., Blanco S., Esteller M. (2018). SnapShot: messenger RNA modifications. Cell 174, 498–498.e1. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., et al. (2016). The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Rechavi G. (2017). Loud and clear epitranscriptomic m1A signals: now in single-base resolution. Mol. Cell 68, 825–826. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J., Czudnochowski N., O’Connell J.D., et al. (2015). Crystal structure of the human tRNA m1A58 methyltransferase-tRNA3Lys complex: refolding of substrate tRNA allows access to the methylation target. J. Mol. Biol. 427, 3862–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann T.M., Jahnel M., Pozniakovsky A., et al. (2018). Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654. [DOI] [PubMed] [Google Scholar]
- Ganassi M., Mateju D., Bigi I., et al. (2016). A surveillance function of the HSPB8–BAG3–HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell 63, 796–810. [DOI] [PubMed] [Google Scholar]
- Ganser L.R., Kelly M.L., Herschlag D., et al. (2019). The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 20, 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Jove Navarro M., Kashida S., Chouaib R., et al. (2019). RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun. 10, 3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. [DOI] [PubMed] [Google Scholar]
- Harcourt E.M., Kietrys A.M., Kool E.T. (2017). Chemical and structural effects of base modifications in messenger RNA. Nature 541, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp M.S., Kasturi P., Hartl F.U. (2019). The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 20, 421–435. [DOI] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N. (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327. [DOI] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., et al. (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Vale R.D. (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., et al. (2002). Evidence that ternary complex (eIF2–GTP–tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., et al. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A., Jain S., Matheny T. et al. (2018). Isolation of mammalian stress granule cores for RNA-Seq analysis. Methods 137, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A., Matheny T., Jain S. et al. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M.A., Bassell G.J. (2006). Neuronal RNA granules: movers and makers. Neuron 51, 685–690. [DOI] [PubMed] [Google Scholar]
- La Spada A.R., Taylor J.P. (2010). Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 11, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. (2015). The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J.J.M., Pyl P.T., Rausch T., et al. (2013). The genomic and transcriptomic landscape of a HeLa cell line. G3 3, 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon E.M., Qiu Y., Ghanbari Niaki A., et al. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xiong X., Wang K., et al. (2016). Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316. [DOI] [PubMed] [Google Scholar]
- Li X., Xiong X., Zhang M., et al. (2017). Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Qian S.-B. (2014). Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA 5, 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Clark W., Luo G., et al. (2016). ALKBH1-mediated tRNA demethylation regulates translation. Cell 167, 816–828.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Macon J.B., Wolfenden R. (1968). 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry 7, 3453–3458. [DOI] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D., Franzmann T.M., Patel A., et al. (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., et al. (2015). 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., He C. (2018). Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52, 349–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerum S., Dégut C., Barraud P., et al. (2017). m1A post transcriptional modification in tRNAs. Biomolecules 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S., Krecic A., Andersland J., et al. (2005). The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer E., Rechavi G., Dominissini D. (2017). Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr. Opin. Chem. Biol. 41, 93–98. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska D., Decatur W.A., Fournier M.J. (2008). The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 36, D178–D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D.S.W., Parker R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Alberti S. (2017). Cell adaptation upon stress: the emerging role of membrane-less compartments. Curr. Opin. Cell Biol. 47, 34–42. [DOI] [PubMed] [Google Scholar]
- Ries R.J., Zaccara S., Klein P., et al. (2019). m6A enhances the phase separation potential of mRNA. Nature 571, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M., Sas-Chen A., Nir R., et al. (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255. [DOI] [PubMed] [Google Scholar]
- Saikia M., Fu Y., Pavon-Eternod M., et al. (2010). Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez de Groot N., Armaos A., Graña-Montes R., et al. (2019). RNA structure drives interaction with proteins. Nat. Commun. 10, 3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K.W., Kleiner R.E. (2020). YTHDF2 recognition of N1-methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chem. Biol. 15, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S.P., Dundr M. (2011). Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173. [DOI] [PubMed] [Google Scholar]
- Singh G., Pratt G., Yeo G.W., et al. (2015). The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 84, 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K.E., Warda A.S., Sharma S., et al. (2017). Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas R.M., Hartl F.U. (2005). Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310, 1960–1963. [DOI] [PubMed] [Google Scholar]
- Vabulas R.M., Raychaudhuri S., Hayer-Hartl M., et al. (2010). Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2, a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B., Protter D.S.W., Matheny T., et al. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl Acad. Sci. USA 115, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigts-Hoffmann F., Hengesbach M., Kobitski A.Y., et al. (2007). A methyl group controls conformational equilibrium in human mitochondrial tRNA Lys J. Am. Chem. Soc. 129, 13382–13383. [DOI] [PubMed] [Google Scholar]
- Wallace E.W.J., Kear-Scott J.L., Pilipenko E.V., et al. (2015). Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tao X., Jacob M.D., et al. (2018). Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep. 24, 1713–1721.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., et al. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Li L., Huang Y., et al. (2017). Readers, writers and erasers of N6-methylated adenosine modification. Curr. Opin. Struct. Biol. 47, 67–76. [DOI] [PubMed] [Google Scholar]
- Xiong X., Li X., Yi C. (2018). N1-methyladenosine methylome in messenger RNA and non-coding RNA. Curr. Opin. Chem. Biol. 45, 179–186. [DOI] [PubMed] [Google Scholar]
- Zhang H., Elbaum-Garfinkle S., Langdon E.M., et al. (2015). RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Kimsey I.J., Nikolova E.N., et al. (2016). m1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol. 23, 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., et al. (2015). Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.