We announce the coding-complete genome sequences of two isolates of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from two coronavirus disease 2019 (COVID-19)-positive samples (RNA isolated from nasopharyngeal swabs) from Belagavi District, Karnataka State, India. Mutational analysis revealed the presence of the D614G substitution in both the isolates.
ABSTRACT
We announce the coding-complete genome sequences of two isolates of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from two coronavirus disease 2019 (COVID-19)-positive samples (RNA isolated from nasopharyngeal swabs) from Belagavi District, Karnataka State, India. Mutational analysis revealed the presence of the D614G substitution in both the isolates.
ANNOUNCEMENT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was first reported in China during December 2019 (1). The virus belongs to the genus Betacoronavirus under the family Coronaviridae. The first case of coronavirus disease 2019 (COVID-19) in India was reported on 30 January 2020 (2). Currently, India has the second highest number of confirmed cases, after the United States. Hence, it is important to understand the spread of virus strains, from district to country level. Here, we report the coding-complete genome sequences of two SARS-CoV-2 isolates, NITMA1086 and NITMA1139, from Belagavi District, Karnataka State, India. First, RNA was extracted from nasopharyngeal swabs (using a Qiagen RNA extraction minikit) from two randomly selected reverse transcriptase PCR (RT-PCR)-positive (DiAGSure nCoV-19 detection assay; GCC Biotech, India) samples.
The libraries for total/viral RNA sequencing were prepared based on the NEBNext RNA Ultra II directional protocol. The whole genomes were sequenced using the HiSeq X (Illumina, USA)-based paired-end (2 × 150-bp) sequencing protocol, which resulted in 12.41 Gb of data with 82 million reads for NITMA1086 and 2.97 Gb of data with 24.6 million reads for NITMA1139. The reads were subjected to adapter trimming using fastq-mcf v1.04.803 (https://github.com/ExpressionAnalysis/ea-utils/blob/wiki/FastqMcf.md). Subsequently, the trimmed reads were aligned to the human genome (hg19) sequence using BWA v0.7.12 (https://github.com/lh3/bwa/releases/tag/0.7.12) to remove the host sequences. This resulted in 28,488,328 host unaligned reads for NITMA1086 and 8,159,678 reads for NITMA1139. These unaligned reads were subjected to de novo assembly using the MetaSPAdes v3.11.1 assembler (3). This assembly resulted in 85,634 scaffolds for NITMA1086 (N50, 1,877 bp) and 52,335 scaffolds for NITMA1139 (N50, 1,035 bp). The assembled scaffolds were oriented to the Wuhan reference strain (GenBank accession number NC_045512.2) using RagTag (https://github.com/malonge/RagTag) (4) analysis, and the whole-genome sequences were retrieved. Subsequently, SNP-Sites (5) was used to perform variant calling of the whole-genome sequences with reference to the Wuhan strain (NC_045512.2). All bioinformatics software used in this study was run with default parameters. NITMA1086 had a sequence length of 29,849 bp (GC content, 38%), with an average sequence depth of 110.57× and 99.81% genome coverage. NITMA1139 had a sequence length of 29,854 bp (GC content, 38%), with an average sequence depth of 3,587.38× and 99.83% genome coverage.
Mutational analysis of NITMA1086 revealed 15 mutations, of which 8 were found to cause amino acid substitutions. In the case of NITMA1139, of the 16 mutations identified, 8 were found to cause substitutions. In both of the isolates, the P314L (ORF1b), D614G (S), R203K (N), G50N (ORF14), and G204R (N) substitutions were found in common (Table 1). Of these variations, D614G is reported to be highly prevalent worldwide and is associated with higher viral load and titers of pseudoviruses (6).
TABLE 1.
List of mutations and amino acid changes observed in the sequenced genomes
| Isolate name | Nucleotide change(s)/SNPa,b | Gene(s) | Variance/amino acid change(s)a |
|---|---|---|---|
| NITMA1086 | C241T | Intergenic | upstream_gene_variant |
| NITMA1086 | C2695T | ORF1ab | synonymous_variant |
| NITMA1086 | C3037T | ORF1ab | synonymous_variant |
| NITMA1086 | G8371T | ORF1a | Q2702H |
| NITMA1086 | C14408T | ORF1b | P314L |
| NITMA1086 | C18877T | ORF1ab | synonymous_variant |
| NITMA1086 | G21468T | ORF1b | M2667I |
| NITMA1086 | A23403G | S | D614G |
| NITMA1086 | A24774T | S | Q1071L |
| NITMA1086 | C24784T | S | synonymous_variant |
| NITMA1086 | C26010T | ORF3a | synonymous_variant |
| NITMA1086 | A28055G | ORF8 | synonymous_variant |
| NITMA1086 | G28881A, G28882A | N | R203K |
| NITMA1086 | G28883C | N, ORF14 | G204R, G50N |
| NITMA1139 | C241T | Intergenic | upstream_gene_variant |
| NITMA1139 | C313T | ORF1ab | synonymous_variant |
| NITMA1139 | A4372G | ORF1ab | synonymous_variant |
| NITMA1139 | C3037T | ORF1ab | synonymous_variant |
| NITMA1139 | A5608G | ORF1ab | synonymous_variant |
| NITMA1139 | C5700A | ORF1ab | A1812D |
| NITMA1139 | C9693T | ORF1ab | A3143V |
| NITMA1139 | G9190T | ORF1ab | synonymous_variant |
| NITMA1139 | C14408T | ORF1ab | P314L |
| NITMA1139 | C16626T | ORF1ab | synonymous_variant |
| NITMA1139 | A18253G | ORF1ab | M15967V |
| NITMA1139 | C18555T | ORF1ab | synonymous_variant |
| NITMA1139 | C23230T | S | synonymous_variant |
| NITMA1139 | A23403G | S | D614G |
| NITMA1139 | G28881A, G28882A | N | R203K |
| NITMA1139 | G28883C | N, ORF14 | G204R, G50N |
Bold indicates the mutations leading to amino acid substitutions commonly observed in NITMA1086 and NITMA1139; italic indicates the other synonymous/intergenic variants commonly observed in NITMA1086 and NITMA1139.
SNP, single nucleotide polymorphism.
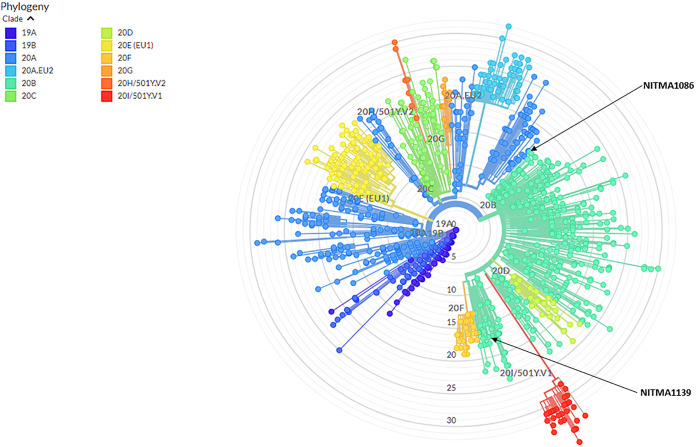
Phylogenetic comparison of these two genome sequences with globally detected viral strains was performed using the Nextstrain tool (with an inbuilt global data set from GISAID) (7). This analysis revealed that both isolates belong to clade 20B, a subclade of 20A, which emerged in the European outbreak and evolved from the 19A Wuhan strain (Fig. 1). In the current scenario, 20B has been reported to be more common in the Indian population and is considered a major basis of disease transmission in India (8). Common sharing and collating of whole-genome sequences during viral outbreaks are essential as a crucial part of outbreak response (9). Thus, the genome sequences of these isolates will aid in studying the evolution and epidemiology of the virus and its transmission dynamics in Belagavi, as well as throughout India.
FIG 1.
Phylogenetic tree displaying sequenced SARS-CoV-2 strains (with reference to GISAID global data set), in accordance to the clades assigned for the virus during phylogenetic analysis using the Augur toolkit (with default parameters) run through the Nextstrain server. Augur implements the FastTree algorithm, which infers the maximum-likelihood method for building phylogenetic trees from alignments of nucleotide or protein sequences (7, 10). Both of the sequenced strains are colored according to the assigned clade, complying with the recently established nomenclature (11). The locations of the two isolates, NITMA1089 and NITMA1139, are marked with black arrows.
Data availability.
These genome sequences have been deposited in the NCBI GenBank database under the accession numbers MW425563.1 and MW425837.1 for NITMA1086 and NITMA1139, respectively. The raw reads were also submitted to the NCBI SRA under the accession numbers SRX9766878 and SRX9766838 for NITMA1086 and NITMA1139, respectively.
ACKNOWLEDGMENTS
We acknowledge the director general of the Indian Council of Medical Research, Balram Bhargava, and the divisions of Basic Medical Sciences (BMS) and Epidemiology and Communicable Diseases (ECD) of ICMR for support and funding. We also acknowledge the Belagavi Institute of Medical Sciences (BIMS) and the district health authority of Belagavi, Karnataka State.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews MA, Areekal B, Rajesh KR, Krishnan J, Suryakala R, Krishnan B, Muraly CP, Santhosh PV. 2020. First confirmed case of COVID-19 infection in India: a case report. Indian J Med Res 151:490–492. doi: 10.4103/ijmr.IJMR_2131_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20:224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, Harris SR. 2016. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom 2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghav S, Ghosh A, Turuk J, Kumar S, Jha A, Madhulika S, Priyadarshini M, Biswas VK, Shyamli PS, Singh B, Singh N, Singh D, Datey A, Avula K, Smita S, Sabat J, Bhattacharya D, Kshatri JS, Vasudevan D, Suryawanshi A, Dash R, Senapati S, Beuria TK, Swain R, Chattopadhyay S, Syed GH, Dixit A, Prasad P, Pati S, Parida A, Odisha COVID-19 Study Group , ILS COVID-19 Team. 2020. Analysis of Indian SARS-CoV-2 genomes reveals prevalence of D614G mutation in spike protein predicting an increase in interaction with TMPRSS2 and virus infectivity. Front Microbiol 11:594928. doi: 10.3389/fmicb.2020.594928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam LT, Hieu NT, Trang NH, Thuong HT, Linh TH, Tien LT, Thao NTN, Loan HTK, Quang PD, Quang LC, Thang CM, Thuong NV, Ha H, Ha CH, Lan PT, Hai TN. 2020. Whole-genome sequencing and de novo assembly of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Vietnam. Vietnam J Biotechnol 18:197–208. doi: 10.15625/1811-4989/18/2/15082. [DOI] [Google Scholar]
- 10.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
These genome sequences have been deposited in the NCBI GenBank database under the accession numbers MW425563.1 and MW425837.1 for NITMA1086 and NITMA1139, respectively. The raw reads were also submitted to the NCBI SRA under the accession numbers SRX9766878 and SRX9766838 for NITMA1086 and NITMA1139, respectively.