Abstract
The brain is composed of a complex web of networks but we have yet to map the structural connections of the human brain in detail. Diffusion MR imaging is a high-throughput method that relies on the principle of diffusion to reconstruct tracts (i.e., pathways) across the brain. Although diffusion MR tractography is an exciting method to explore the structural connectivity of the brain in development and across species, the tractography has at times led to questionable interpretations. There are at present few if any alternative methods to trace structural pathways in the human brain. Given these limitations and the potential of diffusion MR imaging to map the human connectome, it is imperative we develop new approaches to validate neuroimaging techniques. I discuss our recent studies integrating neuroimaging with transcriptional and anatomical variation across humans and other species over the course of development and in adulthood. Developing novel frameworks to harness the potential of diffusion MR tractography provides new and exciting opportunities to study the evolution of developmental mechanisms responsible for variation in connections and bridge the gap between model systems and humans.
Keywords: Cortex, development, human, diffusion MR imaging
Introduction
Pioneering studies in experimental avian embryology combined tissue manipulations with cell‐labeling methods to reveal the developmental origins of cell types, migratory routes, as well as the developmental time course of axon extension and refinement. The use of a quail-specific markers, dyes, and conjugates enabled tracking the fate of cell populations in detail and identified many principles of development (1–3). These studies paved the way for many future studies in developmental and evolutionary biology (4–12).
The present work is focused on the evolution and development of cellular architecture, and in particular the structural connectivity of the human brain. Many of the tools available for study in sauropsid embryology (6–12), are unavailable to the study of the human brain. The lack of appropriate tools in human neuroscience has precluded our ability to visualize connections and trace cell populations in as much detail in humans as in model organisms. Identifying which connections are conserved and which have evolved in the hominid lineage necessitates harnessing methods that only indirectly trace the location and target of axons. The study of connections and their evolution in the human lineage is as much a study on how to study connections as it is a study seeking to identify what connections are variant and invariant across species.
I consider conservation and variation in tractography from diffusion MR imaging, transcription, and neuroanatomy as tools to study connections across species. I illustrate how these approaches enhance our understanding of the developmental mechanisms underlying variation in connectivity in humans relative to model organisms (Figure 1; 13–18). I also discuss our recent efforts capturing temporal variation in transcription to find corresponding time points across humans and model organisms (Figure 2). Developing appropriate norming procedures to compare development permits identifying sources of conservation and variation in developmental programs across species.
Figure 1.
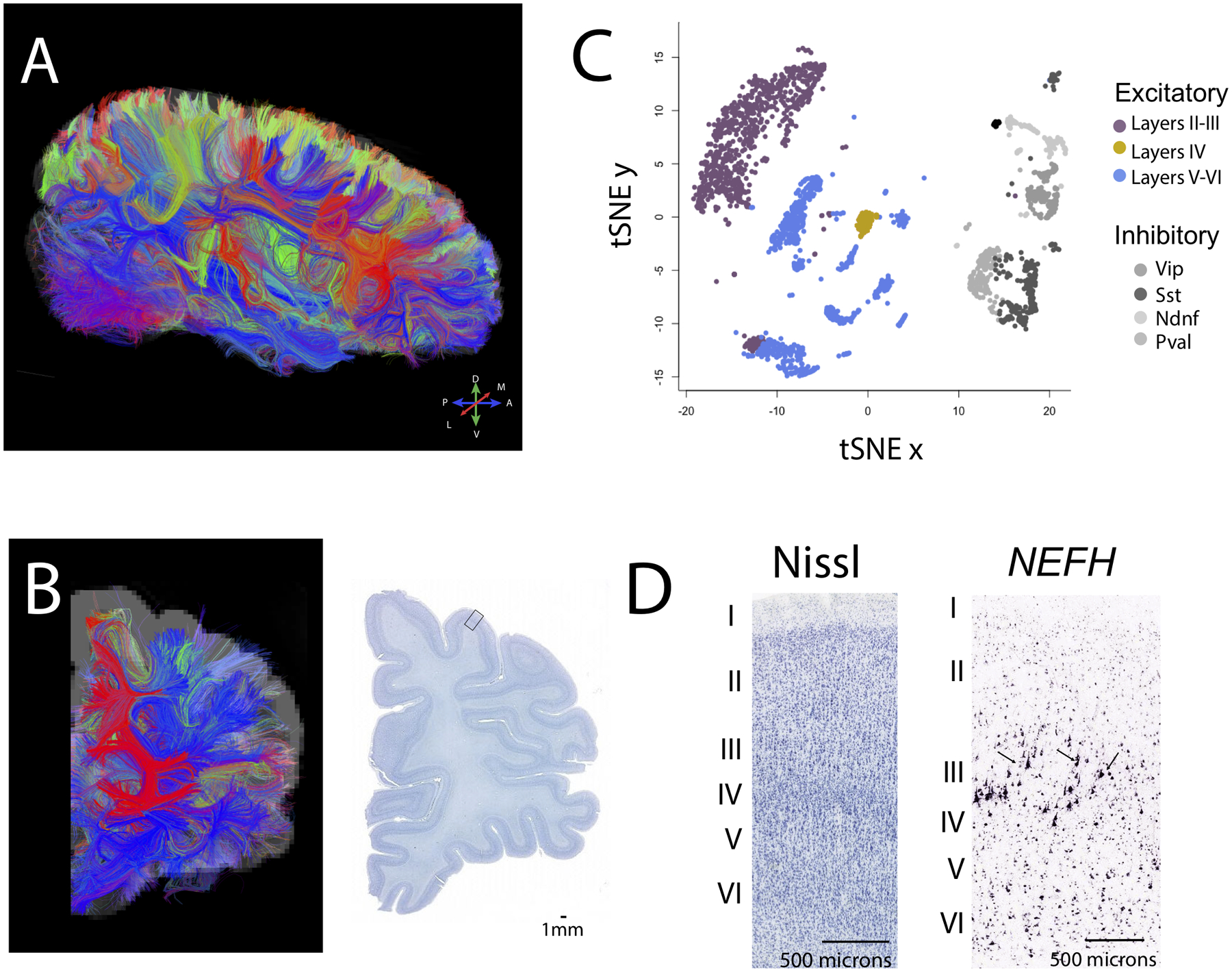
The integration of diffusion MR tractography with transcriptional and anatomical variation can be used to identify conservation and variation in connectivity patterns in humans relative to other species. Diffusion MR tractography viewed from sagittal (A) and (B) coronal slices show fibers (i.e., tracts) that course through the white matter. Color-coding of fibers represent their direction according to the map shown in A (e.g., fibers coursing across the (A) anterior to (P) posterior axis are in blue). (C) A tSNE plot of single cells from methylome sequencing highlights cell populations across layers of the frontal cortex of humans. These data can be used to compare relative populations of cell types across species. (D) Close-up view through the cortex identifies cell populations based on cytoarchitecture (Nissl-stained) and gene expression (NEFH). The expression of NEFH identifies large pyramidal neurons in layer III (arrows). Quantitative comparisons of diffusion MR tractography, single cell sequencing, and neuronal populations based on cytoarchitecture and gene expression can be used to identify conservation and variation in connections across species. Diffusion MR scans and in situ hybridization data are made available on the Allen Brain Institute for science. The diffusion MR scan at 900 μm resolution and Nissl-stained sections are from a 34 year old female brain (150). Single cell methylome sequencing data are from 107. The Nissl-stained section shown in D is taken from the box shown in B. Abbreviations: M: Medial; L: lateral; D: Dorsal; V: ventral.
Figure 2.
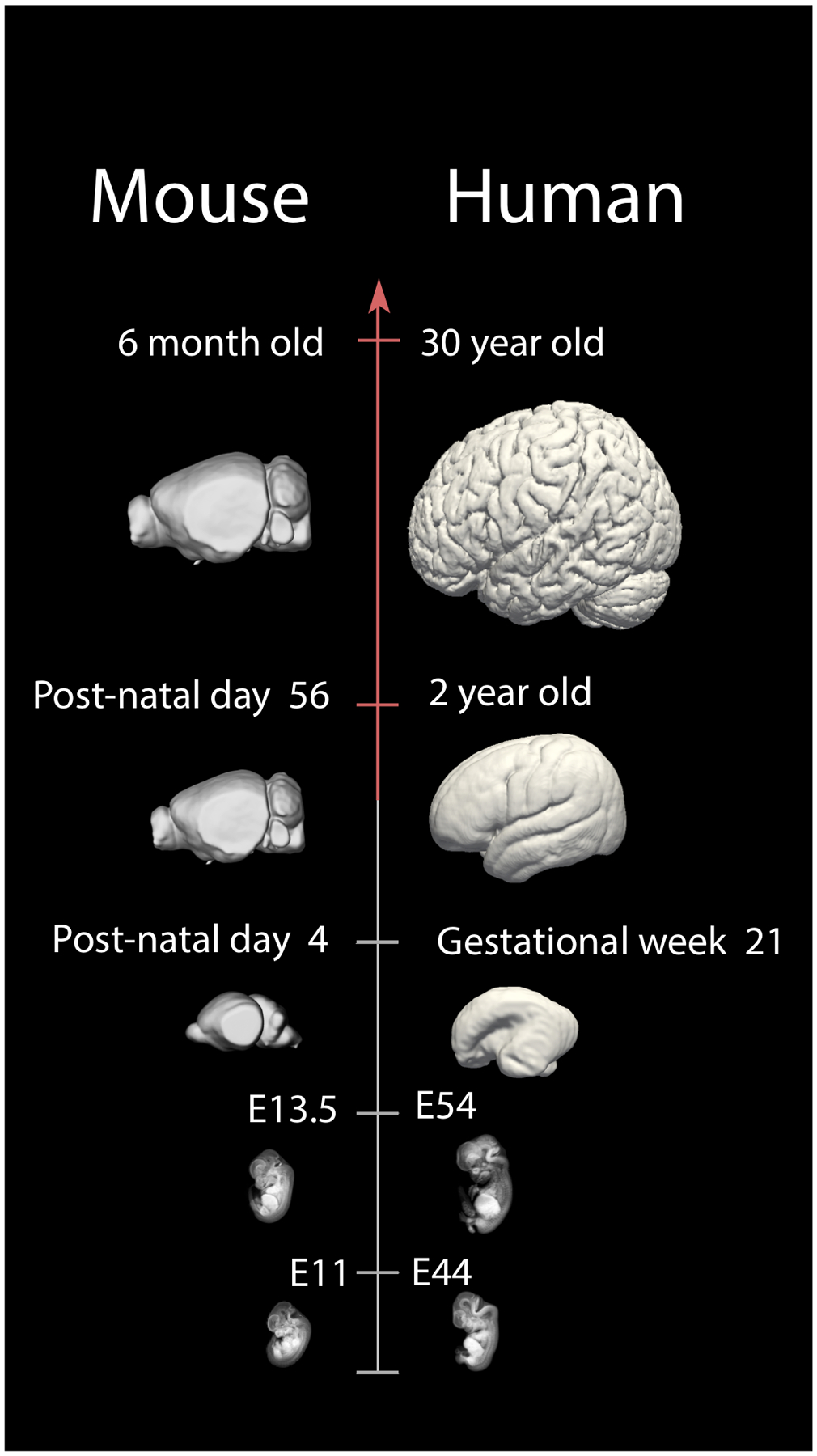
A schematic illustrates examples of corresponding time points between humans and mice throughout fetal and postnatal ages. For example, a mouse at embryonic day (E) 11 is roughly equivalent to a human at E44. A post-natal day 4 mouse equates to a human fetus at gestational week 21, and a 6 month-old mouse is roughly equivalent to a human at 30 years of age. These data will enhance translational work from model systems to humans by finding corresponding ages throughout development and adulthood. Micro CT-scans of prenatal mice are from 151. Structural MR scans of humans at ED 44 and 54 are from the multi-dimensional human embryo project (http://embryo.soad.umich.edu.proxy.library.cornell.edu/index.html). Smooth surfaces were made from human MRI structural scans. Human and mice brains other than those from the Multi-dimensional project and 151 are made available from the Allen Brain Atlas.
Tracing an axon in the human brain: challenges and opportunities
Many of the approaches available to map pathways in model organisms (e.g., avian embryos) are not available to study the human brain. For example, tract-tracers require injections in the living brain, which preclude their study in humans (19–26). Other tracers such as fluorescent dyes, used in embryology, do not diffuse over sufficient distance to enable the reconstruction of axonal projections in the human brain (27). The lack of current tools available to study connections has precluded our ability to develop a map of structural connections in the human brain and to identify how connectivity patterns in humans differ from those of other species.
Among the various methods to map connections in the brain, resting state fMRI measures brain activity via modifications to blood flow and can be used as an indirect means to map connections across the brain (28–30). Identified cortico-cerebellar maps are an interesting and a rare example to emerge from the integration of resting state fMRI with tract-tracers. Whereas tract-tracers provide information about few but detailed localization of connections in model systems, resting state fMRI broadly surveys blood flow and can be used to study the human brain. The integration of these methods at different scales of study resolved the existence and localization of cortico-cerebellar maps. Because there are no monosynaptic projections between the cerebral cortex and the cerebellum, the presence of maps across the cerebral cortex and cerebellum cannot be identified from tract-tracers alone since tract-tracers do not cross synapses (31–36). Resting states fMRIs consider covarying fluctuations in blood flow regardless as to whether these connections are mono or polysynaptic. The use of resting states fMRI and tract-tracers identifies cross-synaptic pathways and cortical representations across the cerebellum. This approach showed that maps of cortical areas are represented multiple times in the cerebellum and are oriented as mirror images of each another (33–36). Although there is uncertainty about the origins of recorded signals from resting state fMRI, it is the combination of tract-tracing techniques with neuroimaging techniques that identified networks across the human brain (37).
Diffusion MR tractography is one of the few methods available to visualizing the structure of pathways in the human brain (Figure 1). Diffusion MR tractography relies on the local diffusion of water to detect the location and orientation of pathways across the brain. In adulthood, diffusion MR tractography identifies long-range projections (i.e., bundles of axons) as fibers (i.e., tracts) across the brain (38–42). Diffusion MR tractography has not only enabled visualizing pathways in the adult brain, it has revealed, with unprecedented detail, the developmental time course of major developmental programs (43–50). Those include radial glia and emerging pathways in brains of humans and other species. For example, during cortical neurogenesis, diffusion MR tractography identifies radial fibers, which act as scaffolds guiding neurons that migrate from the proliferative pool to the cortical plate (Figure 3). As neurogenesis wanes, diffusion MR tractography identifies emerging cross-cortical and subcortical pathways, many of which course through the white matter (45, 46, 51). Diffusion MR tractography is non-invasive and identifies tracts across the entire brain (Figure 1) but, certain limitations imposed by the tractography have precluded its use at the level of detail needed to map connections in the developing and adult brain.
Figure 3.
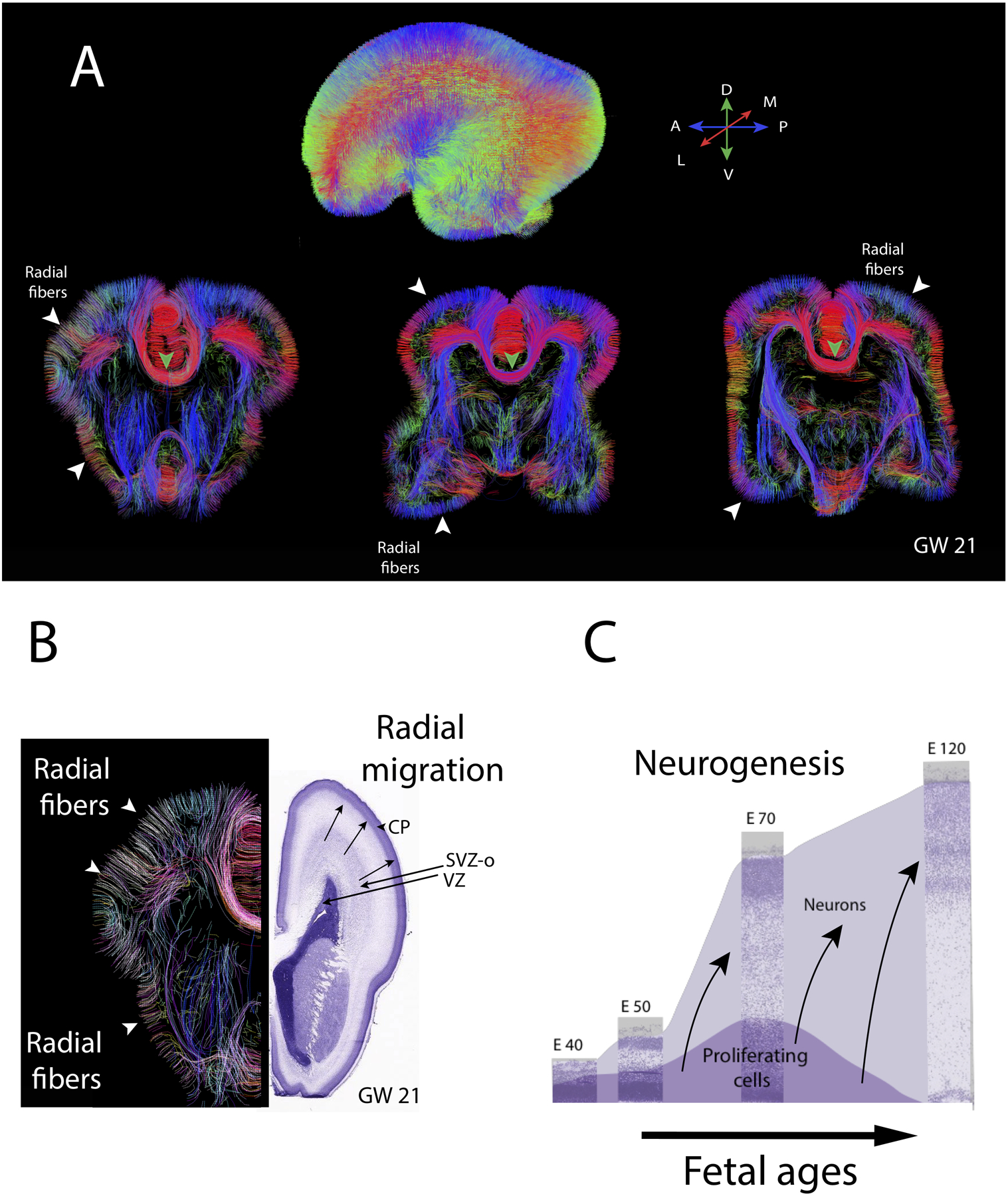
Diffusion MR tractography through the brain of a human fetus at gestational week (GW) 21. (A) Whole brain diffusion MR tractography highlight fibers coursing in different directions with the color-coding highlight the average direction of fibers (see map). Coronal slices through the brain highlight radially aligned fibers (i.e., putative radial glia) coursing from the proliferative zones to the outer surface of the developing cortex (white arrow-heads) as well as emerging pathways such as the corpus callosum (green arrow-heads). (B) Diffusion MR tractography from the left hemisphere show radial fibers coursing from the proliferative zone to the outer surface of the cortex. Nissl-stained sections through the right hemisphere show cell dense proliferative cells. These proliferative zones include the ventricular zone (VZ) and a subventricular zones (inner and outer; SVZO). Over the course of cortical neurogenesis, newly born neurons exiting the proliferative pools migrate radially along radial glia to the developing cortex called the cortical plate (CP). (C) Timetable of cortical neurogenesis at different ages represent the population of proliferative and neurons over the course of development. Neurons are produced over time and migrate radially to the outer surface of the cortex. Nissl-stained sections of sections of the developing cortex of macaques at different ages are in the background from embryonic (E) from E40 to E120. Over the course of fetal neurogenesis, the progenitor pool amplifies concomitant with newly born neurons cells exiting the proliferative zones to migrate to the outer surface of the cortex. These newly born neurons migrate to the outer surface of the cortex, and they are guided by radial glial cells as they migrate from the proliferative pool to the outer surface of the cortex. Neurons generated later migrate previously generated neurons (arrows). The diffusion MR scan of a 21GW fetus is from the Allen Institute for Brain Science. Abbreviations: A: Anterior; P: Posterior; M: Medial; L: lateral; D: Dorsal; V: ventral.
The limitations of diffusion MR tractography have been described in detail. Diffusion MR tractography has difficulty resolving the precise terminations of pathways within the gray matter as it preferentially identifies pathways based on certain characteristics at the expense of others (axon thickness, myelination; 52–53), and does not fully resolve crossing fibers (41; 53–55). It is, therefore, critical that we develop new approaches to ensure the accuracy of tractography emerging from diffusion MR imaging. Validating diffusion MR tractography would enable mapping connectivity patterns of the brain and investigating developmental mechanisms generating variation in connections across species.
Traits that are conserved across mammalian brains can be used to make inferences about the organization of the human brain. This is especially valuable considering that so little is known about the basic circuitry of the human brain relative to what is known in model organisms. I consider conserved properties of the cerebral cortex such as the relationships between birth-order (i.e., when a neuron is born), adult position, neuronal size, gene expression, and stereotypical patterns of connectivity across the depth of the cortex in human, non-human primates, and other mammals (56) to infer the evolution of cortical circuits in the human lineage. Integrating information about cortical structure, stereotypical patterns of connections, gene expression, and diffusion MR tractography enables developing new approaches to enhance our understanding of the evolution and development of connections in the human lineage.
Conservation in cortical properties to the study of the evolution of connections
I first discuss conserved properties of the mammalian cerebral cortex in order to identify how connections evolved across species. I focus on the relationship between birth order (i.e., when a neuron is generated in development), positional information of neuronal soma, as well as stereotypical patterns of connections across the depth of the cortex (1; 19; 57–61). I integrate these data along with the tractography from diffusion MR imaging to tackle the evolution of connections across species (Figure 1).
The grey matter of the cortex is composed of 6 layers or so across mammals and consists of neuronal and glial cell bodies (62). Neurons exhibit stereotypic patterns of projections depending on where their soma are located across the depth of the cortex. A number of studies have employed anatomical tracers to investigate patterns of connectivity across several mammalian species in adults and in development (31, 63–69). The synthesis across these studies have revealed conserved properties in connectivity patterns across the depth of the cortex. Layer II-III preferentially project within and across cortical regions. Layer IV neurons are granular (ie., small) and project locally within the grey matter of the cortex. A number of layer V-VI neurons project to subcortical structures though some layer V-VI neurons project to other cortical areas, many of which are considered feedback projections (57, 68–70). Feedback projections, which originate in layers V and VI, project to layers I-IV. This in contrast with feedforward projections, which originate largely in layer III, and have their axons terminate in layers IV-VI. Feedforward projections originate from cortical areas that are closest to the sensory periphery whereas feedback projections project in the direction opposite to that of feedforward projections. (57, 71–73). Taken together, these observations demonstrate that there is systematic variation in projection patterns across the depth of the cortex in mammals. Given these conserved properties of cortical organization, modifications to neuron numbers within specific cortical layers can be used to make inferences about evolutionary changes in connectivity patterns.
An interesting property of the cortex is the relationship between birth-order and the position of excitatory neuronal soma across the depth of the cortex. In contrast to the “rebel status” that neural crest cells have achieved by virtue of their widespread dispersion during development (3), excitatory cortical pyramidal neurons migrate in a very orderly fashion in the developing cortex. A series of studies employed triated-thymidine in model systems to track lineages of cell populations and demonstrated an inside-out pattern of cortical neurogenesis (74–75). Neurons born (i.e., generated) early in development assume a position at lower layers (i.e., layer VI) and neurons born late during neurogenesis are located in upper layers (e.g., layers II; 74–76). This precise relations between birth-order and soma position across the depth of the cortex has been detailed across a wide range of mammalian species, including eutherian and placental mammals (77–78). Given this conserved inside-out pattern of cortical neurogenesis, varying the rate or duration of cell production in neurogenesis should entail predictable changes in which neuronal population with stereotypical projection patterns amplify (79).
An interesting link between structure and connectivity patterns in vertebrates is the relationship between cell size and projection patterns (15, 80). Neurons that have large somas have a tendency to project over long distances whereas neurons with small somas have a tendency to form local projections. Layer IV neurons are small (i.e., granular) and project locally within the grey matter whereas many layers II-II and V-VI project over long distances. Many neurons with large somas emerging from layer III and V have axons coursing through the white matter to project across cortical areas or subcortical structures. Whether large layer II-III or V neurons project to cortical or subcortical structures depends to a large extent on the position across the depth of the cortex. It is possible to use positional information and cell size across the depth of the cortex to formulate predictions as to whether a given neuron projects locally, over long distances, to cortical, or to subcortical structures.
Cell types possess stereotypical projection patterns and can be classified by the genes they express (81–86). Although a number of genes can be used to identify progenitor subtypes within the presumptive cortex (e.g., PAX6, TBR2), there are relatively fewer markers to distinguish neuronal types based on the projection patterns (87–88). Select genes individually or in combination are expressed by neuronal populations with specific projection patterns (81–86). For example, in mice, Lmo4 has a tendency to be expressed in callosal neurons of layers II/III and V. As another example, Nefh expression from layer V have a tendency to project subcortically in mice (83–84) though Nefh is expressed by cortico-cortical as well as subcortical projections from neurons emerging from layers III and V in macaques (Figure 1D, 4A; 69, 89–90). Of note, many of the genes used as markers to label neurons only partially predict projection patterns.
Figure 4.
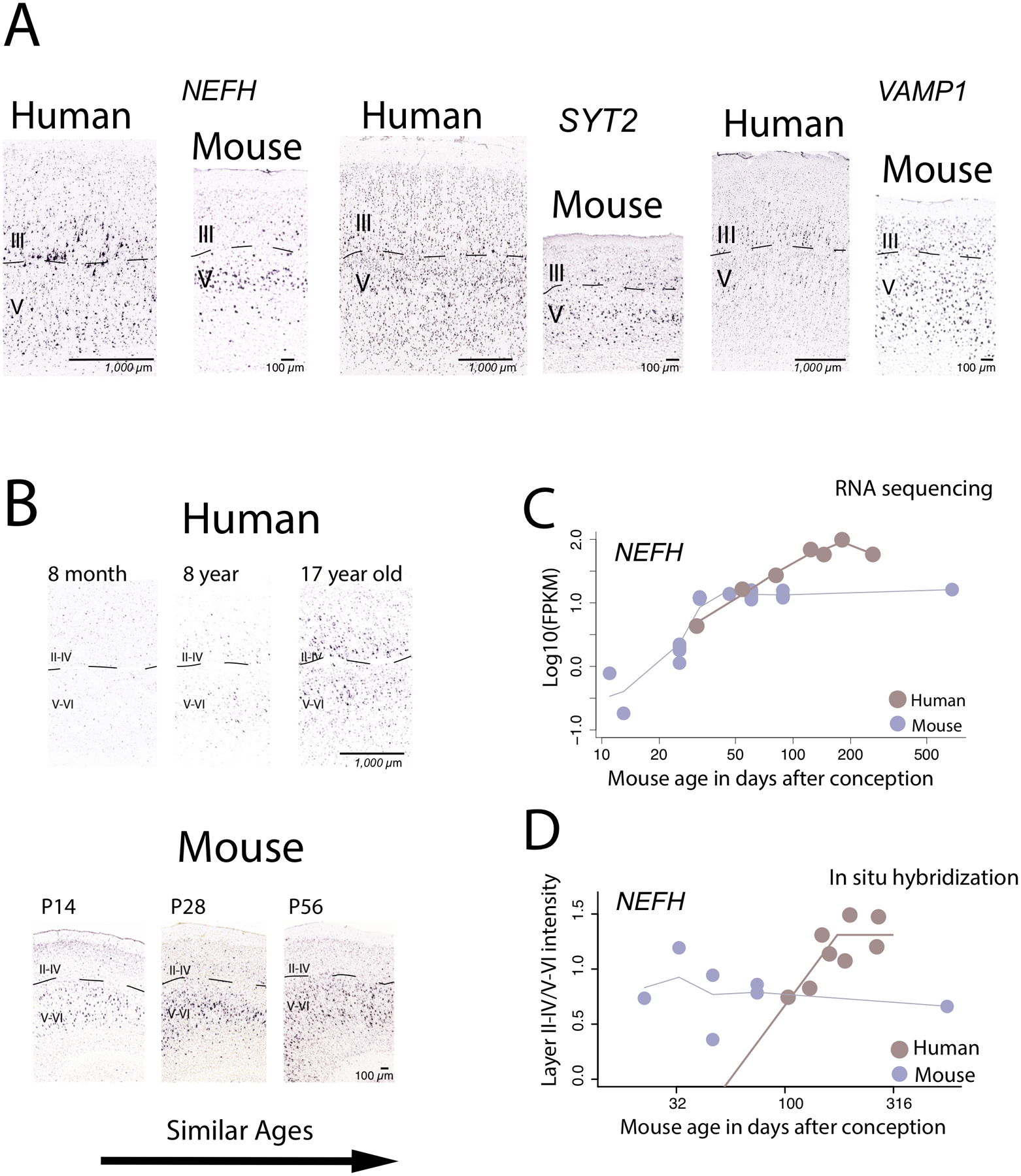
Comparative analysis of supragranular-enriched genes from the cortex of humans and mice in adulthood (A) and in development (B-D). In adulthood, the expression of some supragranular-enriched genes such as NEFH, SYT2, VAMP1, and CRYM are expressed in layers III in humans but are less so in mice. The expression of NEFH in the cortex is protracted in humans relative to mice once variation in developmental schedules are controlled for. This is evident from comparative analyses of (A) NEFH expression from in situ hybridization in the cortex of humans and in mice, (B) RNA sequencing data from the frontal cortex of humans and in mice, as well as (C) and from quantitative investigations of NEFH expression across layers of the cortex in humans and in mice. (A) NEFH expression increases shortly after birth in mice but remains relatively poorly expressed at roughly comparable ages in humans. (B) NEFH expression from an RNA sequencing data from the frontal cortex of humans and mice increase with age in both species but they appear to increase for longer in humans than in mice once variation in developmental schedules are controlled for. (C) Quantitative investigations of NEFH expression intensity between layers II-IV versus V-VI in the cortex of humans and in mice show that the relative expression of NEFH increases for longer in humans than in mice once variation in developmental schedules are controlled for, which suggests that the development of NEFH expression in superficial layers is protracted in humans relative to mice. These findings demonstrate that the protracted time course of NEFH expression from bulk samples in humans relative to mice may be accounted for by the protracted expression of NEFH expression in supragranular layers in humans. These graphs are modified from 14 and 17.
A number of large-scale projects have identified the expression of genes across cortical layers (91–94). The differential expression of genes across cortical layers can further define cell types based on stereotypical patterns of projections. Some genes are expressed across one or multiple layers. For instance, CUX1, CUX2, and CALB1 are expressed across layers II and III. RORβ is primarily expressed by layer IV, and FOXP2 is expressed across of layer VI (83–84, 91, 95). Although there may be subtle variation in the expression of genes across cortical layers in different species, a previous study profiling ~1,000 genes across the primary visual cortex of humans and mice. The study found that the overall expression of most genes are conserved in humans and mice (94). That is, 79% of examined genes display conserved pattern of expression across layers in humans and mice. The remaining 21% vary substantially in their expression across layers of the primary visual cortex between the two species (Figure 4; 94). I consider some of these differentially expressed genes coupled with structural variation across layers in humans and mice as a means to investigate the evolution of cortical connections.
Evolution of cortical connections
Synthesizing information from transcriptional and neuroanatomical variation across cortical layers with diffusion MR tractography enhances the study of evolution of connections (18, 96). I first discuss variation in cortical neurogenesis, neuronal populations, as well as gene expression across cortical layers in different species. It is possible to use these observations to develop a framework with which to study the evolution of connections (13, 97–99). I then discuss how such information can be integrated with comparative analyses of diffusion MR tractography.
Because neuronal populations have a tendency to project stereotypically, variation in subpopulation of neuron numbers can be used to make inferences about variation in connectivity patterns across species (14, 17, 100–103). Neuronal populations can be quantified with stereological methods as well as from sequencing of RNA, methylation, or open chromatin regions at the single cell level (104–110). The use of classical stereological approaches coupled with RNA or DNA sequencing can provide rigorous approaches to compare cell populations across species. This is because statistical procedures used to classify cell populations (i.e., tSNE) from single cell RNA or DNA sequencing can yield varying results (104). The integration of different methods provides rigor and reproducibility in defining and comparing cell populations across groups. We use stereological techniques to quantify the number of cortical neurons across layers of a broad range of mammalian species (59, 100, 105). We grouped layers II-III with IV because a granular layer IV is rather elusive across some of the examined mammalian species (106). Grouping layers II-III with IV captures long-range projecting neurons emerging from layer III as well as local circuit neurons emerging from layer IV. These studies show that layer II-IV neuron numbers are disproportionately amplified in primates compared with other examined species (59).(Figure 1C;). Single cell methylome and RNA sequencing data-set from frontal cortices of humans and mice show that frontal cortex excitatory pyramidal layer III neuron numbers are expanded in humans relative to mice (Figure 1; 107; 14; 17, 110). The amplification of layer II-IV neuronal populations, which may be specific to layer III, coupled with knowledge of stereotypical projection patterns across the depth of the cortex suggest an amplification of neurons projecting cross-cortically is a shared and distinguishing feature of many primates.
The developmental origins responsible for the expansion of layers II-IV neuron numbers have been identified in primates (59, 77, 102). Capturing information about the duration of neuron production, progenitor pools, and cell cycle rates over the course of development as well as quantitative data capturing the number of neuronal populations in adulthood enables building statistical models to identify the developmental basis of variation in brain structure across species. Collectively, this work, which integrates a comparative and experimental approach, has shown that an extension in the duration of cortical neurogenesis, and in particular layer II-IV neurogenesis, accounts for an expansion of layer II-IV neuron numbers in the primate cortex in adulthood (59, 79, 102, 111–116). In birds, a similar approach revealed that extensions in the duration of neurogenesis give rise to an amplification of neuron numbers in the adult avian telencephalon (79, 117–119). Collectively, these studies show that similar developmental processes have evolved within birds and within primates and that extensions in the duration of neurogenesis lead to concomitant amplification of specific population of neurons in adulthood.
Within the cortex, the majority of genes are at least grossly conserved in their spatial expression in rodents and primates (94). I consider both conserved and differentially expressed genes across layers of the cortex in humans and mice. There are 19 supragranular-enriched genes (e.g., NEFH, VAMP1, SYT2, CRYM), which are of particular interest, because these genes encode filaments (e.g., NEFH), synapse (e.g., VAMP1), and voltage-gated channels (e.g., SCN4B). These genes can be used as proxies of long-range projections (Figure 4). The observation that the expression of supragranular-enriched genes can be used as makers of long-range projections is also evident from the fact that the expression of supragranular-enriched genes correlates across long-range distributed association networks across the human cortex (120). Of particular interest is the observation that these supragranular-enriched genes are differentially expressed in humans and mice. In mice, these supragranular-enriched genes are preferentially expressed in layer V whereas these genes appear preferentially expressed in layer IIII in humans (Figure 4A). The differential expression of these supragranular-enriched genes suggests major modifications in long-range cross-connectivity patterns in humans relative to mice. Whether the observed differences in gene expression between human and mice are unique to humans or shared feature among human and non-human primates still remains an open question.
Comparative analyses of differential expressed genes, neuronal populations across layers, and birth-dating studies highlight major modifications to layer III and cortico-cortical connectivity patterns in humans and non-human primates relative to rodents. Primates possess an expanded layer III, an increase in excitatory layer III neuron numbers, and a concomitant increase in the expression of genes linked to long-range cortical projections (e.g., NEFH, 14, 17, 69, 107). Because layer III neurons have a tendency to form cortico-cortical projections, the expansion of layer III and suite of differentially expressed genes across layers suggest an expansion in cortico-cortical projections in humans versus mice. In two recent studies, we used diffusion MR tractography to confirm that primates possess an expansion in cross-cortically projecting pathways relative to rodents (14, 17). We compared the relative proportion of cortico-cortical pathways coursing through the cortical white matter of human and non-human primates relative to rodents. We randomly selected voxels though the white matter of the frontal cortex of humans and mice and quantified the relative percentage of cortico-cortical pathways in humans relative to mice (17). We found that the relative percentage of cortico-cortical pathways coursing through the white matter is significantly greater in humans than it is in mice (14, 17). Cutting across scales of organization from diffusion MR tractography to transcriptional variation enables identifying evolutionary changes in connections across species. It will be of interest to integrate transcriptional information with tractography from diffusion MR imaging within primates to trace the evolution of pathways more closely within the human lineage. I next discuss our efforts to develop new norming procedures to compare brain development at postnatal ages, and how to use this information to integrate neuroimaging with RNA sequencing to identify the developmental processes giving rise to the expansion of cortico-cortical pathways in humans.
Appropriate norming procedures to compare brain development across species
In humans, as in monkeys, cortical connections mature well into adulthood with peak cortical synaptic densities occurring shortly after birth (121–124). The cortical grey and white matter matures well into our 20s (125–129). It is unclear which layers are responsible for this extended development of cortical structure in adulthood in humans. More information on maturation of layers may be instrumental in relating the growth of the grey matter with underlying white matter pathways. Identifying developmental sources of variation in connections requires a systematic approach with which to compare development from fetal stages to adulthood. Species vary widely in their length of development, which makes it a challenge to identify comparable ages throughout development and in different species. I discuss the history of the “translating time” model and our recent efforts to build on this line of work to find corresponding time points at postnatal ages in different species. Developing appropriate norming procedures to compare brain development across species enables identifying developmental sources of conservation and variation in developmental programs across species.
The current database for the “Translating Time” model enables translating equivalent ages in humans and model organisms and can be found at www.translatingtime.org (78). The Translating time project and website is the product of collaborations across multiple individuals over multiple years (130). Initially, Finlay and Darlington, 1995, used birth-dating experiments to capture when neuronal types are generated to find corresponding time points across 6 species. Later work captured the timing of abrupt transformations across a broader range of species and from a broader range of developmental processes (e.g., synaptogenesis, cell death, 78, 131). More recently, we more than doubled the number of transformations and species (102, 132, 133). Dr. Noden was the curator of the Cornell embryological collection and I had the opportunity to meet Dr. Noden when I extracted the timing of developmental transformations from specimens housed in the Cornell embryological collections (e.g., cats, dogs, sheep). The inclusion of specimens from the embryological collection was used to expand the number of transformations (n=271) and species in the data-set. Together, these data enabled finding corresponding time points up to 2 years of age in humans and their equivalent in other species (Figure 2; 132).
Capturing when abrupt changes in developmental processes take place is especially well suited to finding corresponding time points across humans and model organisms at fetal stages of development. The translating time model relied heavily on birth-dating experiments to track when different neuronal populations are born in different species (134). Although a few neural structures such as the hippocampus extend neurogenesis into postnatal ages, the vast majority of neurogenesis occurs during fetal development (135). The low levels of neurogenesis coupled with the poor preservation of human samples have fueled controversy about the mere existence of human hippocampal neurogenesis at postnatal ages (135–138). In an effort to resolve this controversy, we asked whether the temporal profiles of human hippocampal neurogenesis are unusual in humans compared with other species once cross-species variation in developmental schedules are accounted for (133). Based on temporal profiles of hippocampal neurogenesis in model organisms, human hippocampal neurogenesis should drop sharply during childhood to hard to detect levels around adolescence (133), which is consistent with that reported by (135). Although these studies stress conservation in the timetable of hippocampal neurogenesis in humans, these comparative analyses also highlight that methods employed previously (e.g., birth-dating experiments, abrupt changes) are not suitable to find corresponding time points during postnatal development in humans and model organisms.
We developed new approaches to find corresponding time points across during postnatal ages across different species. Most recently, temporal variation in transcription permits finding corresponding time points across humans and model organisms (Figure 2). That is, an RNA sequencing data from the frontal cortex at multiple ages in humans and in mice serves to capture temporal variation in gene expression (14, 17, 107). The timing of peaks in gene expression were used as a basis with which to find corresponding ages across species during postnatal development. Temporal variation in transcription can be used interchangeably with the timing of previously collected transformations to find corresponding ages in humans and mice (Figure 2; 17). Of particular interest, the inclusion of temporal variation in transcription finds corresponding ages up to 30 years of age in humans and 6 months after birth in mice (Figure 2A–B). The use of temporal variation in transcription permits finding corresponding ages at postnatal ages and these norming procedures permit investigating the developmental basis of variation in connections across species.
Evolution of developmental timing in connections across species
Developing appropriate norming procedures to compare brain development at postnatal ages permits testing whether the timing of pathway maturation deviates relative to the timing of other developmental programs in different species. That is, it is now possible to test whether heterochronic changes in pathway maturation account for variation in brain structure in adulthood. A number of hypotheses have focused on the duration of pathway development as a basis with which species differences in connections emerge in adulthood. Many of these hypotheses have focused on timing of axon extension and refinement as a substrate through which connections evolve. However, these hypotheses were not explicitly tested due to the lack of quantitative methods available to study the development of pathways across groups and because of the lack of a rigorous approach with which to find corresponding time points across species during postnatal development. It has been proposed, on the one hand, that early connection formation can outcompete targets (139). Accordingly, evolving projections to target brain regions early relative to the timing of other developmental programs would provide an advantage in gaining territory in the brain (66, 140). It has also been proposed, on the other hand, that extending the duration of axon formation and extension might afford longer time to outcompete targets and gain an expanded territory. The second hypothesis seems intuitive as the protracted development of pathway maturation can expand territory into the postnatal ages, which can subsequently be shaped by environmental input (141). These different hypotheses were proposed several years ago but the limitations of methods available to study the evolution of connections precluded their investigation in much detail.
Integrating neuroimaging with RNA sequencing permits investigating the developmental basis of variation in connectivity across species. Temporal trajectories in the expression of supragranular-enriched genes as well as the growth of the cortical white matter across species can be used as a means to test these two different hypotheses (Figure 4B–D). The temporal pattern in the expression of supragranular-enriched genes is of particular interest because they are markers of long-range projections and variation in the temporal profiles in the expression of these supragranular-enriched genes may reveal whether the timetable of projection development deviates across species (14, 17). Given that the cortical white matter houses axons coursing to or from different cortical areas, comparative analyses of the timetable of white matter growth can also be used as an indirect means to compare the time course of projection development across species. Together, considering temporal trajectories in white matter and gene expression identify deviations in the duration of pathway development account for variation in connectivity patterns across species.
Controlling for variation in developmental schedules in humans and in mice serves to identify developmental programs accounting for the expansion of cortico-cortical pathways in humans. The expression of at least some of the supragranular-enriched genes (e.g., NEFH, VAMP1) increase at postnatal ages and subsequently become invariant in both humans and in mice (14, 17). Of particular interest is the observation that the expression of these genes become invariant much later in humans than in mice once variation in developmental schedules are controlled for. Comparative development of NEFH expression from in situ hybridization further confirms NEFH expression steadily increases for longer in humans than in mice once human age is mapped onto mouse age (Figure 4B–D; 14). These findings demonstrate that temporal trajectories in the expression of genes linked to long-range projection patterns are protracted in humans relative to mice. In addition, the cortical (i.e., frontal cortex, corpus callosum) white matter grows for longer in humans than in mice once age in human age is mapped onto mouse age (14, 17, 50). Together, these findings suggest that the development of long-range projections emerging from the cortex are protracted in humans relative to mice. These few examples illustrate that the integration of neuroimaging with transcription resolves the timetable of pathway development across species and the developmental processes giving rise to the expansion of cortico-cortical pathways in humans.
It will be of interest to integrate transcriptional variation with diffusion MR tractography more broadly across mammals other than in the primate lineage. Some groups, which may be of particular interest are domesticated species (142–143). Integrating transcriptional variation with diffusion MR tractography to study the neurobiological basis of domestication would be of particular interest as it would capture all scales of study from development to brain structure and behavior. Such an approach would provide new opportunities to link modifications to connections with behavior.
Future Directions and applications to neurological disorders
Bridging the gap from transcription to the structural connectome has provided an opportunity to identify evolutionary changes in connectivity patterns across species as well as the developmental basis giving rise to the expansion of cortico-cortical pathways in humans. I have provided just a few examples where integrating across these scales of organization would be of value for understanding the evolution and development of connections. Although the discussion has focused on the evolution and development of connections, these approaches can readily be used to enhance our understanding of the biological basis of disorders.
There is often a focus on identifying neurological abnormalities at a single scale of organization, most commonly at the genetic or at the neuroimaging level (e.g., diffusion MR tractography, fMRI). Yet, the integration of the high-throughput methods in genetics and neuroimaging has the potential to lead to a more complete understanding of neurobiological abnormalities across a wide range of diseases. Abnormalities across layers and pathway development from diffusion MR tractography have been reported across a spectrum of disorders, including schizophrenia and autism spectrum disorder (144–149). For instance, schizophrenia is characterized by abnormalities in layer III and concomitant abnormalities in long range cortico-cortical association pathways (146–148). Identifying neurobiological abnormalities in the development of white matter pathways, transcriptional variation and neuroanatomy together would yield a more complete understanding of the neurobiological basis of disorders. A framework focused on cutting across scales of organization from transcription to diffusion MR tractography provides new venues to map the structural connectivity of the human brain in health and in disease as well as in hominid evolution.
Acknowledgements
Dr. Brad Smith, University of Michigan provided extended access to prenatal human MRI scans available at http://embryo.soad.umich.edu. Imaging was performed at the Center for In-Vivo Microscopy, Duke University, which was funded by NIH N01-HD-6-3257 P/G F003637. In situ hybridization data and associated images from mice and humans were taken from the Allen Institute Website and the Brainspan atlas of the developing human brain. These data are available at http://developingmouse.brain-map.org and at http://www.brainspan.org, which are supported by the National Institute of Health Contract HHSN-271-2008-00 047-C to the Allen Institute for Brain Science. The opinions in this article are not necessarily those of the NIH.
Funding information: Grant sponsor: NIGMS INBRE grant; Grant number: P20GM103446; Grant sponsor: NIGMS COBRE; Grant number: 5P20GM103653.
References
- 1.d’Amico-Martel A, Noden DM. An autoradiographic analysis of the development of the chick trigeminal ganglion. J Embryol Exp Morphol. 1980;55:167–182. [PubMed] [Google Scholar]
- 2.Noden DM. Craniofacial development: New views on old problems. Anat Rec. 1984;208:1–13. [DOI] [PubMed] [Google Scholar]
- 3.Noden DM, Schneider RA. Neural crest cells and the community of plan for craniofacial development: historical debates and current perspectives. Adv Exp Med Biol. 2006;589:1–23. [DOI] [PubMed] [Google Scholar]
- 4.Diogo R, Kelly RG, Christiaen L, et al. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature. 2015;520(7548):466–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziermann JM, Diogo R, Noden DM. Neural crest and the patterning of vertebrate craniofacial muscles. Genesis. 2018;56(6–7):e23097. doi: 10.1002/dvg.23097 [DOI] [PubMed] [Google Scholar]
- 6.Diaz RE Jr, Shylo NA, Roellig D, Bronner M, Trainor PA. Filling in the phylogenetic gaps: Induction, migration, and differentiation of neural crest cells in a squamate reptile, the veiled chameleon (Chamaeleo calyptratus). Dev Dyn. 2019;248(8):709–727. doi: 10.1002/dvdy.38 [DOI] [PubMed] [Google Scholar]
- 7.Newman SA. Physico-genetic determinants in the evolution of development. Science. 2012;338(6104):217–219. doi: 10.1126/science.1222003 [DOI] [PubMed] [Google Scholar]
- 8.Newman SA. Form and function remixed: developmental physiology in the evolution of vertebrate body plans. J Physiol. 2014;592(11):2403–2412. doi: 10.1113/jphysiol.2014.271437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frisdal A, Trainor PA. Development and evolution of the pharyngeal apparatus. Wiley Interdiscip Rev Dev Biol. 2014;3(6):403–418. doi: 10.1002/wdev.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall J, Jheon AH, Ealba EL, et al. Evolution of a developmental mechanism: Species-specific regulation of the cell cycle and the timing of events during craniofacial osteogenesis. Dev Biol. 2014;385(2):380–395. doi: 10.1016/j.ydbio.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young NM, Hu D, Lainoff AJ, et al. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development. 2014;141(5):1059–1063. doi: 10.1242/dev.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallgrímsson B, Katz DC, Aponte JD, et al. Integration and the Developmental Genetics of Allometry. Integr Comp Biol. 2019;59(5):1369–1381. doi: 10.1093/icb/icz105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charvet CJ, Sherwood CC, Takahashi E. Developmental Sequences Predict Increased Connectivity in Brain Evolution: A Comparative Analysis of Developmental Timing, Gene Expression, Neuron Numbers, and Diffusion MR Tractography In Evolution of the Brain, Cognition, and Emotion in Vertebrates. Tokyo, Springer; 2017. [Google Scholar]
- 14.Charvet CJ, Palani A, Kabaria P, Takahashi E. Evolution of Brain connections: integrating Integrating Diffusion MR Tractography With Gene Expression Highlights Increased Corticocortical Projections in Primates. Cereb Cortex. 2019;29:5150–65. pii: bhz054. doi: 10.1093/cercor/bhz054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Cabezas MÁ, Barbas H, Zikopoulos B. Parallel Development of Chromatin Patterns, Neuron Morphology, and Connections: Potential for Disruption in Autism. Front Neuroanat. 2018;12:70. doi: 10.3389/fnana.2018.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burt JB, Demirtaş M, Eckner WJ, et al. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat Neurosci. 2018;21(9):1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendy J, Takahashi E, van der Kouwe AJ, Charvet CJ. Brain wiring and supragranular-enriched genes linked to protracted human frontal cortex development. Cereb Cortex 2020; In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fornito A, Arnatkevičiūtė A, Fulcher BD. Bridging the Gap between Connectome and Transcriptome. Trends Cogn Sci. 2019;23(1): 34–50. [DOI] [PubMed] [Google Scholar]
- 19.Noden DM. Somatotopic and functional organization of the avian trigeminal ganglion: an HRP analysis in the hatchling chick. J Comp Neurol. 1980;190(3):405–428. doi: 10.1002/cne.901900302 [DOI] [PubMed] [Google Scholar]
- 20.Covell DA Jr, Noden DM. Embryonic development of the chick primary trigeminal sensory-motor complex. J Comp Neurol. 1989;286(4):488–503. doi: 10.1002/cne.902860407. [DOI] [PubMed] [Google Scholar]
- 21.Krubitzer LA, Kaas JH. The organization and connections of somatosensory cortex in marmosets. J Neurosci. 1990;10(3):952–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilingoetter CL, Jensen MB. Histological methods for ex vivo axon tracing: A systematic review. Neurol Res. 2016;38:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dooley JC, Krubitzer LA. Alterations in cortical and thalamic connections of somatosensory cortex following early loss of vision. J Comp Neurol. 2019;527(10):1675–1688. doi: 10.1002/cne.24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padberg J, Cooke DF, Cerkevich CM, Kaas JH, Krubitzer L. Cortical connections of area 2 and posterior parietal area 5 in macaque monkeys. J Comp Neurol. 2019;527(3):718–737. doi: 10.1002/cne.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pessoa L, Medina L, Hof PR, Desfilis E. Neural architecture of the vertebrate brain: implications for the interaction between emotion and cognition. Neurosci Biobehav Rev. 2019;107:296–312. doi: 10.1016/j.neubiorev.2019.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timbie C, García-Cabezas MÁ, Zikopoulos B, Barbas H. Organization of primate amygdalar-thalamic pathways for emotions. PLoS Biol. 2020;18(2):e3000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen BK, Miller SM, Mantilla CB, Gross L, Yazemski MJ, Windebank AJ. Optimizing conditions and avoiding pitfalls for prolonged axonal tracing with carbocyanine dyes in fixed rat spinal cords. J Neurosci Methods. 2006;154:256–263. [DOI] [PubMed] [Google Scholar]
- 28.Andreasen NC, O’Leary DS, Cizadlo T et al. Remembering the past: two facets of episodic memory explored with positron emission tomography. Am J Psych. 1995;152(11):1576–85. [DOI] [PubMed] [Google Scholar]
- 29.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med. 1995;34(4):537–41. [DOI] [PubMed] [Google Scholar]
- 30.Buckner RL. The serendipitous discovery of the brain’s default network. Neuroimage. 2012;62(2):1137–1145. doi: 10.1016/j.neuroimage.2011.10.035 [DOI] [PubMed] [Google Scholar]
- 31.Schmahmann JD, Pandya DN. The cerebrocerebellar system. Int Rev Neurobiol. 1997;41:31–60. [DOI] [PubMed] [Google Scholar]
- 32.Stoodley CJ, Schmahmann JD. Functional topography of the human cerebellum. Handbook Clin Neurol. 2018;154:59–70. doi: 10.1016/B978-0-444-63956-1.00004-7. [DOI] [PubMed] [Google Scholar]
- 33.Buckner RL. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron. 2013;80(3):807–15. [DOI] [PubMed] [Google Scholar]
- 34.Krienen FM, Buckner RL. Segregated fronto-cerebellar circuits revealed by intrinsic functional connectivity. Cereb Cortex. 2009;19(10):2485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Reilly JX, Beckmann CF, Tomassini V, Ramnani N, Johansen-Berg H. Distinct and overlapping functional zones in the cerebellum defined by resting state functional connectivity. Cereb Cortex. 2010;20(4):953–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophys. 2011;106(5):2322–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckner RL, Krienen FM, Yeo BT. Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci. 2013;16(7):832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- 38.Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48(4):577–82. [DOI] [PubMed] [Google Scholar]
- 39.Schmahmann JD, Pandya DN, Wang R, et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 2007;130: 630–53. [DOI] [PubMed] [Google Scholar]
- 40.Hagmann P, Kurant M, Gigandet X, et al. Mapping human whole-brain structural networks with diffusion MRI. PloS One. 2007;2(7):e597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wedeen VJ, Wang RP, Schmahmann JD, et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41(4):1267–77. [DOI] [PubMed] [Google Scholar]
- 42.Wedeen VJ, Rosene DL, Wang R, et al. The geometric structure of the brain fiber pathways. Science. 2012;335(6076):1628–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroenke CD, Taber EN, Leigland LA, Knutsen AK, Bayly PV. Regional patterns of cerebral cortical differentiation determined by diffusion tensor MRI. Cereb Cortex. 2009;19(12):2916–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kroenke CD, Van Essen DC, Inder TE, Rees S, Bretthorst GL, Neil JJ. Microstructural changes of the baboon cerebral cortex during gestational development reflected in magnetic resonance imaging diffusion anisotropy. J Neurosci. 2007;27(46):12506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi E, Folkerth RD, Galaburda AM, Grant PE. Emerging cerebral connectivity in the human fetal brain: an MR tractography study. Cereb Cortex. 2012;2:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi E, Guangping D, Wang R, et al. Development of cerebral fiber pathways in cats revealed by diffusion spectrum imaging. Neuroimage. 2010;49:1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolasinski J, Takahashi E, Stevens AA, et al. Radial and tangential neuronal migration pathways in the human fetal brain: anatomically distinct patterns of diffusion MRI coherence. Neuroimage. 2013;79:412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu G, Takahashi E, Folkerth RD, et al. Radial coherence of diffusion tractography in the cerebral white matter of the human fetus: neuroanatomic insights. Cereb Cortex. 2014;24(3):579–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song JW, Mitchell PD, Kolasinski J, Grant EP, Galaburda AM, Takahashi E. Asymmetry of white matter pathways in developing human brains. Cereb Cortex. 2015;25:2883–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charvet CJ, Das A, Song JW, et al. High angular resolution diffusion MRI reveals conserved and deviant programs in the paths that guide human cortical circuitry. Cereb Cortex. 2020;30(3):1447–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson M, Lim AR, Cohen AH, Galaburda AM, Takahashi E. Detection and growth pattern of arcuate fasciculus from newborn to adult. Front Neurosci. 2017;11:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reveley C, Seth AK, Pierpaoli C, et al. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc Natl Acad Sci U S A. 2015;112(21): E2820–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schilling K, Gao Y, Janve V, Stepniewska I, Landman BA, Anderson AW. Confirmation of a gyral bias in diffusion MRI fiber tractography. Hum Brain Mapp. 2018;39:1449–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas C, Frank QY, Irfanoglu MO, et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A. 2014;111(46): 16574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maier-Hein KH, Neher PF, Houde JC, Côté MA, Garyfallidis E, Zhong J, Chamberland M, Yeh FC, Lin YC, Ji Q, Reddick WE. The challenge of mapping the human connectome based on diffusion tractography. Nat Comm. 2017;8(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Cabezas MÁ, Zikopoulos B, Barbas H. The Structural Model: a theory linking connections, plasticity, pathology, development and evolution of the cerebral cortex. Brain Struct Funct. 2019;224,985–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbas H Pattern in the laminar origin of corticocortical connections. J Comp Neurol. 1986;252:415–422. [DOI] [PubMed] [Google Scholar]
- 58.Barbas H, García-Cabezas MÁ. Motor cortex layer 4: less is more. Trends Neurosci. 2015;38(5):259–261. doi: 10.1016/j.tins.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Charvet CJ, Hof PR, Raghanti MA, Kouwe AJ, Sherwood CC, Takahashi E. Combining diffusion magnetic resonance tractography with stereology highlights increased cross‐cortical integration in primates. J Comp Neurol. 2017;525:1075–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goulas A, Zilles K, Hilgetag CC. Cortical gradients and laminar projections in mammals. Trends Neurosci. 2018;41(11):775–88. [DOI] [PubMed] [Google Scholar]
- 61.Goulas A, Majka P, Rosa MG, Hilgetag CC. A blueprint of mammalian cortical connectomes. PLoS biology. 2019;17(3):e2005346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagstyl K, Larocque S, Cucurull G, et al. BigBrain 3D atlas of cortical layers: Cortical and laminar thickness gradients diverge in sensory and motor cortices. PLoS Biol. 2020;18(4):e3000678. doi: 10.1371/journal.pbio.3000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawamura K Corticocortical fiber connections of the cat cerebrum. III. The occipital region. Brain Res. 1973;51:41–60. [DOI] [PubMed] [Google Scholar]
- 64.Kawamura K Corticocortical fiber connections of the cat cerebrum. I. The temporal region. Brain Res. 1973;51:1–21. [DOI] [PubMed] [Google Scholar]
- 65.Kawamura K Corticocortical fiber connections of the cat cerebrum. II. The parietal region. Brain Res. 1973;51:23–40 [DOI] [PubMed] [Google Scholar]
- 66.Kawamura K, Otani K Corticocortical fiber connections in the cat cerebrum: the frontal region. J Comp Neurol. 1970;139:423–448. [DOI] [PubMed] [Google Scholar]
- 67.Striedter GF. Principles of brain evolution. Sunderland, MA: Sinauer associates;2005. [Google Scholar]
- 68.Nudo RJ, Sutherland DP, Masterton RB. Variation and evolution of mammalian corticospinal somata with special reference to primates. J Comp Neurol. 1995;358(2):181–205. doi: 10.1002/cne.903580203. [DOI] [PubMed] [Google Scholar]
- 69.Hof PR, Nimchinsky EA, Morrison JH. Neurochemical phenotype of corticocortical connections in the macaque monkey: Quantitative analysis of a subset of neurofilament protein-immunoreactive projection neurons in frontal, parietal, temporal, and cingulate cortices. J Comp Neurol. 1995;362:109–133. [DOI] [PubMed] [Google Scholar]
- 70.Gilbert CD, Kelly JP. The projections of cells in different layers of the cat’s visual cortex. J Comp Neurol. 1975;163: 81–105. [DOI] [PubMed] [Google Scholar]
- 71.Rockland KS, Pandya DN. Laminar origins and terminations of cortical connections of the occipital lobe in the rhesus monkey. Brain Res. 1979;179(1):3–20. [DOI] [PubMed] [Google Scholar]
- 72.Maunsell JH, van Essen DC. The connections of the middle temporal visual area (MT) and their relationship to a cortical hierarchy in the macaque monkey. J Neurosci. 1983;3(12):2563–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barbas H, Rempel-Clower N. Cortical structure predicts the pattern of corticocortical connections. Cereb Cortex. 1997;7:635–646. [DOI] [PubMed] [Google Scholar]
- 74.Rakic P Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. [DOI] [PubMed] [Google Scholar]
- 75.Rakic P Pre- and post-developmental neurogenesis in primates. Clin Neurosci Res 2002;2:29–39. [Google Scholar]
- 76.Noctor SC, Scholnicoff NJ, Juliano SL. Histogenesis of ferret somatosensory cortex. J Comp Neurol. 1997;387:179–193. [PubMed] [Google Scholar]
- 77.Clancy B, Darlington RB, Finlay BL. Translating developmental time across ‘mammalian species. Neuroscience. 2001;105:7–17. [DOI] [PubMed] [Google Scholar]
- 78.Clancy B, Finlay BL, Darrington RB, Anand KJ. Extrapolating brain development from experimental species to humans. Neurotoxicology. 2007;28(5):931–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cahalane DJ, Charvet CJ, Finlay BL. Modeling local and cross-species neuron number variations in the cerebral cortex as arising from a common mechanism. Proc Natl Acad Sci U S A. 2014;111(49): 17642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cahalane DJ, Charvet CJ, Finlay BL. Systematic, balancing gradients in neuron density and number across the primate isocortex. Front Neuroanat. 2012;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45(2):207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 82.Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29(39):12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Molyneaux BJ, Arlotta P, Macklis JD. Molecular development of corticospinal motor neuron circuitry. Novartis Found Symp. 2007;288:3–98. doi: 10.1016/j.expneurol.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 84.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 85.Lein ES, Belgard TG, Hawrylycz M, Molnár Z. Transcriptomic Perspectives on Neocortical Structure, Development, Evolution, and Disease. Annu Rev Neurosci. 2017;40:629–652. doi: 10.1146/annurev-neuro-070815-013858. [DOI] [PubMed] [Google Scholar]
- 86.Shoemaker LD, Arlotta P. Untangling the cortex: Advances in understanding specification and differentiation of corticospinal motor neurons. Bioessays. 2010;32(3):197–206. doi: 10.1002/bies.200900114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martínez-Cerdeño V, Cunningham CL, Camacho J, et al. Evolutionary origin of Tbr2-expressing precursor cells and the subventricular zone in the developing cortex. J Comp Neurol. 2016; 524(3):433–447. doi: 10.1002/cne.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hevner RF. Intermediate progenitors and Tbr2 in cortical development. J Anat. 2019;235(3):616–625. doi: 10.1111/joa.12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Campbell MJ, Morrison JH. Monoclonal antibody to neurofilament protein (SMI‐32) labels a subpopulation of pyramidal neurons in the human and monkey neocortex. J Comp Neurol. 1989;282(2):191–205. [DOI] [PubMed] [Google Scholar]
- 90.Nimchinsky EA, Hof PR, Young WG, Morrison JH. Neurochemical, morphologic, and laminar characterization of cortical projection neurons in the cingulate motor areas of the macaque monkey. J Comp Neurol. 1996;374(1):136–60. [DOI] [PubMed] [Google Scholar]
- 91.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 92.Belgard TG, Marques AC, Oliver PL, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernard A, Lubbers LS, Tanis KQ, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73(6):1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zeng H, Shen EH, Hohmann JG, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149(2):483–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460(2):266–279. doi: 10.1002/cne.10654 [DOI] [PubMed] [Google Scholar]
- 96.Romero-Garcia R, Whitaker KJ, Váša F, et al. Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex. Neuroimage. 2018;171:256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spocter MA, Hopkins WD, Barks SK, et al. Neuropil distribution in the cerebral cortex differs between humans and chimpanzees. J Comp Neurol. 2012;520(13):2917–2929. doi: 10.1002/cne.23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zikopoulos B, García-Cabezas MÁ, Barbas H. Parallel trends in cortical gray and white matter architecture and connections in primates allow fine study of pathways in humans and reveal network disruptions in autism. PLoS Biol. 2018;16(2):e2004559. doi: 10.1371/journal.pbio.2004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beul SF, Barbas H, Hilgetag CC. A Predictive Structural Model of the Primate Connectome. Sci Rep. 2017;7:43176. doi: 10.1038/srep43176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Charvet CJ, Cahalane DJ, Finlay BL. Systematic, cross-cortex variation in neuron numbers in rodents and primates. Cereb Cortex. 2015;25(1):147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Srinivasan S, Carlo CN, Stevens CF. Predicting visual acuity from the structure of visual cortex. Proc Natl Acad Sci U S A. 2015;112(25):7815–7820. doi: 10.1073/pnas.1509282112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Charvet CJ, Šimić G, Kostović I, et al. Coevolution in the timing of GABAergic and pyramidal neuron maturation in primates. Proc Biol Sci. 2017; 284: pii: 20171169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Srinivasan S, Stevens CF. Scaling Principles of Distributed Circuits. Curr Biol. 2019;29(15):2533–2540.e7. doi: 10.1016/j.cub.2019.06.046. [DOI] [PubMed] [Google Scholar]
- 104.Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Comm. 2019; 10(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Charvet CJ, Stimpson CD, Kim YD, et al. Gradients in cytoarchitectural landscapes of the isocortex: Diprotodont marsupials in comparison to eutherian mammals. J Comp Neurol. 2017;525(8):1811–26. [DOI] [PubMed] [Google Scholar]
- 106.Barbas H General cortical and special prefrontal connections: principles from structure to function. Annu Rev Neurosci. 2015;38:269–289. doi: 10.1146/annurev-neuro-071714-033936 [DOI] [PubMed] [Google Scholar]
- 107.Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Preissl S, Fang R, Huang H, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21(3):432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cusanovich DA, Hill AJ, Aghamirzaie D, et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174(5):1309–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, Shapouri S. A comprehensive transcriptional map of primate brain development. Nature. 2016;535(7612):367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rakic P A small step for the cell — a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. [DOI] [PubMed] [Google Scholar]
- 112.Reillo I, Romero CD, García-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21(7):1674–1694 [DOI] [PubMed] [Google Scholar]
- 113.Martinez-Cerdeno V, Cunningham CL, Camacho J, et al. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS One. 2012; 7(1):e30178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vaccarino FM, Schwartz ML, Raballo R, et al. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat Neurosci. 1999;2(3):246–53. [DOI] [PubMed] [Google Scholar]
- 115.Vaccarino FM, Grigorenko EL, Smith KM, Stevens HE. Regulation of cerebral cortical size and neuron number by fibroblast growth factors: implications for autism. J Autism Dev Disord. 2009;39(3):511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McGowan LD, Alaama RA, Striedter GF. FGF2 delays tectal neurogenesis, increases tectal cell numbers, and alters tectal lamination in embryonic chicks. PloS one. 2013;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Charvet CJ, Striedter GF. Developmental species differences in brain cell cycle rates between northern bobwhite quail (Colinus virginianus) and parakeets (Melopsittacus undulatus): implications for mosaic brain evolution. Brain Behav Evol. 2008;72(4):295–306. [DOI] [PubMed] [Google Scholar]
- 118.Charvet CJ, Striedter GF. Developmental modes and developmental mechanisms can channel brain evolution. Front Neuroanat. 2011;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Olkowicz S, Kocourek M, Lučan RK, et al. Birds have primate-like numbers of neurons in the forebrain. Proc Natl Acad Sci U S A. 2016;113(26):7255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci U S A. 2016;113(4):E469–78. doi: 10.1073/pnas.1510903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bourgeois JP, Goldman-Rakic PS, Rakic P. Synaptogenesis in the prefrontal cortex of rhesus monkeys. Cereb Cortex. 1994;4(1):78–96. [DOI] [PubMed] [Google Scholar]
- 122.Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232(4747):232–5. [DOI] [PubMed] [Google Scholar]
- 123.Petanjek Z, Judaš M, Šimić G, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108(32):13281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Petanjek Z, Judaš M, Kostović I, Uylings HB. Lifespan alterations of basal dendritic trees of pyramidal neurons in the human prefrontal cortex: a layer-specific pattern. Cereb Cortex. 2008;18(4):915–29. [DOI] [PubMed] [Google Scholar]
- 125.Monique JT, Uylings HB. Postnatal maturation of layer V pyramidal neurons in the human prefrontal cortex. A quantitative Golgi analysis. Brain Res. 1995;678(1–2):233–43. [DOI] [PubMed] [Google Scholar]
- 126.Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human life span. Nat Neurosci. 2003;6(3):309–15. [DOI] [PubMed] [Google Scholar]
- 127.Gogtay N, Giedd JN, Lusk L, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101(21):8174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cohen AH, Wang R, Wilkinson M, MacDonald P, Lim AR, Takahashi E. Development of human white matter fiber pathways: from newborn to adult ages. Int J Dev Neurosci. 2016;50:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wilkinson M, Lim AR, Cohen AH, Galaburda AM, Takahashi E. Detection and growth pattern of arcuate fasciculus from newborn to adult. Front Neurosci. 2017;11:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hawkes K, Finlay BL. Mammalian brain development and our grandmothering life. History Physiol Behav. 2018;193(PtA): 55–68. doi: 10.1016/j.physbeh.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 131.Darlington RB, Dunlop SA, Finlay BL. Neural development in metatherian and eutherian mammals: variation and constraint. J Comp Neurol. 1999;411(3):359–368. [PubMed] [Google Scholar]
- 132.Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 2013;33(17): 7368–83. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Charvet CJ, Finlay BL. Comparing adult hippocampal neurogenesis across species: translating time to predict the tempo in humans. Front Neurosci. 2018;12:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995; 268:1578–84. [DOI] [PubMed] [Google Scholar]
- 135.Sorrells SF, Paredes MF, Cebrian-Silla A, et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555(7696):377–381. doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22,589.e5–599.e5. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kozareva DA, Cryan JF, Nolan YM. Born this way: Hippocampal neurogenesis across the lifespan. Aging cell. 2019;18(5):e13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duque A, Spector R. A balanced evaluation of the evidence for adult neurogenesis in humans: implication for neuropsychiatric disorders. Brain Struct Funct. 2019:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Catania KC. Early development of a somatosensory fovea: a head start in the cortical space race? Nat Neurosci. 2001;4(4):353–354. doi: 10.1038/85992. [DOI] [PubMed] [Google Scholar]
- 140.Innocenti GM. Exuberant development of connections, and its possible permissive role in cortical evolution. Trends Neurosci. 1995;18:397–402. [DOI] [PubMed] [Google Scholar]
- 141.Andelin AK, Olavarria JF, Fine I, et al. The Effect of Onset Age of Visual Deprivation on Visual Cortex Surface Area Across-Species. Cereb Cortex. 2019;29(10):4321–33.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Spocter MA, Uddin A, Ng JC, et al. Scaling of the corpus callosum in wild and domestic canids: Insights into the domesticated brain. J Comp Neurol. 2018;526(15):2341–2359. doi: 10.1002/cne.24486 [DOI] [PubMed] [Google Scholar]
- 143.Rosenfeld CS, Hekman JP, Johnson JL, et al. Hypothalamic transcriptome of tame and aggressive silver foxes (Vulpes vulpes) identifies gene expression differences shared across brain regions. Genes Brain Behav. 2020;19(1):e12614. doi: 10.1111/gbb.12614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Benes FM, Davidson J, Bird ED. Quantitative cytoarchitectural studies of the cerebral cortex of schizophrenics. Archives of general psychiatry. 1986;43(1):31–5. [DOI] [PubMed] [Google Scholar]
- 145.Broadbelt K, Byne W, Jones LB. Evidence for a decrease in basilar dendrites of pyramidal cells in schizophrenic medial prefrontal cortex. Schizophr Res. 2002;58(1):75–81. [DOI] [PubMed] [Google Scholar]
- 146.Garey L When cortical development goes wrong: schizophrenia as a neurodevelopmental disease of microcircuits. J Anat. 2010;217(4):324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McIntosh AM, Maniega SM, Lymer GK, et al. White matter tractography in bipolar disorder and schizophrenia. Biological psychiatry. 2008;64(12):1088–92. [DOI] [PubMed] [Google Scholar]
- 148.Ramaker RC, Bowling KM, Lasseigne BN, et al. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017;9(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Velmeshev D, Schirmer L, Jung D, et al. Single-cell genomics identifies cell type–specific molecular changes in autism. Science. 2019;364(6441):685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ding SL, Royall JJ, Sunkin SM, et al. Comprehensive cellular‐resolution atlas of the adult human brain. J Comp Neurol. 2016;524:3127–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wong MD, Dorr AE, Walls JR, Lerch JP, Henkelman RM. A novel 3D mouse embryo atlas based on micro-CT. Development. 2012;139(17):3248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]


