Abstract
Background
The interferon-induced transmembrane (IFITM) protein family consists of interferon-stimulated genes (ISGs) that show potent antiviral capacity against a broad range of viruses. Many studies have been performed to investigate an association between IFITM3 polymorphisms and pandemic influenza A 2009 H1N1 virus infection. However, an association study of IFITM1 polymorphisms with susceptibility to this infection has not been reported thus far.
Objective
To identify an association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms, we compared genotype, allele and haplotype frequencies of the IFITM1 gene between healthy controls and pandemic influenza A 2009 H1N1-infected patients. In addition, we investigated linkage disequilibrium (LD) by Haploview 4.2 and the binding ability of transcription factors according to IFITM1 polymorphism alleles by PROMO. Furthermore, we measured the LD value between the IFITM1 gene and the IFITM3 gene.
Results
We found 3 novel single-nucleotide polymorphisms (SNPs) and did not find an association between IFITM1 SNPs and susceptibility to pandemic influenza A 2009 H1N1 virus infection. We found strong LD among IFITM1 SNPs but did not find a difference in the transcription factor-binding ability according to regulatory IFITM1 SNP alleles. In addition, we found strong LD between IFITM1 SNPs and IFITM3 SNPs.
Conclusion
To the best of our knowledge, this report is the first association study of the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms.
Keywords: IFITM1, IFITM3, Influenza A virus, H1N1, Single nucleotide polymorphism
Introduction
In response to viral invasion, viral sensor proteins of the host immune system, including melanoma differentiation-associated gene-5 (MDA-5), retinoic acid-inducible gene 1 (RIG-1) and Toll-like receptors (TLRs), recognize viral antigens and activate interferon secretion (Uematsu and Akira 2007; Sarkar et al. 2008; Reikine et al. 2014; Brisse and Ly 2019). Secreted interferon binds to interferon receptor (IFNR) and stimulates the expression of interferon-stimulated genes (ISGs) mediated by the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway (Horvath 2004a, b; Schindler et al. 2007). Among various ISGs, previous studies have reported that the clustering of the interferon-induced transmembrane protein (IFITM) protein family showed potent antiviral capacity through blocking the internalization of a broad spectrum of viruses, including Ebola virus (EBOV), Marburg virus (MARV), severe acute respiratory syndrome coronavirus (SARS-CoV), dengue virus (DEV), West Nile virus (WNV), Zika virus (ZIKV) and influenza A virus (IAV) (Feeley et al. 2011; Bailey et al. 2012; Everitt et al. 2012; Diamond and Farzan 2013; Perreira et al. 2013; Bailey et al. 2014; Kim and Jeong 2017; Zani and Yount 2018; Bedford et al. 2019; Kim et al. 2019; Spence et al. 2019; Kim et al. 2020; Kim and Jeong 2020; Kim and Jeong 2021; Lee et al. 2019).
In a recent study, genetic polymorphisms of the IFITM3 gene, a member of the IFITM protein family, were associated with the severity and/or susceptibility to pandemic influenza A 2009 H1N1 virus infection. The rs12252 single-nucleotide polymorphism (SNP), which is located in the splicing receptor site of the IFITM3 gene and induces the production of an N-terminal-truncated form of IFITM3 protein, is associated with the severity of pandemic influenza A 2009 H1N1 infection (Everitt et al. 2012; Zhang et al. 2013; Mills et al. 2014; Xuan et al. 2015; Gaio et al. 2016; Pan et al. 2017). In addition, the rs34481144 and rs6598045 SNPs, which are located in the transcriptional regulatory region of the IFITM3 gene, are also strongly related to the severity of and susceptibility to pandemic influenza A 2009 H1N1 virus infection, respectively (Allen et al. 2017; David et al. 2018; Kim et al. 2020). The IFITM1 protein, a member of the IFITM protein family, plays a pivotal role in the antiviral response of the host innate immune system. Unlike the IFITM3 protein, which is mainly expressed in the late endosome and prevents viral fusion, the IFITM1 protein mainly expressed in the cytosolic leaflet and blocks entry of viruses to the early endosome (Perreira et al. 2013). However, there are no association studies of genetic polymorphisms of the IFITM1 gene with the susceptibility to pandemic influenza A 2009 H1N1 virus infection.
To identify an association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms, we investigated the genotype, allele and haplotype frequencies of the IFITM1 gene. In addition, we compared the genotype, allele and haplotype frequencies of the IFITM1 gene between healthy controls and pandemic influenza A 2009 H1N1-infected patients and estimated an association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and the IFITM1 polymorphisms. Furthermore, we investigated linkage disequilibrium (LD) by Haploview 4.2 and the binding ability of transcription factors according to IFITM1 polymorphism alleles by PROMO. Last, we measured the LD value between the IFITM1 and IFITM3 genes.
Materials and methods
Ethics statements
Detailed information on all participants was described in a previous study (Kim et al. 2020). All samples were provided with informed consent under institutional review board-approved protocols. All procedures performed in the present study were approved according to guidelines of the institutional review board (IRB) of Jeonbuk National University and in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards (approval number: JBNU 2017-08-009). All the samples and related data were anonymized prior to study.
Genomic material
Genomic DNA was extracted from 200 μl of whole blood using a Blood Genomic DNA Isolation Kit (Qiagen, Valencia, California, USA) following the manufacturer’s recommendations.
Amplification of the IFITM1 gene and genetic analysis
The human IFITM1 gene was amplified from genomic DNA using sense and antisense gene-specific primers. The sequences of the primers were as follows: IFITM1-F (AGTGAGGTGAGGGCTTTTGG) and IFITM1-R (CACAGTCACAGGGACACACA). Polymerase chain reaction (PCR) was carried out using GoTaq® DNA Polymerase (Promega, Fitchburg, Wisconsin, USA). The PCR mixture contained 20 pmol of each primer, 5 μl of 10 × Taq DNA polymerase buffer, 1 μl of 10 mM dNTP mixture and 2.5 units of Taq DNA polymerase. The PCR conditions for the IFITM1-F and IFITM1-R primers were 94 °C for 2 min to denature; 35 cycles of 94 °C for 45 s, 71 °C for 45 s, and 72 °C for 1 min 30 s; and then 1 cycle of 72 °C for 10 min to extend the reaction. PCR was performed using an S-1000 Thermal Cycler (Bio-Rad, Hercules, California, USA). The PCR products were eluted by a PCR Purification Kit (Thermo Fisher Scientific, Bridgewater, New Jersey, USA) and directly sequenced with an ABI 3730 automatic sequencer (ABI, Foster City, California, USA). Sequencing results were read by Finch TV software (Geospiza Inc, Seattle, USA), and genotyping was performed.
Statistical analysis
Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc., USA). The differences in the distribution of genotypes, alleles and haplotypes of the IFITM1 gene between cases and control populations were analyzed using the χ2 test. The Hardy–Weinberg equilibrium (HWE) test and haplotype analysis were performed using Haploview 4.2 (Broad Institute, Cambridge, MA, USA). LD analysis was performed using coefficient r2 values by the program Haploview version 4.2 (Broad Institute, Cambridge, MA, USA).
Prediction of transcription factors of the IFITM1 gene
Binding sites and transcription factors were predicted by PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). The maximum matrix dissimilarity rate was entered as a “0” value. Four major haplotypes of proximal promoter sequences of the IFITM1 gene were identified and analyzed in this study.
Results
Investigation of polymorphisms of the IFITM1 gene
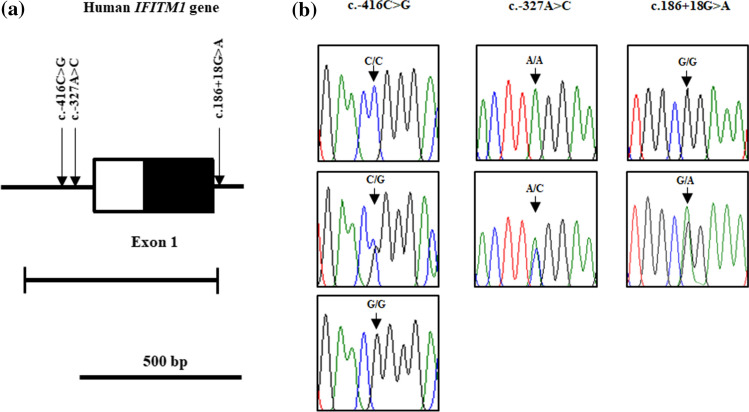
To investigate the genotype and allele frequencies of IFITM1 gene polymorphisms in the Korean population, we performed direct sequencing in 175 healthy controls and 30 H1N1 pandemic influenza 2009 virus-affected patients. Detailed information on the participants is described in Table 1. The sequenced products were homologous to the IFITM1 gene of Homo sapiens registered in GenBank (Gene ID: 8519). We found a total of 3 novel SNPs, c.-416C > G, c.-327A > C and c.186 + 18G > A (Fig. 1). The c.-327A > C and c.186 + 18G > A SNPs were found in only healthy controls (Table 2).
Table 1.
Detailed information on the study population
| Characteristics | Cases | Controls |
|---|---|---|
| Number | 30 | 175 |
| Age | 55.27 ± 17.88 | 62.43 ± 8.96 |
| Sex (n, %) | ||
| Male | 11 (36.67) | 61 (34.86) |
| Female | 19 (63.33) | 114 (65.14) |
| No. of *ICU admissions | 1 | **NA |
*ICU intensive care unit
**NA not applicable
Fig. 1.
Gene map and polymorphisms identified in the human interferon-induced transmembrane protein 1 (IFITM1) gene on chromosome 11. a The black block indicates the open reading frame (ORF), and the white block indicates the 5′ untranslated region (UTR). Horizontal bar with edges indicates the region sequenced. Arrows indicate the novel polymorphisms found in this study. b Electropherogram of 3 novel single-nucleotide polymorphisms (SNPs), c.-416C > G, c.-327A > C and c.186 + 18G > A, identified in this study. The four colors indicate individual bases of DNA sequence determined using an ABI 3730 automatic sequencer (blue: cytosine, red: thymine, black: guanine, green: adenine)
Table 2.
Comparison of genotype and allele frequencies of the IFITM1 polymorphisms between healthy controls and H1N1 pandemic influenza 2009 virus-affected patients in the Korean population
| Polymorphisms | Total, n | Genotype frequencies, n (%) | P value | Allele frequencies, n (%) | P value | HWE | |||
|---|---|---|---|---|---|---|---|---|---|
| c.-416C > G | CC | CG | GG | C | G | ||||
| Control | 175 | 101 (57.7) | 61 (34.9) | 13 (7.4) | 263 (75.2) | 87 (24.8) | 0.376 | ||
| Case | 30 | 16 (53.3) | 13 (43.3) | 1 (3.4) | 0.5980 | 45 (75) | 15 (25) | 0.9811 | 0.394 |
| c.-327A > C | AA | AC | CC | A | C | ||||
| Control | 175 | 174 (99.4) | 1 (0.6) | 0 (0) | 349 (99.7) | 1 (0.3) | 0.969 | ||
| Case | 30 | 30 (100) | 0 (0) | 0 (0) | 1.0 | 60 (100) | 0 (0) | 1.0 | – |
| c.186 + 18G > A | GG | GA | AA | G | A | ||||
| Control | 175 | 174 (99.4) | 1 (0.6) | 0 (0) | 349 (99.7) | 1 (0.3) | 0.969 | ||
| Case | 30 | 30 (100) | 0 (0) | 0 (0) | 1.0 | 60 (100) | 0 (0) | 1.0 | – |
Evaluation of an association between IFITM1 polymorphisms and susceptibility to H1N1 influenza 2009 pandemic virus infection in the Korean population
To examine an association between the genetic distribution of the IFITM1 gene and susceptibility to H1N1 pandemic influenza 2009 virus infection, we compared the genotype, allele and haplotype frequencies of the IFITM1 gene polymorphisms between the healthy controls and H1N1 pandemic influenza 2009 virus-affected patients. Interestingly, genotype and allele frequencies of all 3 SNPs of the IFITM1 gene showed no association between healthy controls and H1N1 pandemic influenza 2009-affected patients (Table 2). Detailed information on the genotype and allele frequencies of SNPs of the IFITM1 gene is described in Table 2. To analyze the haplotype frequencies, we investigated the distribution of haplotypes using Haploview 4.2. Three major haplotypes were found in the SNPs of the human IFITM1 gene. The detailed degree of haplotype distribution is described in Table 3. In brief, a statistical comparison of 3 major haplotypes showed no significant distribution (P > 0.9) between healthy controls and H1N1 pandemic influenza 2009 virus-affected patients. We also investigated the LD among the three IFITM1 SNPs with r2 values. Notably, all the IFITM1 SNPs showed weak LD (r2 < 0.3, Table 4).
Table 3.
Comparison of haplotype frequencies of IFITM1 polymorphisms between healthy controls and H1N1 pandemic influenza 2009 virus-affected patients in the Korean population
| Haplotypes | Frequency | P value | |
|---|---|---|---|
| Control (350) | Case (60) | ||
| CAG | 263 (0.751) | 45 (0.75) | 0.9811 |
| GAG | 85 (0.243) | 15 (0.25) | 0.9052 |
| Others | 2 (0.006) | 0 (0) | 1.0 |
Table 4.
Linkage disequilibrium (LD) among 3 polymorphisms of human IFITM1 gene in the Korean populations
| r2 | c.-416C > G | c.-327A > C | c.186 + 18G > A |
|---|---|---|---|
| c.-416C > G | – | – | – |
| c.-327A > C | 0.009 | – | – |
| c.186 + 18G > A | 0.009 | 0 | – |
Transcription factor-binding ability according to alleles of SNPs in the promoter region of the IFITM1 gene
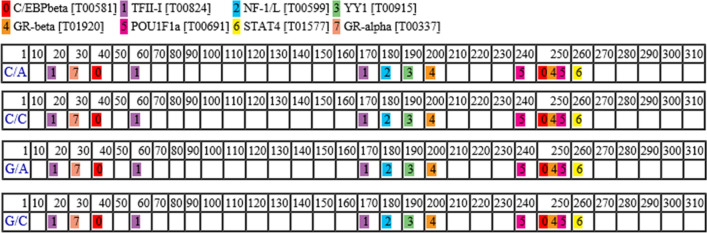
We analyzed the binding capacity of transcription factors according to alleles of regulatory SNPs using PROMO. The proximal promoter located approximately 300 base pairs upstream of the transcription start site was classified into 4 major haplotypes, and the binding ability of transcription factors was predicted according to these haplotypes. Interestingly, there was no difference in the binding ability of transcription factors according to the 4 haplotypes of promoter sequences (Fig. 2).
Fig. 2.
Analysis of the binding ability of transcription factors among 4 haplotypes of proximal promoter sequences in the human IFITM1 gene. C/A consists of the C allele of c.-416C > G and the A allele of c.-327A > C. C/C consists of the C allele of c.-416C > G and the C allele of c.-327A > C. G/A consists of the G allele of c.-416C > G and the A allele of c.-327A > C. G/C consists of the G allele of c.-416C > G and the C allele of c.-327A > C
Analysis of the genetic linkage between the IFITM1 and IFITM3 SNPs
To examine whether the IFITM1 SNPs have genetic linkage with IFITM3 SNPs, LD analysis was carried out among SNPs of these two genes in heathy controls and H1N1 pandemic influenza 2009 virus-affected patients. Detailed values are described in Tables 5, 6. In healthy controls (Table 5), the IFITM1 c.-416C > G SNP showed strong LD with 5 IFITM3 SNPs, c.-204G > T (0.422), c.-181T > C (0.851), c.-178A > C (0.866), c.-175T > C (0.866) and c.42C > T (0.321). The remaining IFITM1 SNPs showed weak LD with IFITM3 SNPs (r2 < 0.3). In H1N1 pandemic influenza 2009 virus-affected patients (Table 6), the IFITM1 c.-416C > G SNP showed strong LD with 4 IFITM3 SNPs, c.-204G > T (0.536), c.-181T > C (0.5), c.-178A > C (0.5) and c.-175T > C (0.5).
Table 5.
Linkage disequilibrium (LD) between single nucleotide polymorphisms (SNPs) of the IFITM1 and IFITM3 genes with r2 values in the healthy controls
| IFITM3 | IFITM1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| c.-204G > T | c.-188T > C | c.-181T > C | c.-178A > C | c.-175T > C | c.42C > T | c.669T > C | c.-416C > G | c.-327A > C | c.186 + 18G > A | |
| IFITM3 | ||||||||||
| c.-204G > T | – | – | – | – | – | – | – | – | – | – |
| c.-188T > C | 0.145 | – | – | – | – | – | – | – | – | – |
| c.-181T > C | 0.478 | 0.085 | – | – | – | – | – | – | – | – |
| c.-178A > C | 0.469 | 0.084 | 0.983 | – | – | – | – | – | – | – |
| c.-175T > C | 0.469 | 0.084 | 0.983 | 1.0 | – | – | – | – | – | – |
| c.42C > T | 0.758 | 0.316 | 0.349 | 0.343 | 0.343 | – | – | – | – | – |
| c.699T > C | 0.039 | 0.192 | 0.019 | 0.019 | 0.019 | 0.048 | – | – | – | – |
| IFITM1 | ||||||||||
| c.-416C > G | 0.422 | 0.084 | 0.851 | 0.866 | 0.866 | 0.321 | 0.019 | – | – | – |
| c.-327A > C | 0.005 | 0.001 | 0.01 | 0.01 | 0.01 | 0.004 | 0 | 0.01 | – | – |
| c.186 + 18G > A | 0.005 | 0.001 | 0.01 | 0.01 | 0.01 | 0.004 | 0 | 0.01 | 0 | – |
Bold texts indicate strong LD between IFITM1 SNP and IFITM3 SNPs (r2 > 0.3)
Table 6.
Linkage disequilibrium (LD) between single nucleotide polymorphisms (SNPs) of the IFITM1 and IFITM3 genes with r2 values in pandemic influenza A 2009 H1N1-affected individuals
| IFITM3 | IFITM1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| c.-204G > T | c.-188 T > C | c.-181 T > C | c.-178A > C | c.-175 T > C | c.42C > T | c.669 T > C | c.-416C > G | c.-327A > C | c.186 + 18G > A | |
| IFITM3 | ||||||||||
| c.-204G > T | – | – | – | – | – | – | – | – | – | – |
| c.-188T > C | NA | – | – | – | – | – | – | – | – | – |
| c.-181T > C | 0.932 | NA | – | – | – | – | – | – | – | – |
| c.-178A > C | 0.932 | NA | 1.0 | – | – | – | – | – | – | – |
| c.-175T > C | 0.932 | NA | 0.863 | 0.863 | – | – | – | – | – | – |
| c.42C > T | NA | NA | NA | NA | NA | – | – | – | – | – |
| c.699T > C | NA | NA | NA | NA | NA | NA | – | – | – | – |
| IFITM1 | ||||||||||
| c.-416C > G | 0.536 | NA | 0.5 | 0.5 | 0.5 | NA | NA | – | – | – |
| c.-327A > C | NA | NA | NA | NA | NA | NA | NA | NA | – | – |
| c.186 + 18G > A | NA | NA | NA | NA | NA | NA | NA | NA | NA | – |
Bold texts indicate strong LD between IFITM1 SNP and IFITM3 SNPs (r2 > 0.3)
Discussion
The IFITM1 protein is a member of the IFITM protein family, which is composed of IFITM 1, 2, 3, 5 and 10. IFITM family proteins are highly homologous with each other and contain a well-conserved CD225 domain, which consists of two major structures, transmembrane 1 and a conserved intracellular loop. The IFITM family in vertebrates can be divided into three subfamilies. IFITM1, 2 and 3 are crucial antiviral effectors, so these proteins can be classified into the immunity-related IFITM (IR-IFITM) subfamily. The IFITM1 gene is located 4.8 kb downstream of the IFITM3 gene, and the regulatory region of the IFITM3 gene affects translational regulation of the IFITM1 gene (Diamond and Farzan 2013; Smith et al. 2014; Zhao et al. 2018; Yanez et al. 2020). Since there apparently is a structural and functional relationship between IFITM1 and IFITM3 genes and the IFITM3 gene showed a strong association with the severity of and susceptibility to pandemic influenza A 2009 H1N1 virus infection, we investigated an association of the IFITM1 gene with the susceptibility to pandemic influenza A 2009 H1N1 virus infection.
Since IFITM1 polymorphisms have not been investigated thus far, we first investigated the polymorphisms of the IFITM1 gene in the present study. We found a total of 3 novel SNPs, c.-416C > G, c.-327A > C and c.186 + 18G > A (Fig. 1, Table 2). In a previous study, we found 15 polymorphisms of the IFITM3 gene in Korean populations. However, in the present study, we found only 3 SNPs in the same test groups (Kim et al. 2020). Of these 3 IFITM1 SNPs, 2 SNPs are extremely rare SNPs, with a minor allele frequency of < 1% (Table 2). The difference in the number of SNPs between the IFITM1 and IFITM3 genes indicates that the IFITM1 gene is more relatively well conserved than the IFITM3 gene. However, this result could also be influenced by ethnic features or small sample sizes; thus, further investigation is needed to confirm polymorphic characteristics of the IFITM1 gene in larger and different ethnic groups.
We compared genotype, allele and haplotype frequencies of the IFITM1 gene between healthy controls and pandemic influenza A 2009 H1N1 virus-infected patients and evaluated an association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and the IFITM1 polymorphisms. Notably, there was no association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms (Table 2). Previous studies have reported that the specificity of protection afforded by the IFITM3 protein against influenza A virus is higher than that for the IFITM1 protein, which may be the reason that no association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms was observed (Perreira et al. 2013). However, we annotated IFITM1 SNPs and found that 2 regulatory SNPs did not affect transcription factor-binding ability (Fig. 2). In addition, the remaining SNP did not disrupt splicing regulatory elements. Thus, since the IFITM1 SNPs has been shown to have no effect on the phenotype of the IFITM1 gene, nonfunctional IFITM1 SNPs found in this study can also be responsible for the lack of association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms. Further investigation of novel functional IFITM1 polymorphisms can be helpful to confirm this interpretation in the future. We found a strong genetic linkage between IFITM3 SNPs and IFITM1 c.-416C > G SNP. However, the degree of strong genetic linkage is different between healthy controls and influenza A H1N1-infected patients and the genetic LD of pandemic influenza A H1N1-infected patients showed weaker than that of healthy controls (Tables 5, 6). Thus, the weak genetic linkage between IFITM1 and IFITM3 SNPs is associated with various genotypes of these two genes and may be related to the various phenotypes of these two genes. Further analysis is needed to elucidate the association between the degree of LD value and the susceptibility of pandemic influenza A H1N1 virus infection. In addition, although the IFITM2 gene, another member of the IR-IFITM subfamily, is related to antiviral function, the IFITM2 protein showed a weaker antiviral effect than the IFITM3 protein in previous study (Perreira et al. 2013). Further study of the relationship between the susceptibility of the influenza H1N1 virus infection and IFITM2 SNPs is highly desirable in the future.
In conclusion, we identified 3 novel SNPs of the IFITM1 gene. In addition, we investigated the genotype, allele and haplotype frequencies of the IFITM1 gene, analyzed the LD of this gene and evaluated an association between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and the IFITM1 polymorphisms. Furthermore, we analyzed the binding ability of transcription factors according to the alleles of the IFITM1 SNPs and measured the LD values between the IFITM1 and IFITM3 genes. To the best of our knowledge, this report is the first association study between the susceptibility to pandemic influenza A 2009 H1N1 virus infection and IFITM1 polymorphisms.
Acknowledgements
The biospecimens and data used in this study were provided by the Biobank of Chonbuk National University Hospital, a member of the Korea Biobank Network, which is supported by the Ministry of Health, Welfare and Family Affairs. All samples derived from the Korea Biobank Network were obtained with informed consent under institutional review board-approved protocols. This research was supported by the Basic Science Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education, Science and Technology (2018R1D1A1B07048711). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876). Min-Ju Jeong was supported by the BK21 Plus Program in the Department of Bioactive Material Sciences. This work was supported by NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Author contributions
YCK, SYW and BHJ conceived and designed the experiments. YCK and SYW performed the experiments. YCK and BHJ analyzed the data. YCK, SYW and BHJ wrote the paper. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All procedures performed in the present study were accredited by the institutional review board of the Jeonbuk National University and were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards (Approval number: JBNU 2017-08-009).
Contributor Information
Yong-Chan Kim, Email: kych@jbnu.ac.kr.
Sae-Young Won, Email: gkfh32@jbnu.ac.kr.
Byung-Hoon Jeong, Email: bhjeong@jbnu.ac.kr.
References
- Allen EK, et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8(9):e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford JG, O'Keeffe M, Reading PC, Wakim LM. Rapid interferon independent expression of IFITM3 following T cell activation protects cells from influenza virus infection. PLoS ONE. 2019;14:e0210132. doi: 10.1371/journal.pone.0210132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse M, Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol. 2019;10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S, et al. Population genetics of IFITM3 in Portugal and Central Africa reveals a potential modifier of influenza severity. Immunogenetics. 2018;70(3):169–177. doi: 10.1007/s00251-017-1026-2. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13(1):46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AR, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley EM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaio V, et al. Hospitalization risk due to respiratory illness associated with genetic variation at IFITM3 in patients with influenza A(H1N1)pdm09 infection: a case-control study. PLoS ONE. 2016;11:e0158181. doi: 10.1371/journal.pone.0158181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM. The Jak-STAT pathway stimulated by interferon alpha or interferon beta. Sci STKE. 2004;2004:tr10. doi: 10.1126/stke.2602004tr10. [DOI] [PubMed] [Google Scholar]
- Horvath CM. The Jak-STAT pathway stimulated by interferon gamma. Sci STKE. 2004;2004:tr8. doi: 10.1126/stke.2602004tr8. [DOI] [PubMed] [Google Scholar]
- Kim YC, Jeong BH. No correlation of the disease severity of influenza A virus infection with the rs12252 polymorphism of the interferon-induced transmembrane protein 3 gene. Intervirology. 2017;60(1–2):69–74. doi: 10.1159/000479087. [DOI] [PubMed] [Google Scholar]
- Kim YC, Jeong BH. Ethnic variation in risk genotypes based on single nucleotide polymorphisms (SNPs) of the interferon-inducible transmembrane 3 (IFITM3) gene, a susceptibility factor for pandemic 2009 H1N1 influenza A virus. Immunogenetics. 2020;72(9–10):447–453. doi: 10.1007/s00251-020-01188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Jeong BH. Strong correlation between the case fatality rate of COVID-19 and the rs6598045 single nucleotide polymorphism (SNP) of the interferon-induced transmembrane protein 3 ( IFITM3) gene at the population-level. Immunogenetics. 2021;12(1):42. doi: 10.3390/genes12010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Jeong MJ, Jeong BH. Genetic characteristics and polymorphisms in the chicken interferon-induced transmembrane protein (IFITM3) gene. Vet Res Commun. 2019;43(4):203–214. doi: 10.1007/s11259-019-09762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Jeong MJ, Jeong BH. Strong association of regulatory single nucleotide polymorphisms (SNPs) of the IFITM3 gene with influenza H1N1 2009 pandemic virus infection. Cell Mol Immunol. 2020;17(6):662–664. doi: 10.1038/s41423-019-0322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kang JH, Yun MH, Lee SB. Curcumin inhibits poly(dA:dT)-induced IL-18 secretion by inhibiting the ASC oligomerization in human keratinocytes. Mol Cell Toxicol. 2019;15(4):399–406. doi: 10.1007/s13273-019-0043-7. [DOI] [Google Scholar]
- Mills TC, et al. IFITM3 and susceptibility to respiratory viral infections in the community. J Infect Dis. 2014;209(7):1028–1031. doi: 10.1093/infdis/jit468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, et al. IFITM3 Rs12252-C variant increases potential risk for severe influenza virus infection in Chinese population. Front Cell Infect Microbiol. 2017;7:294. doi: 10.3389/fcimb.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreira JM, Chin CR, Feeley EM, Brass A. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikine S, Nguyen JB, Modis Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Desalle R, Fisher PB. Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting. Proc Natl Acad Sci USA. 2008;105:17040–170405. doi: 10.1073/pnas.0804956105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–2005963. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Smith S, Weston S, Kellam P, Marsh M. IFITM proteins-cellular inhibitors of viral entry. Curr Opin Virol. 2014;4:71–77. doi: 10.1016/j.coviro.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JS, et al. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol. 2019;15:259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- Xuan Y, et al. IFITM3 rs12252 T>C polymorphism is associated with the risk of severe influenza: a meta-analysis. Epidemiol Infect. 2015;143:2975–2984. doi: 10.1017/S0950268815000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez DC, Ross S, Crompton T. The IFITM protein family in adaptive immunity. Immunology. 2020;159:365–372. doi: 10.1111/imm.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani A, Yount JS. Antiviral protection by IFITM3 in vivo. Curr Clin Microbiol Rep. 2018;5:229–237. doi: 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li J, Winkler CA, An P, Guo JT. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front Microbiol. 2018;9:3228. doi: 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]