Abstract
Acireductone dioxygenase (ARD) is an intriguing enzyme from the methionine salvage pathway that is capable of catalysing two different oxidation reactions with the same substrate depending on the type of the metal ion in the active site. To date, the structural information regarding the ARD–acireductone complex is limited and possible reaction mechanisms are still under debate. The results of joint experimental and computational studies undertaken to advance knowledge about ARD are reported. The crystal structure of an ARD from Homo sapiens was determined with selenomethionine. EPR spectroscopy suggested that binding acireductone triggers one protein residue to dissociate from Fe2+, which allows NO (and presumably O2) to bind directly to the metal. Mössbauer spectroscopic data (interpreted with the aid of DFT calculations) was consistent with bidentate binding of acireductone to Fe2+ and concomitant dissociation of His88 from the metal. Major features of Fe vibrational spectra obtained for the native enzyme and upon addition of acireductone were reproduced by QM/MM calculations for the proposed models. A computational (QM/MM) study of the reaction mechanisms suggests that Fe2+ promotes O—O bond homolysis, which elicits cleavage of the C1—C2 bond of the substrate. Higher M3+ /M2+ redox potentials of other divalent metals do not support this pathway, and instead the reaction proceeds similarly to the key reaction step in the quercetin 2,3-dioxygenase mechanism.
Keywords: acireductone dioxygenase, EPR spectroscopy, Mössbauer spectroscopy, protein structures, reaction mechanisms
Introduction
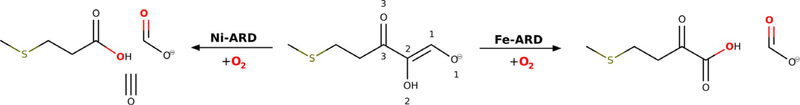
Metalloenzymes that can bind different transition-metal ions in their active centres either proceed with the same reaction regardless of the identity of the bound ion or the non-native metal ion is incapable of supporting catalysis and inhibits the enzyme. An exception to this general trend is acireductone dioxygenase (ARD). ARD from Klebsiella oxytoca (kARD) presented two different activities on the same substrate (5-methylthio-3-oxo-1-penten-1,2-diol; common name: acireductone) controlled by the metal ion in the active centre.[1] The Fe–kARD isoform yields the α-keto-acid (2-oxo-4-(methyl-thio)-butyrate) and formate by oxidative cleavage of the C1—C2 bond, whereas Ni–kARD generates methylthiopropionate, formate and carbon monoxide, which is consistent with cleavage of both the C1—C2 and C2—C3 bonds[2] (Figure 1).
Figure 1.
Reactions catalysed by Fe–ARD and Ni–ARD.[65]
This metal-dependent dual chemistry of ARD has recently been confirmed for the homologue from Mus musculus (MmARD).[3]
Isotopic-labelling studies confirmed that the carbon atom in the carbon monoxide product originates from the C2 atom of acireductone,[4,5] whereas experiments with 18O2 showed that oxygen atoms derived from dioxygen are incorporated into both organic acid products, but not into carbon monoxide in the case of Ni–ARD.[4]
The reactivity of Fe–ARD has a confirmed biological role in cells—it is a part of the 5’-methylthioadenosine (MTA) cycle that is used to synthesise (S)-adenosylmethionine (SAM). The penultimate reaction in the MTA cycle is catalysed by ARD and yields an α-keto-acid that is an immediate precursor of methionine (Met). Every mammalian cell metabolises methionine, but around 50% of its processing takes place in liver cells where Met is converted to SAM. The fact that the ARD enzyme family is evolutionarily highly conserved, from bacteria to mammals, proves the importance of the MTA cycle. Accordingly, silencing of the ARD gene causes fertility problems in mammals.[6]
Up to now it is not clear if Ni–ARD has any biological role, yet one of the reaction products—carbon monoxide—is a signalling molecule that acts as neurotransmitter, possible physiological regulator of cyclic guanosine monophosphate (cGMP)[7] and it prevents apoptosis.[8]
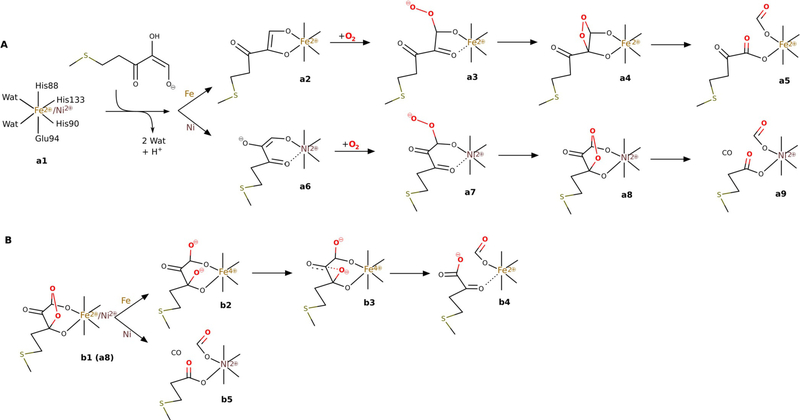
ARD belongs to a cupin superfamily of proteins that feature a conservatively preserved β-barrel fold with a jelly-roll motif.[1] In the resting form of ARD, both Fe2+ and Ni2+ have octahedral coordination geometry with a somewhat less-typical binding motif for dioxygenases: 3His-1-Acid supplemented by two water ligands. Among dioxygenases the most common binding motif is a 2His-1-Acid facial triad.[9] Metal coordination was previously studied with the extended X-ray absorption fine structure (EXAFS) method, which determined that three or four nitrogen atoms originating from His residues coordinate to the metal and one of the His residues may dissociate from the metal upon substrate binding.[10] The particular mode of substrate binding is very relevant; it was previously suggested that Fe–kARD and Ni–kARD bind the substrate differently, and hence direct the reaction into two different channels.[5] The substrate could bind to the metal ion with its O1 and O2 oxygen atoms or via atoms O1 and O3 (a2 and a6, respectively, in Figure 2A).
Figure 2.
Previously suggested mechanisms for ARD enzymes: A) the chelate hypothesis; B) the suggested mechanism based on previous DFT studies (see ref. [65]).
There are no substantial differences in the geometry of the metal cofactors for the resting states of the two isoforms of kARD, only slight differences in the bond length between the His ligands and the metal ion (2.07 Å for Ni–kARD and 2.18 Å for Fe–kARD).[11] However, NMR spectroscopic studies of kARD suggested structural differences for the C-terminal fragment, which might affect the active-site geometry, and hence the substrate binding mode (via O1 and O2 in Fe–kARD (a2); via O1 and O3 in Ni-kARD (a6); Figure 2A).[12,13] Further reaction steps would involve reaction with dioxygen and yield different endoperoxide intermediates, and therefore different products.[14,15] Notably, Fe is the only transition metal that supports the reaction leading to the ketoacid and formate, that is C1—C2 bond cleavage, whereas Ni is not unique because Mn2+ and Co2+, when bound in the ARD active site, also support C1—C2 and C2—C3 cleavage.[14]
Recently, X-ray crystal structures were obtained for Ni–MmARD and Co–MmARD with various bound ligands, however only the Ni–MmARD-d-lactic acid structure (PDB code: 5I8T) features a ligand bound directly to the metal ion.[3]
In previous computational studies on the ARD catalytic mechanism[16] it was suggested that ARD binds acireductone in the same bidentate geometry with oxygen atoms O1 and O3 coordinated to either metal and the negatively charged O2 oxygen atom stabilised by a hydrogen bond to the Arg154 residue. Then, O2 can attack the C1 and C3 carbon atoms (note these reaction steps were not studied) and afford species b1 with a five-membered endoperoxide ring (Figure 2B). The proposed reaction pathway for Fe–ARD goes through an unprecedented intermediate (b2), in which the O—O bond has been cleaved heterolytically to produce two O− ions bound to the C1 and C3 carbon atoms. It is worth noting that formally this species should feature Fe4+ otherwise some amino acid group(s) would have to be oxidised. The C3-bound O− is then proposed to migrate to the C2 carbon atom and, only then, inserts itself into the C1—C2 bond, which eventually leads to the ketoacid and formate. To the best of our knowledge the migration b2→b3→b4 is unprecedented; more commonly the C3-bound O− ion will attack and cleave the nearest bond, that is C2—C3.
In light of the results summarised above it is apparent that further studies, both experimental and computational, are needed to bring us closer to an answer to the intriguing question about the origin of the metal-dependent reaction specificity of ARD enzymes. In this work we report the results of joint structural, spectroscopic and computational studies undertaken with this aim in mind. The results, in short, reveal that ARD from Homo sapiens in a complex with selenomethionine binds dianionic acireductone to Fe2+ via the O1 and O3 atoms, which triggers dissociation of the His88 residue from the metal, and that the unique reactivity of Fe–ARD stems from the low Fe2+/Fe3+ redox potential that facilitates O—O bond homolysis coupled to concerted, but asynchronous, C1—C2 bond cleavage.
Results and Discussion
Structure model
The structural model of human ARD (hARD) consists of 178 residues. Only the last residue, Ala179, and the N-terminal linker (GAAA) could not be fitted into the electron-density map. Additionally, the model contains four sulphate ions and one acetate ion. The central part of the protein is composed of two anti-parallel β-sheets (A and B) that form a cup-shaped β-sandwich jelly roll, in which each sheet is built by antiparallel β-chains perpendicular to the barrel axis and connected by short loops. Both sheets, A (β1, β7, β6, β9, β4, β11) and B (β2, β8, β5, β10, β3), have 14352(6) topology. The protein also contains four α-helices (H1–H4), which account for 20% of the ternary structure according to the STRIDE server,[17] whereas the β-sheets account for 33%. α-Helix H4 at the C-terminus of hARD is placed along the barrel cleft.
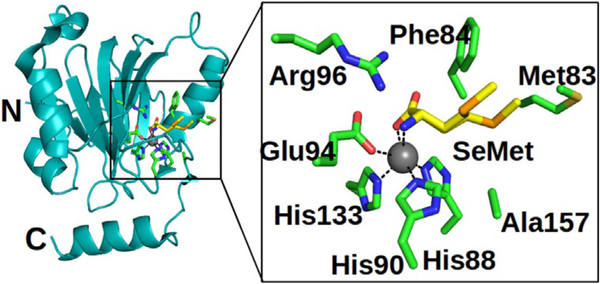
Comparison of their protein structures shows high similarity between the hARD and MmARD structures, including in the substrate-binding niche. The RMSD calculated for the backbones of hARD (PDB: 4QGN) and MmARD (PDB: 5I91) is 0.8 Å. The active centre inside the β-barrel consists of an Fe2+ ion (shown by Mössbauer analysis, and despite the enzyme being crystallised aerobically) coordinated by the side chains of the His88, His90, His133 and Glu94 residues (Figure 3), and it is open to the solvent.
Figure 3.
Cartoon representation of the overall structure of hARD and details of the enzyme active site; the N- and C-terminal residues are labelled. Residues that form active and SeMet binding sites are shown by stick representation (green). SeMet (yellow) binds to the enzyme with two distinct side chain conformations.
The bond lengths of Fe2+ to the coordinated ligands are 2.2, 2.2, 2.2 and 2.3 Å for His88, His90, His133 and Glu94, respectively. Moreover, l-selenomethionine, which was added to the crystallisation solution, coordinates the metal ion with its carboxyl and amino groups, and is trans to His90 and His133. For this ligand, which might be considered as an analogue of the natural substrate, the bond lengths to the metal are both 2.2 Å. The side chain of the l-selenomethionine ligand is surrounded by the Met83, Phe84, Ala157 and Arg96 residues.
The positions of the external ligands are different in the structures of human and mouse ARD. In Ni–MmARD (5I91) and Co–MmARD (5I8Y), 2-keto-4-(methylthio)-butyric acid (the product of the Fe–ARD reaction) is bound in a hydrophobic niche formed by the Ile98, Ile67, Ile69, Phe84, Phe105, Phe135, Val143, and Ala145 residues, and is hydrogen bonded to the Ne and Nη2 groups of Arg96 via its O2 and O5 oxygen atoms, whereas the side chain of bound l-selenomethionine in Fe–hARD is exposed to the solvent. The quality of the map and ligands can be verified by MOLSTACK.[18]
EPR spectroscopy for the NO adducts of Fe–hARD
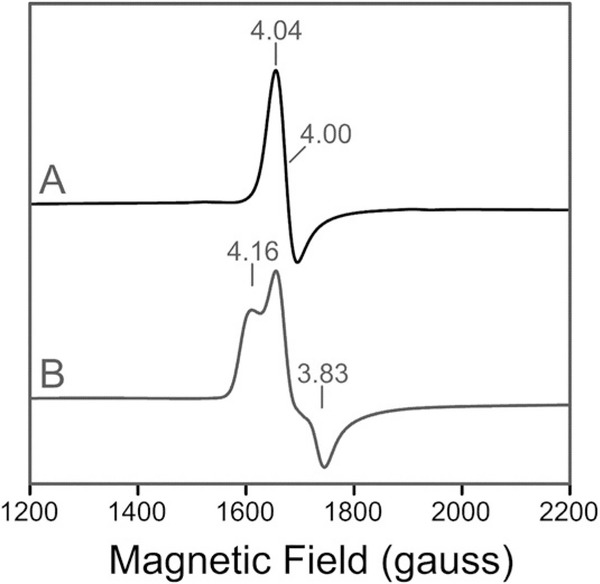
EPR spectroscopy of an anaerobic preparation of Fe–hARD revealed only a small concentration of high-spin ferric iron at g ≈4.23. Quantification of this species by double integration and comparison to a spin standard indicated that it accounts for less than 5% of the total iron in the sample. In accordance with Mössbauer spectroscopy (see below), this suggests that most of the enzyme prepared in this fashion is in the ferrous state. The 10 K EPR spectrum of the anaerobic enzyme upon exposure to NO is a highly axial spectrum (E/D ≈0.007) with g values at 4.04, 3.95 and 2.00 (Figure 4a). This type of signal is characteristic of the nitrosyl adducts of other O2 activating non-heme Fe2+ enzymes,[19] and arises from a complex with an electronic spin of S=3/2. Addition of the enolic form of model acireductone [1,2-dihydroxy-hex-1-en-3-one] (ACI) to the Fe–hARD–NO complex resulted in partial conversion (≈35%) of the axial signal to a new form with higher rhombicity (E/D=0.026) and new g values at 4.16 and 3.83 (Figure 4b). This type of perturbation, which has been observed in studies of the nitrosyl complexes of several Fe2+-dependent oxygenases,[20] demonstrates that acireductone most likely coordinates Fe directly, rather than displacing the NO ligand. Based on the yield of this new signal, a relatively high dissociation constant (≈5.4 mm) is estimated at low temperature.
Figure 4.
10 K X-band EPR spectra for the NO moiety of the nitrosyl adducts in the native form of Fe2+–hARD in the A) absence or B) presence of the acireductone substrate.
Mössbauer and nuclear inelastic scattering (NIS) spectroscopy for the native and substrate-bound forms of Fe–hARD
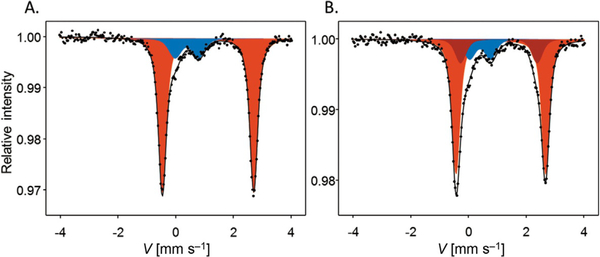
Mössbauer spectra were recorded for hARD in its native form and upon addition of a model acireductone substrate (Figure 5a and b, respectively; Table 1). For the native form the spectrum is dominated (85% of the total spectrum intensity, red component in Figure 5a) by a doublet with isomer shift (IS)=1.215(3) mms−1 and quadrupole splitting (QS)=3.165(5) mms−1, for which the hyperfine parameters are typical of high-spin Fe2+ species of the resting form of Fe–hARD. The second component (blue component in Figure 5a) accounts for 15% of the signal, and its parameters (IS=0.48(2) mms−1, QS=0.81(4) mms−1) are typical for high-spin Fe3+ species. Upon addition of ACI, the signal becomes more complex (Figure 5b) but it can be very well deconvoluted into three doublets. Two of them (red and blue as in Figure 5a) have spectral parameters very similar to those observed for the native sample (70% red: IS=1.217(7), QS=3.105(12); 14% blue: IS=0.49(2), QS=0.73(5)). The third component (dark-red, 16%) has significantly different spectral parameters (IS=1.15(2) mms−1, QS=2.66(10) mms−1), and it is ascribed to the substrate-bound high-spin Fe2+ form of Fe–hARD. The dissociation constant estimated for ACI from this Mössbauer data is 6.3 mm, which compares well with the value estimated from EPR signal quantification (5.4 mm).
Figure 5.
Mössbauer spectra recorded for hARD. A) Native form (as isolated). B) Anaerobic complex with ACI. Points are the experimental data and solid lines show the best fit for the resting Fe2+ (red shading), substrate-bound high-spin Fe2+ (dark red) and Fe3+ (blue shading) species.
Table 1.
Experimental Mössbauer parameters [isomer shift (IS) and quadruple splitting (QS)] ascribed to native and substrate-bound forms of Fe2+–hARD and the difference between the two spectral Fe2+ components (Δ) in Fe–hARD–ACI. The signal assigned to Fe3+ is not reported in the table because it is not related to an active form of the holoenzyme.
| Fe2+–hARD | Fe2+–hARD–ACI | |||
|---|---|---|---|---|
| red | red | dark-red | Δ | |
| IS [mm s−1] | 1.215(3) | 1.217(7) | 1.15(2) | −0.06(1) |
| QS [mm s−1] | 3.165(5) | 3.105(12) | 2.66(10) | −0.45(10) |
To derive some structural information regarding the coordination geometry around the Fe2+ ion, especially in the Fe–hARD–substrate complex, we constructed a model for the native state of the Fe2+–hARD (M0; see the Supporting Information) and four other models for the Fe2+–hARD–ACI complex (M1–M4; see the Supporting Information). Consistent with the results of previous EXAFS studies of Fe–kARD,[12] in M0 two water molecules complete the coordination shell of Fe2+. In M1 the water ligands are replaced by ACI, which binds to the metal as a dianion through its O1 and O2 atoms. In M2, ACI2− coordinates Fe2+ with its O1 and O3 atoms. In models M3 and M4 the substrate binds in the same way as in M1 and M2, respectively, but the side chain of the His88 residue is no longer coordinated to the metal, but is instead rotated away and exposed to the solvent. The hypothesis that the His88 residue might dissociate from the metal upon substrate binding is based on several clues. Firstly, previous EXAFS studies on Fe-kARD and Ni–kARD[10] indicated that one nitrogen ligand most likely dissociates from the metal upon substrate binding. Secondly, backbone atoms of the His88 residue are part of the protein edge at the entrance to the active site and its side chain has enough space to adapt to an alternative conformation (rotamer), in which it is placed in the metal’s second coordination shell. Thirdly, the His88 residue can be considered as an additional protein ligand to the most common 2-His-1-carboxylate facial triad binding motif, commonly found in non-heme Fe enzymes.[9] Finally, the EPR results described above for NO adducts strongly suggest that in the Fe–hARD–ACI complex the NO molecule can bind directly to the metal, which is most naturally accounted for by assuming that the coordination shell of Fe in the complex has an empty site.
The model of the native ferrous state of Fe–hARD (M0) closely reproduces the experimentally determined Mössbauer spectral parameters (Table 1 versus Table 2). However, the relative differences can benefit from error cancellation therefore the discussion below concentrates on the experimental and calculated shifts of the parameter values induced by substrate binding. Comparison of the calculated shifts (Table 2) with the experimental shifts (Table 1) revealed that, among the models considered in this work, M4 reproduces the experimental data best. Models M1 and M2 give far too small IS shifts, whereas M2 and M3 have QS shifted in the opposite direction relative to the experimental data. The QS shift for M1 is in the right direction, yet it is rather small. Hence, in the computational studies on the reaction mechanism reported below we have used model M4 for both Fe–hARD and Ni–hARD. The choice to use the same coordination model for the Fe and Ni forms was motivated by the results of the previous EXAFS studies on kARD,[10] which suggested similar changes of the metal coordination shells upon substrate binding, and the commonly observed fact that metal substitution at the enzyme active site is most often isostructural.
Table 2.
Computed Mössbauer parameters [isomer shift (IS) and quadrupole splitting (QS)] for the model of the native state (M0) and relative shifts of the parameters computed for four different models of the Fe2+–hARD–ACI complex (M1–M4) with respect to M0.
| Fe2+–hARD model (M0) | Δ(M1–M0) | Δ(M2–M0) | Δ(M3–M0) | Δ(M4–M0) | |
|---|---|---|---|---|---|
| IS [mm s−1] | 1.107 | −0.039 | −0.029 | −0.086 | −0.076 |
| QS [mm s−1] | 3.107 | −0.114 | 0.243 | 0.447 | −0.385 |
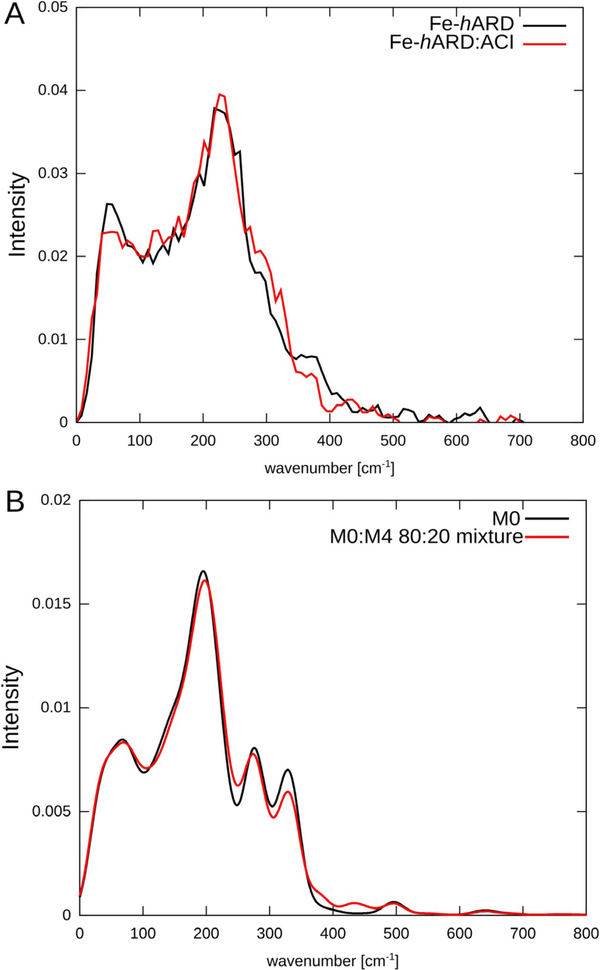
The experimental Fe vibrational densities of states (DOS) obtained for the native form of Fe–hARD and of Fe-hARD upon addition of ACI (Figure 6a) have two maxima at and 230 cm−1, with two shoulders at and 380 cm−1. The two spectra show statistically significant differences in three regions: around the maximum at and around the two shoulders at and 380 cm−1. The simulated vibrational DOS based on results of harmonic frequency calculations with our own n-layered integrated molecular orbital and molecular mechanics (ONIOM) method qualitatively reproduces these features (Figure 6b). The positions of the first two maxima are fairly well reproduced, and in the region where there are two shoulders in the experimental spectra the computational model shows two weak maxima. The intensity shift in the shoulder region that was observed upon addition of the acireductone substrate is also qualitatively reproduced. Unfortunately, due to the high dissociation constant, we were not able to prepare samples with a greater content of the Fe–hARD–ACI complex, which could reveal more differences in the spectra and possibly allow for discrimination between the various models for the complex (suggested by simulations only).
Figure 6.
A) Experimental and B) simulated iron vibrational DOS for hARD.
ONIOM(B3LYP:AMBER) studies on the reaction mechanisms
C1—C2 and C2—C3 oxidative cleavage of acireductone
The oxidative cleavage reaction catalysed by M–ARD (M=Ni, Co or Mn), which involves C1—C2 and C2—C3 bond cleavage and CO release, bears close resemblance to the reaction catalysed by quercetin 2,3-dioxygenase (QDO). In both cases the substrates are electron-rich enolates with a keto group conjugated to the enol group, two adjacent C—C bonds are cleaved with release of CO and various divalent metal ions can support the enzymatic reaction.[21,22] Thus, it is not surprising that the reaction mechanisms for hARD and QDO found with the use of computational methods are very much alike (compare Figure 7 (black arrows) and Figure S3 in the Supporting Information).
Figure 7.
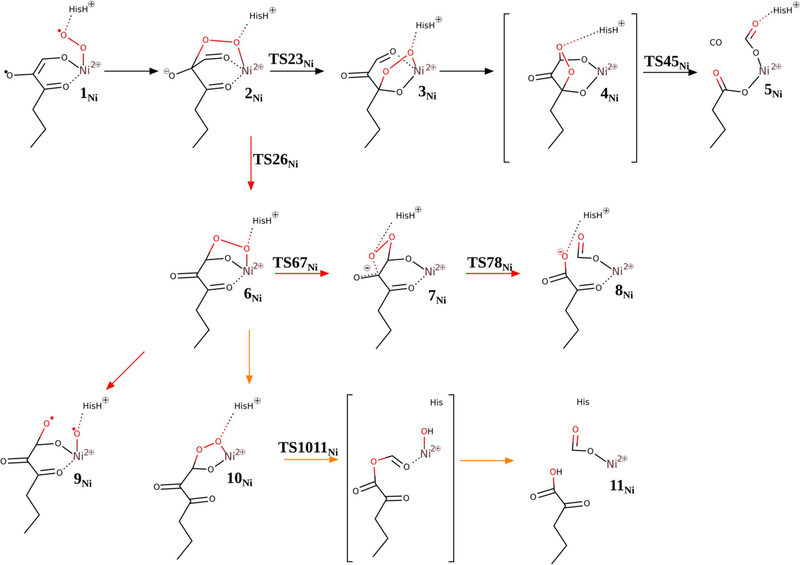
Reaction mechanisms for Ni–hARD (see ref. [65]: suggested mechanism (black arrows), analogous mechanism to Fe–hARD proposed mechanism (red arrows), Baeyer–Villiger type mechanism (yellow arrows).
Acireductone, which exists as a monoanion at physiological pH, binds to the divalent metal ion in the ARD active site as a dianion,[5] and presumably delivers a proton to the His88 residue, which most likely dissociates from the metal upon substrate binding. ACI2− coordinates the metal through its O1 and O3 atoms. Similar to QDO, dioxygen binds to the metal and accepts one electron from the electron-rich substrate (1Ni). For this species, with unpaired electrons on three centres (two on triplet Ni2+, one on O2− and one on the substrate), several spin states are close in energy; the quintet and singlet states are predicted to be 4.3 and 3.1 kcal mol−1 more stable than the triplet state, respectively. However, from species 2Ni onwards the reaction takes place on the triplet potential-energy surface (PES), therefore the triplet state of 1Ni is taken as an energy reference point (Figure 8).
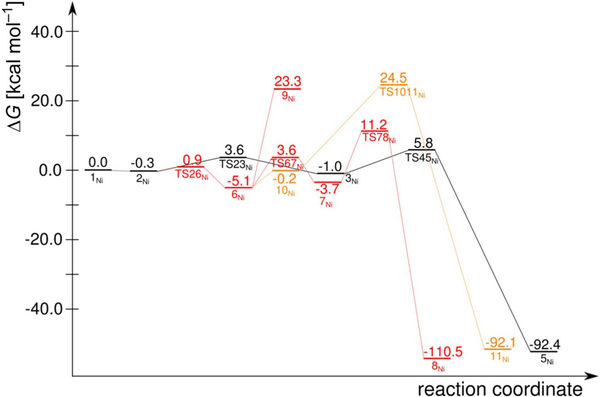
Figure 8.
Computed reaction free-energy profiles for Ni–hARD: suggested mechanism (black), mechanism analogous to the Fe–hARD proposed mechanism (red), Baeyer–Villiger type mechanism (yellow). See Figure S4 in the Supporting Information for the (Eel+ZPE) profile.
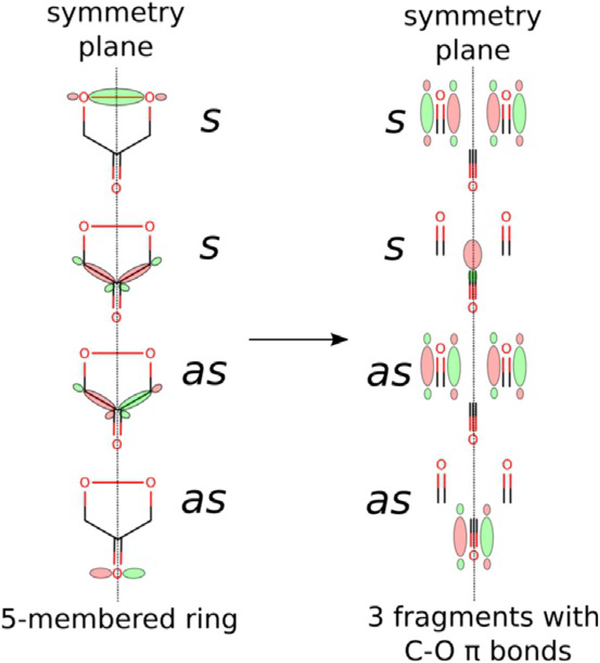
A peroxo intermediate forms easily as the distal oxygen atom forms a bond with the central carbon atom of the ketoenolate group (C2). This step (1Ni→2Ni) was predicted to proceed without any barrier; potential-energy scans of the triplet PES show a monotonous drop in energy as the two atoms get closer. Thus, species 1Ni might be stable in the quintet and singlet spin states, in which the spins located on the superoxide and substrate-derived radical are parallel (hence, not prepared for coupling into a chemical bond), and metastable on the triplet PES, in which the two spins are opposite and ready to form the bond. From 2Ni the reaction continues by migration of the C2-bound oxygen atom to the C3 keto carbon atom (2Ni→3Ni), which involves a low barrier (ΔΔG=4.8 kcalmol−1) via the transition state TS23Ni. Once the peroxo group is bound to the C3 carbon atom, the endoperoxide species with the O—O bridge between C1 and C3 can easily form. This species (4Ni) is not a stationary point for Ni–hARD, yet it lies on the reaction coordinate leading to the key transition structure TS45Ni. Just like in the QDO reaction (Figure S3 in the Supporting Information), the endoperoxide intermediate (4Ni) decays in a concerted and synchronous process of O—O, C1—C2 and C2—C3 bond cleavagel in a single step. Importantly, this reaction (3Ni→TS45Ni→5Ni) does not engage the metal ion (neither Ni2+ nor Fe2+), which is evident from the unaffected spin populations throughout this step. Moreover, no unpaired spin densities were observed for TS45Ni, which showed that the bond cleavage is heterolytic. The barrier for this process is small, which is in accordance with previous computational results.[16,23] All the above facts are consistent with the simplified orbital evolution picture that shows the process is symmetry allowed (Figure 9) because the total symmetry of the ground state, with respect to the symmetry plane passing through the middle of the O—O bond and C2 carbon atom, is preserved during the process.
Figure 9.
Simplified orbital diagram for symmetry-allowed concerted and synchronous heterolytic cleavage of the O—O, C1—C2 and C2—C3 bonds.
Alternative mechanisms we tested for Ni–hARD involved Baeyer–Villiger type rearrangement (2Ni→6Ni→10Ni→11Ni; yellow in Figures 7 and 8), homolytic O—O cleavage (2Ni→6Ni→9Ni) and a mechanism that passes through a dioxethane-like intermediate (2Ni→6Ni→7Ni→8Ni; red in Figures 7 and 8). These alternatives involve significantly higher barriers than the suggested mechanism, and hence are considered unlikely.
Because the key steps of the reaction that lead to CO release and C1—C2 and C2—C3 bond cleavage proceed without any change of oxidation state of the metal ion or its ligands, it is not surprising that very similar results were obtained for the Fe–hARD model (red in Figures 10 and 11).
Figure 10.
Reaction mechanisms considered for Fe–hARD (see ref. [65]: suggested mechanisms (black and grey arrows), mechanism analogous to that proposed for Ni–hARD (red arrows).
Figure 11.
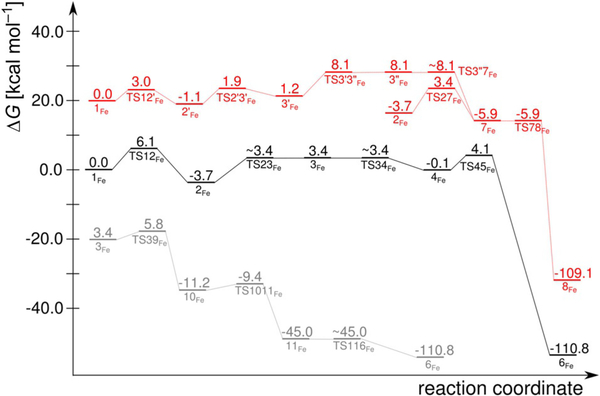
Reaction free-energy profiles for Fe–hARD [for clarity profiles for alternative reaction channels have been shifted with respect to the profile for the suggested mechanism (black)]: suggested mechanisms (black and grey) and the mechanism analogous to that proposed for Ni–hARD (red). See Figure S5 in the Supporting Information for the (Eel+ZPE) profile.
Thus, this reaction channel is characterised by a low barrier, and it is present on the PES irrespective of the identity of the metal ion, very much like the enzymatic reaction of QDO, which is supported by various divalent metal ions (e.g., Cu2+, Mn2+, Co2+ and Fe2+).[23–25] Therefore, the reaction channel unique to Fe–ARD, that is, the one leading to C1—C2 bond cleavage alone, must be fast enough to surpass this “default” symmetry-allowed process. Two plausible mechanisms for the Fe–ARD reaction are proposed below.
C1—C2 oxidative cleavage of acireductone
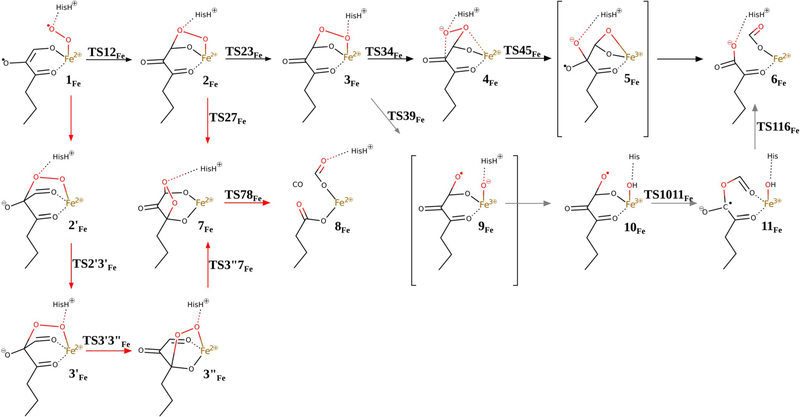
Two efficient (i.e. low barrier) reaction channels in the present study led to exclusive C1—C2 bond cleavage, which is a reaction characteristic for Fe–ARD. They feature barriers only slightly higher than that found for the CO release process and these differences (ΔΔG=0.7 and 2.4 kcalmol−1, respectively) are certainly within the error margin of the methods that can be applied today to such systems and research problems. The entrance to the two channels is hydrogen-bond rearrangement between the His88 residue and the peroxo bridge between the C1 atom and the Fe centre. This hydrogen bond migrates from the oxygen atom that is distal to Fe in 2Fe to the proximal oxygen atom in species 3Fe, which is a slightly endoergic process. From species 3Fe, the two channels diverge (black and grey in Figures 10 and 11).
One reaction channel that leads to C1—C2 cleavage (grey in Figures 10 and 11) parallels a consensus mechanism for Fe2+-dependent extradiol dioxygenases (Figure S6 in the Supporting Information).[26–29] Hence, the O—O bond+ of species 3Fe is cleaved homolytically with the help of Fe2+, which provides one electron to reduce the proximal oxygen atom. In extradiol dioxygenases the proximal oxygen atom (of species equivalent to 3Fe) accepts a proton (like here, from the second shell histidine) before the O—O bond cleavage step. However, for Fe–ARD we could not optimise a species of this type (3Fe with H+ migrated from the His88 residue to the proximal oxygen atom) because the proton always moved back to the His88 residue. Hence, for Fe–ARD O—O bond cleavage precedes proton transfer, yet the process leads to the same type of product (10Fe) because the proton jumps with no barrier once the O—O bond is broken. The following reaction steps are also very similar to those suggested for extradiol dioxygenases.[29] More specifically, the oxyl radical of species 10Fe is very reactive and easily inserts into the C1—C2 bond to form mixed anhydride radical intermediate 11Fe. The final step of the chemical reaction is attack of the OH ligand on the mixed anhydride, which yields the final product complex 6Fe. The highest free-energy point along this reaction channel is TS39Fe, which has a calculated free energy of +5.8 kcalmol−1 (cf. +3.4 kcalmol−1 for TS27Fe).
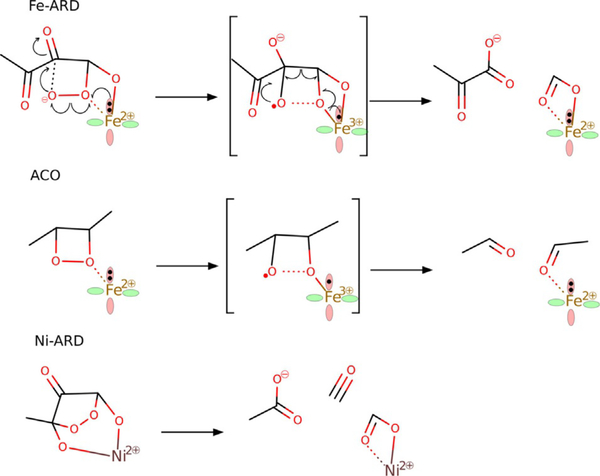
The second reaction channel that leads to C1—C2 cleavage (black in Figures 10 and 11) starts with a reorganisation of the structure of 3Fe; the proximal oxygen atom departs from Fe and makes contact with the C2 carbon atom, and the distal oxygen establishes direct contact with Fe in 4Fe. Subsequently, O—O bond cleavage takes place and, as in the “extradiol-like” channel described above, it is facilitated by electron transfer from Fe2+ to the nearby oxygen atom of the peroxo bridge. The resultant metastable intermediate features an oxyl radical bound to the C2 atom, and finally C1—C2 bond cleavage and electron transfer back to Fe complete the reaction and yield the product complex 6Fe. The highest free-energy point along this reaction channel is TS45Fe (for the structure see Figure S7 in the Supporting Information) with a calculated free energy of +4.1 kcalmol−1 (cf. +3.4 kcalmol−1 for TS27Fe). Interestingly, this reaction channel is closest to the originally suggested mechanism for Fe–ARD, which involved a dioxethane intermediate (O—O bridge between atoms C1 and C2).[5] The important difference, however, is that in the mechanisms proposed here the His88 residue departs from the metal as the substrate binds and hence provides a coordination site to bind the C1-bound oxygen atom of the peroxo intermediate. With such direct contact between the peroxo group and Fe2+ the O—O bond can be very easily cleaved; similar Fe2+-catalysed cleavage of dioxethane-like species was previously proposed in the context of the reaction mechanism of apocarotenoid dioxygenases (ACO, Figure 12).[30]
Figure 12.
Mechanism of redox catalysis by Fe2+ for O—O homolytic bond cleavage in Fe–ARD and apocarotenoid dioxygenase (ACO) compared to “uncatalysed” heterolytic cleavage of the 1,3-endoperoxo intermediate of Ni–ARD.[65]
Notably, an uncatalysed concerted O—O and C—C bond cleavage of dioxethane-like species is symmetry forbidden (see Figure S8 in the Supporting Information). Hence, the unique ability of Fe2+ to promote exclusive C1—C2 bond cleavage in acireductone is attributed to its low Fe3+/Fe2+ redox potential and the ease with which the latter can be utilised in catalysis when there is direct contact between the Fe2+ ion and the peroxo group (made possible by dissociation of the His88 residue from the metal upon substrate binding).
The alternative mechanism for Fe-hARD leads to CO release via the 1,3-endoperoxide intermediate (7Fe), which is the “default” mechanism without the redox activity of the metal cofactor. Two variants of this mechanism were identified, and they only differ in the way the 1,3-endoperoxide intermediate is reached. One involves species 2Fe, 7Fe and 8Fe and has a barrier of 3.4 kcalmol−1, and the other passed through 2′Fe, 3′Fe, 3″Fe, 7Fe and 8Fe and has a barrier of 8.1 kcalmol−1 (red arrows, Figure 10).
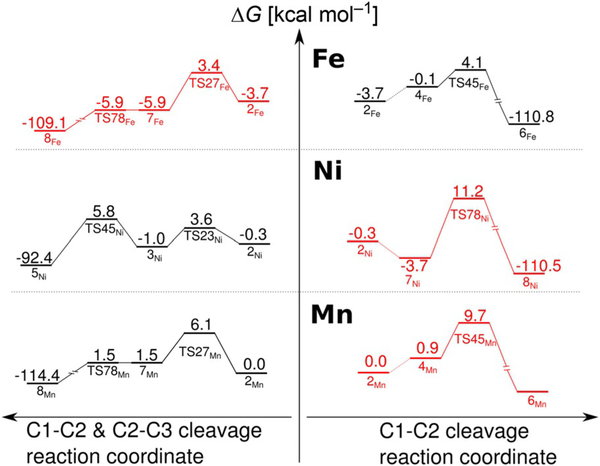
To further substantiate the explanation of the metal-dependent reaction specificity of ARD proposed above we performed additional calculations for Ni–hARD and Mn–hARD. From the range of metals that support the CO-release mechanism of ARD (i.e., Mn, Co and Ni), Mn has the M3+/M2+ redox potential most similar to that of Fe (E=1.51 and 0.77 eV for Mn and Fe, respectively).[31] Substituting Fe2+ with Mn2+ hardly changes the reaction free-energy profile for the CO-release mechanism (Figure 13 left; the barrier drops by 1.0 kcalmol−1), whereas the barrier for exclusive C1—C2 cleavage (Figure 13 right) increases by 1.9 kcalmol−1.
Figure 13.
Key fragments of the reaction-energy profiles computed for M2+–hARD that explain the unique ability of Fe2+ to promote exclusive C1—C2 oxidative bond cleavage. Experimentally observed reactions (black); alternative reaction (red).
Ni, unlike Fe and Mn, is not oxidised to M3+ during the O—O cleavage (for the structure of TS78Ni, see Figure S9 in the Supporting Information), thus the difference between the activation barriers for the two reaction channels is even larger (Figure 13). Therefore, the proposed mechanisms successfully reproduce the observed reaction specificity for Mn, Ni, and, within the uncertainty of the model and methods used here, also for Fe–hARD.
Other mechanisms considered for Ni-hARD
Taking into account the high redox potential of the Ni3+/Ni2+ couple it was unsurprising that for Ni-hARD the O—O bond homolysis starting from species 6Ni is a difficult process (see Figure 7 and Figure 8). Similarly a high barrier was found for a Baeyer–Villiger type rearrangement to give the C1—C2 bondcleavage products (6Ni→10Ni→11Ni; Figure 8). Considering the high barriers computed for these two processes they are reasonably considered to be very improbable mechanisms.
Conclusion
hARD was crystallised in complex with Se-methionine, which can be viewed as a model for the natural enzyme substrate acireductone. The structure of the complex was used as a starting point for computational studies that helped to interpret the Mössbauer spectra obtained for the native and substrate-bound forms of Fe–hARD. Four different models were considered for the Fe–hARD–ACI complex, and the model with ACI bound to Fe2+ through atoms O1 and O3 with the His88 residue dissociated from the metal best reproduced the parameters of the observed spectra. EPR spectra obtained for NO adducts with native and ACI-bound Fe–hARD support the model with one of the protein ligands dissociating upon substrate binding, which is also in agreement with previous EXAFS studies on Fe- and Ni-kARD.[10]
ONIOM studies of the reaction mechanisms, employing the enzyme—substrate model consistent with the spectroscopic results, revealed that the reaction channel leading to CO release after C1—C2 and C2—C3 oxidative cleavage is present irrespective of the identity of the divalent metal ion bound in the active site because the metal is not redox active throughout the reaction. This reaction mechanism parallels that suggested for QDO, which—like ARD—can accept different metal ions in the active site and catalyse the cleavage of two C—C bonds with concomitant CO release from an enolate substrate. This process is symmetry allowed and involves a low activation barrier that hardly varies with the metal identity. The second reaction channel is only available in the case of Fe2+, and can rival the first because it involves one-electron oxidation of the metal at the key step (O—O bond cleavage). In this process the symmetry forbidden concerted O—O and C1—C2 bond cleavage is avoided. Instead, the O—O bond is cleaved first by Fe redox catalysis, which is made possible by dissociation of the His88 residue from the metal upon substrate binding. We hope the results presented here will provoke further experimental studies on this intriguing metal-dependent oxidation reaction.
Experimental Section
Gene cloning and expression
The human gene of ARD was cloned into modified pET15b vector, in which the gene for thioredoxin was first cloned into the NcoI/NdeI sites then a linker containing HisTag and Tobacco Etch Virus (TEV) protease cleavage site (ENLYFQ/G) followed by a restriction site for NotI was introduced into NdeI/XhoI sites.[32] The gene of hARD was amplified by using primers (forward: 5′-CG GCGGCCGC T ATG GTT CAG GCG TGG TAC ATG-3′; reverse: 5′-ATA CTCGAG TTA TGC CGT CTG AGC CAG GAA TTT AAC G-3′), and then cloned into the NotI/XhoI sites of modified pET15b. The clones were confirmed by DNA sequencing and transformed into E. coli strain BL21 (DE3) Codon Plus RIPL (Stratagene). Bacteria were grown at 37°C to OD600=0.6 in LB medium and incubated with shaking overnight after induction with isopropyl β-D-1-thiogalactopyranoside (0.25 mm) in 12°C, and then harvested and stored at −80°C.
To obtain an enzyme with a specific metal ion (57Fe2+), the bacterial culture was prepared by using a minimal medium (1 L) that consisted of Na2HPO3 (15 g), KH2PO4 (3 g), NaCl (0.5 g) and NH4Cl (1 g). The appropriate metal salt (20 μm) was added 10 min after induction. The culture was additionally supplied with citrate (25 μm).
Protein purification
The thawed cells were resuspended in buffer A [Tris (50 mm) pH 7.8, NaCl (500 mm), glycerol (5%), inidazole (2.5 mm)] and lysed by a cell disruptor after addition of the protease inhibitor cocktail cOmplete (Roche). Recombinant hARD protein was purified by affinity chromatography with Ni-NTA resin (Qiagen) as previously published.[33] The supernatant after centrifugation was applied to the Ni-NTA resin and washed with buffer A containing imidazole (10 mm). The protein was eluted with buffer A containing imidazole (350 mm). Thioredoxin with a HisTag was cleaved by treatment with His-tagged TEV protease overnight in dialysis buffer [Tris (50 mm) pH 7.8, NaCl (500 mm), glycerol (5%)]. The hARD protein was obtained by flowing the mixture through a second Ni-NTA column. Pure hARD protein was obtained after size-exclusion chromatography with a HiLoad16/60 Superdex 200 prep grade column attached to an AKTA fast protein liquid chromatography (FPLC) system (GE Healthcare) in crystallisation buffer [Tris (50 mm) pH 7.8, NaCl (150 mm)] and concentrated by using an Amicon 10 kDa molecular weight cut-off concentrator (Millipore).
Protein crystallisation, data collection and structure determination
hARD protein (0.92 mm) was crystallised with l-selenomethionine (2.5 mm) by sitting-drop vapour diffusion under the following crystallisation conditions: 1:0.8 sodium acetate (0.1m), ammonium sulphate (2.0m pH 4.5) from Index screen (HamptonResearch) at 4°C. Diffraction data were collected at the 19-ID beam line at the Advanced Photon Source (APS) at Argonne National Laboratory (Argonne, IL, USA) at 100 K.[34] Data processing and the initial model were obtained with the HKL-3000 package[35] which interacts with MOLREP[36] and other programs from the CCP4 package.[37] The structure was solved by using the molecular-replacement method with a structure for Mus musculus ARD (PDB ID: 1VR3)[1] used as a starting model. The obtained model was further rebuilt and refined with CCP4, REFMAC5[38] and COOT[39] as implemented in HKL-3000. The MOLPROBITY[40] and ADIT[41] programs were used for structure validation. The protein crystallised in the P64 space group with one protein chain in the asymmetric unit. Data set parameters and the structure refinement statistics are presented in Table S1 (see the Supporting Information). The structure was determined to 3.05 Å resolution and deposited in the PDB (identifier: 4QGN).
Substrate synthesis
The model dethio ARD substrate, which features an n-propyl chain instead of the CH3—S—CH2—CH2— unit of the natural substrate acireductone, was synthesised in its keto form [1,2-dihydroxy-5-(methylthio)pent-1-en-3-one] as described previously.[42] Throughout the text the enolic form of the model substrate is abbreviated as ACI.
EPR Spectroscopy
The substrate powder (1 mg) was resuspended in H2O (2.29 mL) and allowed to stir overnight in an anaerobic chamber. Subsequently, the substrate solution (150 μL) was mixed with N2 purged KPi buffer (50 mm, 350 μL, pH 7.5) and heated for 2 min at 90°C. Immediately after heating, a UV/Vis spectrum of the substrate was recorded that showed a maximum absorbance at λ=307 nm, which confirmed conversion to the enol form. The extinction coefficient (ε307=16400m−1 cm−1) was used to calculate the concentration of the final substrate stock used to prepare the enzyme–substrate complexes.
The anaerobically purified protein (300 mm) was thawed, made anaerobic by degassing with N2 and transferred to a Coy anaerobic chamber. As isolated ARD was transferred directly to a sealed quartz EPR tube and frozen in liquid N2. Nitrosyl complexes were prepared by the addition of MAHMA NONOate (3 equiv) from a 10 mm stock solution prepared in NaOH (10 mm). The NO-bound enzyme was either transferred to the EPR tube and frozen, or incubated with ACI (10 equiv) prior to freezing. EPR spectroscopy was performed by using an X-band Bruker EMXplus spectrometer equipped with an Oxford Instruments ESR900 liquid helium continuous flow cryostat. Spectra were recorded at 10 K with a 0.6 mT modulation amplitude and 5 mW microwave power.
57Fe Mössbauer and NIS spectroscopy
The protein was purified according to the procedure described above, but under anaerobic conditions in a glovebox without the final size-exclusion chromatography step. The protein purity was confirmed by SDS-PAGE electrophoresis. The enzyme was concentrated to 1.07 mm in a glovebox. The 57Fe content in the hARD sample was 97.7±0.3%, which was measured by using the CrackSet10 (Merck) for the sample mineralisation and Spectroquant Iron Test (Merck) for metal analysis according to an available manual adjusted for a 0.5 mL sample volume.
Firstly, the Mössbauer data was collected for the native form of Fe–hARD (1 mL). Next, Fe–hARD (0.71 mm) was mixed with a solution of ACI (1.64 mm). The Mössbauer spectra were recorded at 85 K by using a home-made liquid nitrogen stationary cryostat and a conventional-transmission constant-acceleration spectrometer. A Co/Rh (100 mCi) was kept at room temperature. The spectra were fitted by using a Voigt-line based method for the description of a static quadrupole interaction distribution.[43] The isomer shifts are given with respect to metallic iron at RT.
The NIS experiment (also known as a nuclear resonance vibrational spectrum—NRVS)[45] of synchrotron radiation (SR) was performed at the Nuclear Resonance beamline ID 18[44] at the European Synchrotron Radiation Facility in Grenoble, France, for the native form of Fe–hARD and upon anaerobic addition of ACI. In a NIS experiment, incident SR-photons that are slightly detuned from the 57Fe nucleus excitation energy can be resonantly absorbed by the nucleus if the energy is exchanged with Fe atom vibrations. The experimental NIS spectrum represents a probability of the absorption as a function of detuning the SR energy from the resonance. In the present experiment the energy of SR was monochromatised to 2 meV and tuned around the transition energy of 57Fe (14.412 keV) from −20 meV to 100 meV. NIS spectra were recorded at 30 K by using a flow cryostat. The intensity of the nuclear absorption was measured by counting the delayed 6.4 keV KFe fluorescence by using a large-area fast-avalanche photodiode. Along with the NIS spectra, coherent nuclear forward scattering was measured by a second detector, and provided simultaneous control of the instrumental function (from the energy spectrum) and the sample hyperfine pattern (from the time spectrum). A statistically meaningful NIS spectrum was obtained after approximately 20 h. After subtraction of the elastic contribution by using the instrumental function, DOS was calculated from the inelastic part of the NIS spectrum in a parameter-free procedure, described by Kohn and Chumakov.[46] More details of the experimental method and data evaluation can be found in the literature.[47]
Computational details
Classical MD simulations:
Molecular dynamics simulations were performed under periodic boundary conditions with the use of the Amber ff03 force field for standard amino acid residues,[48] GAFF force field for model acireductone[49] and TIP3P explicit model for water.[50] Atomic charges for the metal cofactor and its first shell ligands were derived from the electrostatic potential (ESP) calculated with the B3LYP exchange-correlation functional and basis set combining the lacv3p+ basis for Fe and the cc-pVTZ basis for all other atoms. A solvent model with dielectric constant of 4.0 and probe radius 1.4 a was used in these ESP calculations. The models and procedures of charge fitting were the same as those proposed previously.[51] The bonded terms of the force field that are due to explicit bonds between Fe and its ligands were projected from the Hessian matrix calculated at the B3LYP/lacvp level.[52]
The simulation cell included either hARD–Fe2+–(H2O)2, which is a resting state of the enzyme, or one of four models of the hARD–Fe2+–ACI2− complex, water molecules filling the rectangular box with at least 10 Å between the protein surface and the wall of the box, and an appropriate number of Na+ ions to neutralize the electric charge of the protein–substrate complex. The protonation state of the protein residues was predicted by the propKa 3.1 program.[53]
Prior to the MD runs, the structures were minimised. The SHAKE algorithm was used to constrain bonds formed by the hydrogen atoms, the integration time was 2 fs and standard cut-off values were used for non-bonding interactions. All simulations were at T=300 K and p=1 bar and their length (each) exceeded 180 ns. Snapshots were saved every 10 ps, and the last 10 ns of a stable “production” period of the simulations were used for clustering based on the geometry of the protein residues within 10 Å of the metal ion. A representative structure of a dominating cluster was chosen for subsequent QM/MM optimisation.
The four models of the hARD–Fe2+–ACI2− complex differed in the coordination around the metal. ACI coordinated Fe either via the O1 and O2 or O1 and O3 oxygen atoms. Moreover, the possibility that binding of ACI to Fe2+ can trigger dissociation of the His88 residue from the metal centre was considered.
Classical MD simulations were performed with the Amber12 package[54] by using the GPU version of the PMEMD program.[55]
ONIOM(B3LYP:AMBER):
The real system includes the whole protein and all water molecules located within 20 Å of Fe. All residues with atoms within 15 Å from Fe were allowed to optimize their geometry, whereas coordinates of the remaining part of the system were fixed (see Figure S1 in the Supporting Information).
The QM region included Fe2+, ACI2− or two water ligands, amino acid side chains coordinating the metal centre and amino acid side chains and water molecules forming hydrogen bonds with the first shell ligands. Two basis sets were employed: the lacvp basis (BS1) for most optimisations, frequency and ESP calculations and a basis set that combined the lacv3p+ basis set for the metal ion with 6–311G(d,p) basis set for all other atoms (BS2). BS2 was used for geometry optimisation of the models to calculate Mössbauer parameters and for single-point energy calculations along a reaction coordinate. Mechanical embedding was used for geometry optimisation, yet the atomic charges of the QM atoms were updated for each stationary point optimised. To this end, an initial model for a given stationary point was first optimised with the atomic charges for the QM region as for the preceding structure. Then a new set of charges was derived from ESP calculated in the presence of point charges of the MM region (ONIOM electronic embedding scheme). Finally, the geometry was reoptimised with the new set of QM charges. Previous works have shown that mechanical embedding with a charge-update scheme converges very quickly[56] and yields reaction-energy profiles very similar to those obtained with electronic embedding.[57] A D3(BJ) van der Waals correction was computed for the QM region with the DFTD3 program.[58,59] The final reported energies combine the ONIOM electronic-embedding energies with the BS2 basis set, D3(BJ) corrections for van der Waals interactions within the QM region and the zero-point energy (ZPE) term plus thermal corrections to the Gibbs free energy computed within the harmonic approximation and ONIOM mechanical embedding with the BS1 basis set (reaction-energy profiles without the thermal correction, but with ZPE included, are presented in the Supporting Information). All ONIOM calculations were performed with the Gaussian 09 software package.[60]
Computational Mössbauer spectroscopy:
Mössbauer parameters were calculated with ORCA[61] with the B3LYP functional and a basis set that combined the CP(PPP) basis set for Fe, TZVP basis set for O and N atoms and SV(P) basis set for C and H atoms. Molecular mechanics atoms were represented by their atomic charges (ONIOM electronic-embedding Scheme). Regression parameters to calculate isomer shifts were taken from the literature.[62]
For comparison with the NIS experiment, the Fe DOS values were simulated with the Q-Spector program, which we obtained courtesy of Dr. Vladimir Pelmentschikov.[63,64] Uniform Gaussian line broadening with full width at half maximum (FWHM)=14 cm−1, which corresponds to the experimental energy resolution of 2 meV, was used to model DOS.
Optimised structures have been deposited in the ioChem database and are freely available at: https://doi.org/10.19061/iochem-bd-4-4 (Fe–hARD Mössbauer models) https://doi.org/10.19061/iochem-bd-4-6 (Fe–hARD) https://doi.org/10.19061/iochem-bd-4-5 (Ni–hARD).
Supplementary Material
Acknowledgements
This research project was supported by grant no. UMO-2011/03/B/NZ1/04999 from the National Science Centre, Poland. This research was supported in part by PL-Grid Infrastructure. Computations were performed in the AGH Cyfronet Supercomputer Centre. The structural work was supported by National Institutes of Health (grants: GM117325 and HG008424). We wish to thank the EU COST association for support through COST action CM1305. XRD results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The NIS spectra reported in this report are derived from work performed at European Synchrotron Radiation Facility, Grenoble, France.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information, including animated .gif files that present the imaginary frequency normal modes for TS45Fe, TS45Ni, TS78Fe and TS78Ni, and the ORCID numbers for the authors of this article can be found under https://doi.org/10.1002/chem.201704617.
References
- [1].Xu Q, Schwarzenbacher R, Sri Krishna S, McMullan D, Agarwalla S, Quijano K, Abdubek P, Ambing E, Axelrod H, Biorac T, Canaves JM, Chiu H-J, Elsliger M-A, Grittini C, Grzechnik SK, DiDonato M, Hale J, Hampton E, Han GW, Haugen J, Hornsby M, Jaroszewski L, Klock HE, Knuth MW, Koesema E, Kreusch A, Kuhn P, Miller MD, Moy K, Nigoghossian E, Paulsen J, Reyes R, Rife C, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, White A, Wolf G, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA, Proteins Struct. Funct. Bioinforma. 2006, 64, 808–813. [DOI] [PubMed] [Google Scholar]
- [2].Dai Y, Wensink PC, Abeles RH, J. Biol. Chem. 1999, 274, 1193–1195. [DOI] [PubMed] [Google Scholar]
- [3].Deshpande AR, Wagenpfeil K, Pochapsky TC, Petsko GA, Ringe D, Biochemistry 2016, 55, 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wray JW, Abeles RH, J. Biol. Chem. 1995, 270, 3147–3153. [DOI] [PubMed] [Google Scholar]
- [5].Dai Y, Pochapsky TC, Abeles RH, Biochemistry 2001, 40, 6379–6387. [DOI] [PubMed] [Google Scholar]
- [6].Chou H-Y, Lin Y-H, Shiu G-L, Tang H-Y, Cheng M-L, Shiao M-S, Pai L-M, J. Biomed. Sci. 2014, 21, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Verma A, Hirsch D, Glatt C, Ronnett G, Snyder S, Science 1993, 259, 381–384. [DOI] [PubMed] [Google Scholar]
- [8].Kim KM, Pae H-O, Zheng M, Park R, Kim Y-M, Chung H-T, Circ. Res. 2007, 101, 919–927. [DOI] [PubMed] [Google Scholar]
- [9].Kovaleva EG, Neibergall MB, Chakrabarty S, Lipscomb JD, Acc. Chem. Res. 2007, 40, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chai SC, Ju T, Dang M, Goldsmith RB, Maroney MJ, Pochapsky TC, Biochemistry 2008, 47, 2428–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Al-Mjeni F, Ju T, Pochapsky TC, Maroney MJ, Biochemistry 2002, 41, 6761–6769. [DOI] [PubMed] [Google Scholar]
- [12].Ju T, Goldsmith RB, Chai SC, Maroney MJ, Pochapsky SS, Pochapsky TC, J. Mol. Biol. 2006, 363, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ragsdale SW, J. Biol. Chem. 2009, 284, 18571–18575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pochapsky TC, Pochapsky SS, Ju T, Mo H, Al-Mjeni F, Maroney MJ, Nat. Struct. Biol. 2002, 9, 966–972. [DOI] [PubMed] [Google Scholar]
- [15].Boer JL, Mulrooney SB, Hausinger RP, Arch. Biochem. Biophys. 2014, 544, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sparta M, Valdez CE, Alexandrova AN, J. Mol. Biol. 2013, 425, 3007–3018. [DOI] [PubMed] [Google Scholar]
- [17].Heinig M, Frishman D, Nucleic Acids Res. 2004, 32, W500–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Porebski PJ, Sroka P, Zheng H, Cooper DR, Minor W, Protein Sci. 2017, 10.1002/pro.3272 [DOI] [PMC free article] [PubMed]; b) http://molstack.bioreproducibility.org/project/view/8v2Rtgx5lMHcTEeImc2h.
- [19].Arciero DM, Lipscomb JD, Huynh BH, Kent TA, Münck E, J. Biol. Chem. 1983, 258, 14981–14991. [PubMed] [Google Scholar]
- [20].Arciero DM, Lipscomb JD, J. Biol. Chem. 1986, 261, 2170–2178. [PubMed] [Google Scholar]
- [21].Schaab MR, Barney BM, Francisco WA, Biochemistry 2006, 45, 1009–1016. [DOI] [PubMed] [Google Scholar]
- [22].Wojdyła Z, Borowski T, J. Biol. Inorg. Chem. 2016, 21, 475–489. [DOI] [PubMed] [Google Scholar]
- [23].Siegbahn PEM, Inorg. Chem. 2004, 43, 5944–5953. [DOI] [PubMed] [Google Scholar]
- [24].Gopal B, Madan LL, Betz SF, Kossiakoff AA, Biochemistry 2005, 44, 193–201. [DOI] [PubMed] [Google Scholar]
- [25].Adams M, Jia Z, J. Biol. Chem. 2005, 280, 28675–28682. [DOI] [PubMed] [Google Scholar]
- [26].Fielding AJ, Lipscomb JD, Que L, JBIC J Biol. Inorg. Chem. 2014, 19, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Borowski T, Wójcik A, Miłaczewska A, Georgiev V, Blomberg MRA, Siegbahn PEM, J. Biol. Inorg. Chem. 2012, 17, 881–890. [DOI] [PubMed] [Google Scholar]
- [28].Borowski T, Georgiev V, Siegbahn PEM, in J Mol. Model, Springer-Verlag, 2010, pp. 1673–1677. [DOI] [PubMed] [Google Scholar]
- [29].Siegbahn PEM, Haeffner F, J. Am. Chem. Soc. 2004, 126, 8919–8932. [DOI] [PubMed] [Google Scholar]
- [30].Borowski T, Blomberg MRA, Siegbahn PEM, Chem. Eur. J. 2008, 14, 2264–2276. [DOI] [PubMed] [Google Scholar]
- [31].CRC Handbook of Chemistry and Physics, 87th ed. Ed: Lide David R. (National Institute of Standards and Technology). CRC Press/Taylor and Francis Group,2006. Boca Raton, FL. [Google Scholar]
- [32].Petkowski JJ, NRMT: An α-N-Methyl Transferase, Ph. D. Thesis, University of Virginia, 2012. [Google Scholar]
- [33].Zhang R-G, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Sevchenko A, Structure 2001, 9, 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang R-G, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A, Synchrotron Radiat J. 2006, 13, 30–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, Acta Crystallogr. Sect. D 2006, 62, 859–866. [DOI] [PubMed] [Google Scholar]
- [36].Vagin A, Teplyakov A, Acta Crystallogr. Sect. D 2010, 66, 22–25. [DOI] [PubMed] [Google Scholar]
- [37].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Acta Crystallogr. Sect. D 2011, 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, Acta Crystallogr. Sect. D 2011, 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Emsley P, Cowtan K, Acta Crystallogr. Sect. D 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- [40].Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, Richardson DC, Acta Crystallogr. D. 2010, 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD, Acta Crystallogr. Sect. D 2004, 60, 1833–1839. [DOI] [PubMed] [Google Scholar]
- [42].Trzewik B, Chruszcz-Lipska K, Miłaczewska A, Opalińska-Piskorz J, Karcz R, Gryboś R, Oszajca M, Luberda-Durnaś K, Łasocha W, Fitch A, Sulikowski B, Borowski T, New J Chem. 2016, 40, 9291–9303. [Google Scholar]
- [43].Rancourt DG, Ping JY, Nucl. Instrum. Methods Phys. Res. Sect. B 1991, 58, 85–97. [Google Scholar]
- [44].Rüffer R, Chumakov AI, Hyperfine Interact. 1996, 97–98, 589–604. [Google Scholar]
- [45].Alp EE, Sturhahn W, Toellner TS, Zhao J, Hu M, Brown DE, Hyperfine Interact. 2002, 144, 3–20. [Google Scholar]
- [46].Kohn VG, Chumakov AI, Hyperfine Interact. 2000, 125, 205–221. [Google Scholar]
- [47].Chumakov AI, Rüffer R, Leupold O, Sergueev I, Struct. Chem. 2003, 14, 109–119. [Google Scholar]
- [48].Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P, J. Comput. Chem. 2003, 24, 1999–2012. [DOI] [PubMed] [Google Scholar]
- [49].Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA, J. Comput. Chem. 2004, 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- [50].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- [51].Peters MB, Yang Y, Wang B, Füsti-Molnár L, Weaver MN, Merz KM Jr, J. Chem. Theory Comput. 2010, 6, 2935–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Seminario JM, Int. J. Quantum Chem. 1996, 60, 1271–1277. [Google Scholar]
- [53].Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA, Nucleic Acids Res. 2004, 32, W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Hayik S, Roitberg A, Seabra G, Swails J, Gøtz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA, AMBER 12, 2012. [Google Scholar]
- [55].Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC, Chem J. Theory Comput. 2013, 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- [56].Dutta D, Mishra S, Phys. Chem. Chem. Phys. 2014, 16, 26348–26358. [DOI] [PubMed] [Google Scholar]
- [57].Kawatsu T, Lundberg M, Morokuma K, Chem J. Theory Comput. 2011, 7, 390–401. [DOI] [PubMed] [Google Scholar]
- [58].Grimme S, Antony J, Ehrlich S, Krieg H, J. Chem. Phys. 2010, 132, 154104. [DOI] [PubMed] [Google Scholar]
- [59].Grimme S, Ehrlich S, Goerigk L, J. Comput. Chem. 2011, 32, 1456–1465. [DOI] [PubMed] [Google Scholar]
- [60].Gaussian 09, Revision D.01, Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ, Gaussian, Inc., Wallingford CT, 2009. [Google Scholar]
- [61].Neese F, WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar]
- [62].Römelt M, Ye S, Neese F, Inorg. Chem. 2009, 48, 784–785. [DOI] [PubMed] [Google Scholar]
- [63].Lauterbach L, Gee LB, Pelmenschikov V, Jenney FE, Kamali S, Yoda Y, Adams MWW, Cramer SP, Dalton Trans. 2016, 45, 7215–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pelmenschikov V, Guo Y, Wang H, Cramer SP, Case DA, Faraday Discuss. 2011, 148, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].MarvinSketch was used for drawing structures and reactions; MarvinSketch (14.11.10.0), ChemAxon, 2014, http://www.chemaxon.com/products/marvin/marvinsketch/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.