Abstract
Purpose of Review
Human race is currently facing the wrath of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a highly transmittable and pathogenic RNA virus, causing coronavirus disease 2019 (COVID-19), the worst ever global pandemic. Coronaviruses (CoVs) have emerged as a major public health concern. Urgent global response to COVID-19 outbreak has been to limit spread of SARS-CoV-2 via extensive monitoring and containment. Various treatment regimens have been adopted to manage COVID-19, with known drugs and drug combinations used to decrease the morbidity and mortality associated with COVID-19. Intensive research on various fronts including studying molecular and structural aspects of these viruses and unraveling the pathophysiology and mechanistic basis of COVID-19 aimed at developing effective prophylactic, therapeutic agents and vaccines has been carried out globally.
Recent Findings
No approved antiviral treatment except remdesivir exists for SARS-CoV-2 till date though novel drug targets have been identified. However, worldwide frantic and competitive vaccine development pharmaceutical race has borne fruit in the form of a number of promising candidate vaccines, out of which few have already received emergency use authorization by regulatory bodies in record time.
Summary
This review highlights the painstaking efforts of healthcare workers and scientific community to successfully address the COVID-19 pandemic—though damage in the form of severe illness, loss of lives, and livelihood has left a serious mark. Focusing on extensive research on various therapeutic options and antiviral strategies including neutralizing antibodies, potential drugs, and drug targets, light has been shed on various diagnostic options and the amazing vaccine development process as well.
Keywords: COVID-19, Coronaviruses, SARS-CoV-2, Vaccines, RT-PCR
Introduction
The disease called “COVID-19” has been declared as a global pandemic on March 11, 2020, by the World Health Organization (WHO). COVID-19 is moderately infectious with a relatively high mortality rate, but the information available in public reports and published literature is rapidly increasing. It is caused by a virus initially known as “novel coronavirus 2019” (nCoV-2019) which was renamed as “SARS-CoV-2” by the international committee of the Coronavirus Study Group (CSG) [1]. Although origin of the virus is not known, it is thought to be originated from bats [2]. SARS-CoV-2 is an enveloped RNA beta-coronavirus, with phylogenetic similarity to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) [3]. The US Food and Drug Administration issued an emergency use authorization for investigational antiviral drug remdesivir for the treatment of suspected or laboratory-confirmed COVID-19 in adults and children hospitalized with the severe disease [4]. Although many medications are being tested, there are still limited therapeutic options to treat COVID-19 patients. Angiotensin receptor blockers, such as losartan, have also been suggested for the treatment of COVID-19 [5]. So, the best solution for controlling the pandemic will be the simultaneous application of sensitive diagnostic approaches, using current available drugs while still developing novel treatments, and above all the development of vaccines for long-term prevention of the disease [6]. COVID-19 is a public health concern, and ongoing changes in the climate make upcoming occurrence of such pandemics more probable [7••]. As per the US CDC criteria, epidemiological factors are used to assess the requirement of testing for persons under investigation (PUI) [8] which include close contact with a laboratory-confirmed patient within 14 days of symptoms or travel history to an infected area within 14 days of symptom onset [8]. This study presents the latest information about COVID-19 diagnostics, potential therapeutic options, and vaccine development to summarize the current understanding of COVID-19.
COVID-19 Therapeutics
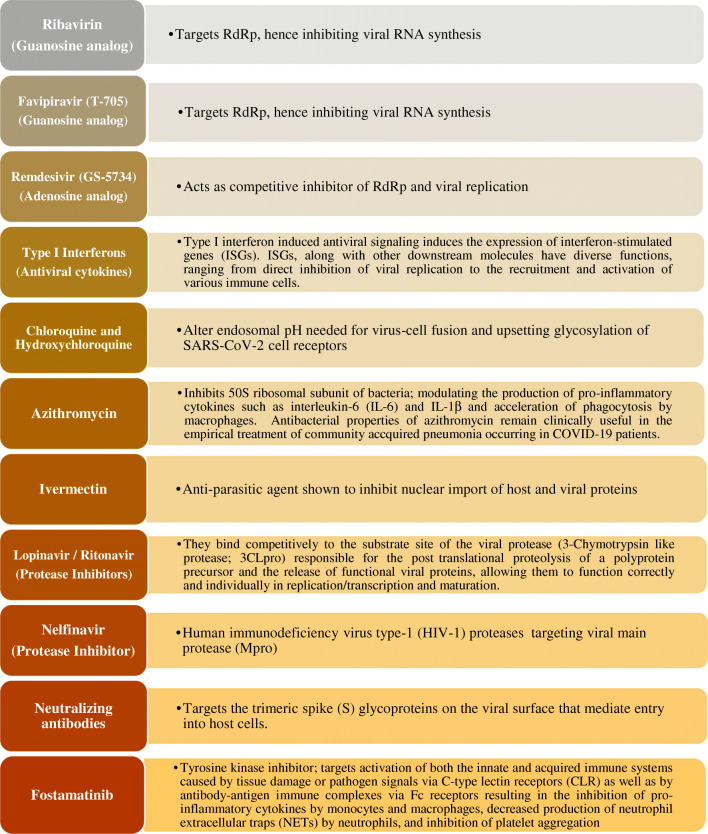
Acute respiratory distress syndrome (ARDS) is the most common complication in COVID-19 patients [9], followed by anemia, acute cardiac injury, and secondary infections. Therefore, antiviral drugs, antibiotics, and systemic corticosteroids are used as treatment. Likewise, the treatment is symptomatic in nature. Apart from the already discovered treatments being applied to combat COVID-19 and its associated complications, scientists are trying hard to develop new potential therapeutic strategies, consisting of monoclonal antibodies, vaccines, peptides, interferon-based therapies, protease inhibitors, and small-molecule drugs to conquest the COVID-19 pandemic. Nevertheless, it might take several months to test their efficacy in vitro and in vivo and even longer in the case of clinical trials [10]. Therapeutic weapons and their targets for combating COVID-19 are contained in Fig. 1. The various therapeutic modalities used and present innovations in drug discovery to battle COVID-19 are discussed in detail.
Fig. 1.
Therapeutic weapons and their targets for combating COVID-19
Antiviral Agents: Nucleoside Analogs and Interferons in the Treatment of COVID-19
Various combination of nucleoside analogs, approved antiviral agents, have been evaluated for the treatment of COVID-19. Nucleoside analogs are viral RNA synthesis inhibitors, hindering viral RNA replication by targeting RNA-dependent RNA polymerase (RdRp), inducing mutations and inhibiting nucleotide synthesis [11]. Type I interferons are antiviral cytokines of various subtypes (α, β, ω, ε, and κ), hindering viral replication [12], IFN-α being more potent than IFN-β [12]. COVID-19 is being managed by nucleoside analog combinations with interferons.
Ribavirin, a nucleoside analog alone or in combination, has been used to treat hepatitis C virus, respiratory syncytial virus infections, and SARS. In vitro, along with IFN- β, it acts synergistically to inhibit SARS-CoV [13, 14]. Along with pegylated interferon, it stimulates the innate antiviral reaction against SARS-CoV-2. COVID-19 patients in China were managed by administration of IFN-α in combination with ribavirin via vapor inhalation in patients twice a day [15]. Favipiravir (T-705) is a guanine analog approved in Japan for influenza treatment, suppression of replication of Ebola, enterovirus, and norovirus [16]. Favipiravir in combination with other antiviral drugs like baloxavir has been used in COVID-19 patients [17]. Glenmark Pharmaceuticals Ltd. in May 2020 has initiated phase 3 clinical trials on antiviral favipiravir for which it received approval from India’s drug regulator DCGI in late April 2020, estimated to be completed by July/August 2020 [18].
Lopinavir and ritonavir, approved anti-HIV drugs, have been found to possess antiviral activity against SARS and MERS as well. Nelfinavir, which is a choosy HIV protease inhibitor, has been shown to suppress SARS-CoV, signifying a substitute healing choice for COVID-19 [19]. Remdesivir (GS-5734) is an adenine analog acting as competitive inhibitor of RdRp and viral replication; it shows exceptional antiviral activity compared to lopinavir and ritonavir, in vitro [20].
Clinically, prophylactic efficacy of remdesivir was observed. Remdesivir in amalgamation with lopinavir, ritonavir, and interferon β was more operational in reducing viral load of MERS-CoV and treatment of symptoms [21]. In the USA, remdesivir administered to the first reported case of SARS-CoV-2 infection witnessed improvement after only 1 day of drug administration [22]. Subsequently, data from two global clinical trials—The National Institute for Allergy and Infectious Diseases’ placebo-controlled phase 3 study in patients with moderate to severe symptoms of COVID-19 including those who were critically ill and Gilead Sciences, Inc., USA’s phase 3 global study evaluating 5-day and 10-day dosing durations of remdesivir in patients with severe disease—have revealed that remdesivir shortened the recovery times and lowered the viral load in treatment group. The US Food and Drug Administration (FDA) recently gave emergency use authorization (EUA) for the use of investigational antiviral remdesivir in the treatment of COVID-19 patients to Gilead Sciences, Inc., USA [4, 23, 24].
Repurposing of Various Established Drugs to Manage COVID-19
Chloroquine, a drug for treatment of malaria and autoimmune diseases, has been established as prospective broad-spectrum antiviral agent—acting by altering endosomal pH needed for virus-cell fusion and upsetting glycosylation of SARS-CoV cell receptors [25]. Studies reveal efficacy of chloroquine phosphate against COVID-19-associated pneumonia and infection [17, 26]. Hydroxychloroquine, an analog of chloroquine, is more effective in treating SARS-CoV-2 infections [27]. Further, both chloroquine and hydroxychloroquine inhibit the COVID-19-associated “cytokine storm” [28]. Chinese hospitals and Oxford University have initiated 21 clinical studies to evaluate their efficacy in COVID-19 infection. However, in the face of poorly evaluated safety and benefit of these treatment regimens, their use in COVID-19 is raging with controversies, ranging from some countries recommending hydroxychloroquinone use, some allowing, and others banning its use in COVID-19 management. The WHO recently halted ongoing clinical trials of hydroxychloroquinone for COVID-19 treatment. These drugs alone or in combination may increase the risk of cardiac arrhythmia, needing urgent medical attention [29].
Azithromycin is a macrolide antibiotic with a 15-membered lactone ring. It has outstanding tissue penetration and inhibits 50S ribosomal subunit that prevents the synthesis of proteins in a wide range of Gram-positive and Gram-negative bacteria imparting antimicrobial tag to it. It modulates the manufacture of interleukin-6 (IL-6), IL-1β thereby acting as anti-inflammatory drug. It hastens phagocytosis by macrophages, classifying azithromycin as a senolytic drug that selectively attacks and kills senescent cells with an efficiency of almost 97% [30]. Although there is no direct evidence of the efficacy of azithromycin in treating COVID-19, some scientific bodies have suggested that the antibacterial properties of azithromycin remain clinically useful in the empirical treatment of community-acquired pneumonia occurring in COVID-19 patients [31]. Azithromycin can significantly increase the efficacy of hydroxychloroquine in treating severe COVID-19 [32]; a regimen of hydroxychloroquine with azithromycin can be a prospective alternative to remdesivir in COVID-19 disease management.
Ivermectin, an FDA-approved antiparasitic agent, which in recent years has shown antiviral activity against a broad range of viruses in vitro [33–35], inhibits the replication of SARS-CoV-2 also in vitro [36]. Researchers at Monash Biomedicine Discovery Institute, Australia, are actively trying to establish the role of ivermectin in managing COVID-19. A popular Indian financial news platform Livemint recently reported that physicians from Bangladesh Medical College found that combination of ivermectin with antibiotic doxycycline yielded promising results against SARS-CoV-2 [36]. However, the use of ivermectin to treat COVID-19 will depend on results of in vivo preclinical testing and ultimately human clinical trials. Phase 2/3 interventional double-blind randomized controlled parallel study entitled “Clinical trial evaluating safety and efficacy of ivermectin and nitazoxanide combination as adjuvant therapy in COVID-19 newly diagnosed Egyptian patients” has started at Tanta University Egypt in May 2020, not yet concluded [37].
Proteins in the Virus: Human Interactome as Potential Antiviral Targets
Specifically targeting key proteins in the virus-human interactome is a very attractive clinical research and treatment strategy to be adopted toward COVID -19 [38];
Spike, Envelope, Membrane Proteins, and Associated Cellular Proteases: Targets for Drugs and Neutralizing Antibodies
SARS-CoV-2, the virus which causes COVID-19, is diverse with single-stranded RNA as its genetic material. Therapeutics targeting spike (S) protein and its receptor-binding domain (RBD) could be designed to combat COVID-19 [39]. Extensive studies on the identification of putative protease cleavage sites in SARS-CoV-2 S protein, their relative importance for SARS-CoV-2 S protein activation, viral pathogenesis, and cellular tropism have led to effective development of neutralizing antibodies and vaccines which the human world was badly in need of at this time.
The trimeric S protein of CoVs is cleaved by host cell proteases during infection to expose the fusion peptide of the S2 domain, which induces fusion of viral and cellular membranes. Cleavage of S protein occurs at different sites, identified in various CoVs to be between the S1 and S2 domains (S1/S2 site) and within the S2 domain proximal to the fusion peptide (S20 site). It is believed that cleavage at both sites is required for viral entry [40]. Different proteases that cleave S1/S2 site in various coronaviruses have been identified; their inhibitors termed protease inhibitors (PIs) are antiviral agents having therapeutic potential. The S1/S2 site (RSVR#SV) of MERS-CoV S protein is cleaved by furin after its biosynthesis during viral replication [41], whereas SARS-CoV S1/S2 site has been shown to be cleaved by cathepsin L following receptor binding and during viral entry in late endosomes [42]. SARS-CoV-2 has an S1/S2 site (AYT#M) that is identical to the one in SARS-CoV [43], and it is propounded that the SARS-CoV-2 S1/S2 site may be cleaved by cathepsin L as in SARS-CoV. Other proteases such as trypsin, elastase, cathepsin L, and TMPRSS2 have been shown to cleave SARS-CoV S protein at other sites between S1 and S2 domains during viral entry [44, 45]. This is supported by several studies which showed that cathepsin L and TMPRSS2 promote SARS-CoV entry, while their inhibition suppressed infection of permissive cells [46]. Given that SARS-CoV-2 has the same S20 site as SARS-CoV, it is proposed that processing of SARS-CoV-2 S protein at S20 site is similar to SARS-CoV. The membrane protein is pivotal to viral entry in the cell as reflected by in silico studies of the interactome, as TMPRSS2 is being downregulated by viral infection. The TMPRSS2 node is a potentially interesting region of the interactome and a target for SARS-CoV-2 antiviral therapy. It has been experimentally demonstrated that inhibition of TMPRSS2 with a protease inhibitor (camostat mesylate) inhibits SARS-CoV-2 replication [47], making this protein an extremely promising target for antiviral therapy.
In addition to the previous S1/S2 sites, SARS-CoV-2 has a furin-like protease cleavage S20 site (KR#SF) that is identical to that in SARS-CoV, though there is no evidence that SARS-CoV S protein is cleaved by furin-like proteases at the S20 site during viral egress. Further, SARS-CoV-2 has an additional furin-like protease cleavage site (RRAR#SV) existing N terminus to the S1/S2 site (AYT#M) which is absent in SARS-CoV [48]. The presence of this furin-like cleavage site in SARS-CoV-2 suggests its cleavage by furin during viral egress. Similarly, MERS-CoV has an S20 site (RXXR#SA) that is less efficiently cleaved by furin and most probably cleaved by TMPRSS2 or cathepsin L during viral entry. Antiviral small molecules that inhibited cathepsin L were able to inhibit SARS-CoV infections in vitro and that of other viruses that depend on cathepsin L for entry, such as Ebola, Hendra, and Nipah viruses [49, 50]. As the putative protease cleavage sites in SARS-CoV-2 S protein have been identified, the presence of a cathepsin L cleavage site in SARS-CoV-2 S protein (S1/S2 site) implies that cathepsin L inhibitors may be valuable in inhibiting SARS-CoV-2 infections as well. Thus, inhibitors of the proteases that prime S for fusion possess antiviral activity [51]; mediators directly aiming at highly conserved S2 subunit may be possible therapeutic candidates.
The E (envelope) protein is the smallest transmembrane structural protein of coronaviruses with the charged cytoplasmic tail and the hydrophobic domain [52]. The E proteins are involved in assembly process based on its interaction with M protein; production of virions, i.e., release of infectious viruses that require the hydrophobic domain (HD) of the E protein; and lastly formation of ion channels, which is mainly associated with pathogenesis [53, 54]. Studies have shown that the recombinant CoVs lacking E protein exhibit significantly reduced viral titer or incompetent virus propagation [55]. On the other hand, the main function of the membrane (M) protein is conservation of the shape of the viral envelope. It does this activity by cooperating with other coronavirus proteins, integrating Golgi complex into new virions, and stabilizing of N protein [56]. Therefore, E and M proteins can be targeted for developing anti-COVID-19 drugs. Many antiviral, antibacterial, anti-asthmatic, anti-,inflammatory and antitumor drugs were found to have relatively good affinity to these proteins, thereby preventing viral replication in host cells. Besides, small interfering RNAs (siRNAs) showed strong binding affinity for M protein mRNA [57]. A recent study observed rutin (bioflavonoid) and doxycycline (antibacterial and antiviral agent) as potential inhibitors for E protein and caffeic acid and ferulic acid (phenylpropanoids) as a potential inhibitor for M protein. Besides that, rutin (natural antiviral) and caffeic acid and ferulic acid (hydrocinnamicacids) were found to interact with all the structural proteins of SARS-CoV-2 as well, revealing a strong tendency and efficiency of natural antiviral compounds against SARS-CoV-2 proteins [58].
Neutralizing antibodies target spike protein domains: Antibodies against RBD and S2 domain of SARS-CoV and MERS-CoV S proteins have been found effective in neutralizing infections of permissive cell lines in vitro [59–62]. In addition, neutralizing antibodies were capable of treating infections in experimental animals and in infected patients during these major outbreaks [63]. Several studies in vitro or in vivo suggest that monoclonal antibodies (mAbs) against the S1 subunit of receptor-binding domain (RBD) and fusion inhibitors targeting the S2 subunit possess potential anti-SARS-CoV-2 ability [64]. Varying studies regarding binding of SARS-CoV RBD-specific monoclonal antibodies to SARS-CoV-2 S protein have revealed that several such monoclonal antibodies did not bind to SARS-CoV-2 S protein [65], whereas the SARS-CoV-specific monoclonal antibody, CR3022, bound with high affinity to SARS-CoV-2 RBD being a therapeutic candidate for COVID-19 either alone or in combination with other neutralizing mAbs [66] suggesting that differences between the two RBDs can impact the cross-reactivity of many neutralizing antibodies. However, neutralizing antibodies against SARS-CoV-2, once developed, could be promising in controlling the current SARS-CoV-2 outbreak. Further, neutralizing antibodies against other pro-inflammatory cytokines (TNF, IL-6) may be useful to decrease the severe inflammatory response (cytokine storm) during COVID-19 [67].
Host ACE2 Protein Receptor as Therapeutic Target
One of the SARS-CoV-2-interacting protein networks most downregulated by beta-coronaviruses as per a keymaster regulator analysis is ACE2, an experimentally validated molecular target used by SARS-CoV-2 for entry in the cell [47]. Administration of recombinant ACE2 to compensate for this protein loss has been shown to be effective in treating acute respiratory distress syndrome, which is often caused by severe pulmonary infections [68]. Recombinant ACE2 has also been proposed as a potential cure for SARS-CoV-2’s most severe symptoms, and in vitro studies have shown that soluble forms of ACE2 are beneficial to SARS patients, likely because they act as competitive binders of SARS-CoV spike proteins, preventing binding to the host cell ACE2 [69]. Unpublished experimental results report recombinant ACE2 to also be a molecular neutralizer of SARS-CoV-2 [70]. In addition, mAbs or molecules targeting the host receptor ACE2 (angiotensin-converting enzyme 2) are effective anti-SARS-CoV-2 drugs [70].
Other Therapeutic Target Proteins
Further, a vast number of host cell proteins can be targets for viruses. In a recent in silico study, the spike protein of COVID-19 was modeled using solved structures in a protein data bank. After model validation, molecular docking was performed to test its binding affinity against glucose-regulating protein 78 (GRP78), a master chaperone protein, as it happens in the case of MERS-CoV. A study suggests the existence of COVID-19 spike protein-GRP78 binding site, thus paving route for drug designers to prevent binding and the infection [38]. Another, recent in silico study highlights several other proteins that could be directly repressed or enhanced by specific drugs including a mild virus-activated protein, the serine/threonine kinase MARK2, which is one of the targets of the drug fostamatinib, a tyrosine kinase inhibitor with potential anti-inflammatory and immune-modulating activities currently used in the treatment of immune thrombocytopenic purpura [71].
Recently, Dai and co-workers have designed a structural analog binding the substrate-binding pocket of Mpro, the enzyme facilitating SARS-CoV-2 replication and transcription giving rise to a promising antiviral drug contestant [72].
Clinical Intervention by Downregulating Apoptotic Mechanisms Induced by the Virus
As a general viral mechanism to control the host cell metabolism, mitochondrial proteins, interacting with SARS-CoV-2, appear to be downregulated in beta-coronavirus infections, ultimately steering apoptosis, thus preventing mitochondria-induced apoptosis [73]. Upregulation of antiapoptotic BCL-2 family member, MCL1, in viral replication occurs across many RNA-based viruses; MCL1 activation in both SARS-CoV and MERS-CoV infections has been reported [74], possibly due to impaired protein synthesis eliciting ER stress-induced apoptosis via unfolded protein response (UPR) [75]. Since beta-coronavirus-induced UPR has been shown to modulate innate immunity [76], targeting aberrant monocyte/macrophage activation may be another possible intervention strategy to reduce the severity of coronavirus-induced symptomatology.
Convalescent Plasma (CP) Transfusion Therapy
CP therapy is around a century old, having shown some benefit in treating measles, chickenpox, and rabies. Convalescent plasma has been injected in patients with viral infections including SARS, H5N1, H1N1, and Ebola virus [77]. Small studies have shown faster clearance of virus in the case of MERS and SARS if given earlier in the course of disease, though no benefit was seen in 2015 on some Ebola patients treated with CP. Antibodies from CP may inhibit viremia; studies showed that SARS-CoV-2 could be neutralized by the serum of several previously infected patients [78]; improvement in clinical conditions following such plasma transfusions has been observed [77].
In the absence of any effective treatment or a vaccine, the COVID-19 pandemic provides an opportunity to ascertain the clinical benefits of CP therapy through randomized controlled trials. The US FDA and ICMR India have presently approved the use of plasma from recovered COVID-19 patients for trial purposes only. Several uncontrolled case series of convalescent plasma use in patients with COVID-19 have suggested a possible benefit [79, 80]. An open-label, multicenter, randomized clinical trial was performed in 7 medical centers in Wuhan, China, with laboratory-confirmed COVID-19 patients. In this study, among patients with severe or life-threatening COVID-19, CP therapy added to standard treatment, compared with standard treatment alone, did not result in a statistically significant improvement in time to clinical improvement within 28 days. CP use in the study was associated with some clinical improvement in severely ill patients, but not in critically ill patients [81]. Greater efficacy in less ill individuals is expected because antibody therapies generally work best when administered earlier in the disease [82]. It is not surprising that patients with COVID-19 who had tachypnea and hypoxia might benefit more from CP than those who required mechanical ventilation. However, any indication of possible benefit in the severely ill group is noteworthy because these individuals had advanced disease, which is not considered optimal for antibody therapy [83]. An open-label, parallel-arm, phase II, multicenter, randomized controlled trial was conducted by the Indian Council of Medical Research (ICMR), across thirty-nine public and private hospitals in India (CTRI/2020/04/024775). The study concluded that CP was not associated with reduction in mortality or progression to severe COVID-19 [84].
Mesenchymal Stem Cell Therapy
Stem cell therapy too is making its way into COVID-19 management. Of late, investigators have confirmed that intravenous transplantation of mesenchymal stem cells (MSCs) was safe and effective for COVID-19 pneumonia, particularly for clinically ill patients [85]. Cytokine storm is combated by MSC-based immunomodulation treatment, and endogenous repair is promoted via the stem cells [86].
Diagnostic Strategies for SARS-CoV-2: Current and Future Prospects
The COVID-19 pandemic has put a huge burden on the public health system of the world. Sensitive and specific detection of SARS-CoV-2 is urgently needed for timely diagnosis, treatment, and prevention of COVID-19 transmission. Orthodox methods for detection of virus such as virus culture and microscopic analysis are generally time-consuming and labor-intensive with limited sensitivity [87]. In contrast, the last decade witnessed emergence of molecular and serologic approaches, including real-time reverse transcription-polymerase chain reaction (RT-PCR), enzyme-linked immunosorbent assay (ELISA), colloidal gold immunochromatography, and chemiluminescence immunoassay (CLIA), which can be developed into a rapid and effective tool for detection of respiratory pathogens and their infections [88, 89].
There are two broad categories of SARS-CoV-2 tests including those that detect the virus itself and those detecting the host’s response to the virus [90].
Tests for Viral RNA
RT-PCR for Detection of SARS-CoV-2
Since SARS-CoV-2 is a RNA virus, most of the tests currently identify viral RNA by nucleic acid amplification, using RT-PCR. These tests detect viral RNA present in the sample collected; the most common sample types being tested are swabs taken from the nasopharynx and/or oropharynx, the former being a bit more sensitive [91], and both can be collected, combined, and tested in a single reaction to conserve reagents. Viral RNAis extracted and amplified by RT-PCR. Usually, patients with pneumonia have negative nasal or oropharyngeal samples but positive lower airway samples [92]. True clinical sensitivity of any of these tests is unknown, and possibility of false negative results remains. False positive is very unlikely though stray viral RNA that makes its way into the testing process as a result of specimen cross-contamination or testing performed by a laboratory worker infected with SARS-CoV-2 could conceivably result in falsely positives [93].
The assay is composed of two main steps: extraction of RNA from patient specimens followed by one-step reverse transcription and PCR amplification with SARS-CoV-2-specific primers and real-time detection with SARS-CoV-2-specific probes. Virus isolate is the gold standard for establishment and standardization of assay performance. Since SARS-CoV-2 virus isolate was not available earlier, based on genetic sequence of SARS-CoV-2 and closely related SARS-CoV (2002–2003), the WHO shared protocols for screening and confirmation of probable cases by targeting various regions of viral genome including E, N, RdRp, and S genes [94]. The primers for the detection of SARS-CoV-2 have been designed against various assay target regions including viralnucleocapsid (N) gene (N1, N2, and N3) by the US CDC; China CDC; Hong Kong University; National Institute of Infectious Diseases, Japan; and National Institute of Health, Thailand [95–99]. Primers for viral envelope (E) gene and Rdbp gene have been designed by Charite, Germany [100]. TaqMan probe-based assays are being used for quantitative RT-PCR, and the absence of nonspecific PCR inhibition is ensured by an internal positive amplification control (IPC) included with each specimen. Viral RNA-based tests are gold standard especially in setting of acute illness [87, 88].
Sample Pooling Strategy: Boosting Test Efficiency for SARS-CoV-2
In the current COVID19 pandemic crisis, national healthcare systems are overburdened with demand coupled with acute shortage of tests for SARS-CoV-2; in these settings sample pooling has been efficiently used to amplify testing. The strategy is to pool samples taken from several subjects and test this combined sample; if negative, all subjects are negative, and if test is positive, individual tests are rerun. This was recently suggested by Dina Berenbaum of the Technion Israel Institute of Technology [101] and has been increasingly implemented in other countries. It has been purported that around 30 samples can be pooled to significantly reduce the testing burdens at hospitals and airports [102].
CRISPR Gene Editing for Detection of SARS-CoV-2
CRISPR (clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea [103]. Cas9 or “CRISPR-associated protein 9” is one of the enzymes that uses CRISPR sequences as a guide to recognize and cleave specific strands of DNA complementary to the CRISPR sequence forming the basis of a technology known as CRISPR-Cas9 that can be used to edit genes within organisms [104]. CRISPR is like a molecular scissor allowing scientists to target any particular gene; it can very quickly and precisely identify a gene corresponding to SARS-CoV-2 as well; this much-lauded gene editing technology CRISPR having wide variety of applications in the diagnosis and treatment of diseases [105, 106] is now proffering novel assays for SARS-CoV-2.
The CRISPR-Cas12-based SARS-CoV-2 DETECTR (DNA endonuclease-targeted CRISPR trans reporter) assay for detection of SARS-CoV-2 from respiratory swab RNA that can deliver results in ~40 min has been established [107, 108]. This assay performs simultaneous reverse transcription and isothermal amplification using loop-mediated amplification (RT–LAMP) of RNA extracted from nasopharyngeal or oropharyngeal swabs, followed by Cas12 detection of predefined SARS-CoV-2 sequences, after which cleavage of a reporter molecule confirms detection of the virus by releasing a signal visualized on a lateral flow strip [109]. This test detects the E and N genes on SARS-CoV-2 and uses human RNase P gene as a control; N gene is highly expressed in viral infections and the easiest to detect. This test has been approved by the WHO and CDC, USA, being recently cleared for emergency use authorization (EUA) by the Food and Drug Administration (FDA), USA.
CRISPR-based testing approach is reliable for nucleic acid-based molecular diagnostics. The SARS-CoV-2 DETECTR assay has distinct advantages over RT-PCR-based ones being easy to implement, cheaper, and accurate, providing visual and faster alternative to RT-PCR assay, and having 95% positive predictive value and 100% negative predictive value. It is slightly less sensitive as the limit of detection (LoD) is 10 copies per μl input as compared to RT-PCR which has an LoD of 1 copy per μl input. Moreover, the synthetic in vitro-transcribed (IVT) SARS-CoV-2 RNA gene targets in nuclease-free water, thus CRISPR-Cas12-based detection can distinguish SARS-CoV-2 with no cross-reactivity for related CoV strains [110]. Unlike RT-PCR-based methods, CRISPR-based nucleic acid detection methods can work at a constant temperature without the need for an expensive thermal cycler. Further, this assay uses different set of reagents offering a solution to a dearth of RT-PCR-based assays that has arisen due to sudden spurt in their demand.
Serum-Based Tests
Although, RT-qPCR is a highly sensitive test for SARS-CoV-2, it has its limitations. RT-qPCR requires high-quality nasopharyngeal swabs containing sufficient amounts of viral RNA which could vary tremendously between and within the same patients depending on various factors including timing of the test, the onset of infection, and/or symptoms. In addition, nasopharyngeal swabs are very unpleasant for the patient. Advanced trained personnel are required, and generally, RT-qPCR leads to delays in these testing times of global pandemic with potentially millions to be tested putting immense burden on healthcare system. Both false negative and false positive tests are reported, though rarely. IgG/IgM serological tests offer several gains over RT-qPCR. Firstly, serological tests detect human antibodies which are known to be much more stable than viral RNA. Secondly, because antibodies are typically uniformly distributed in the blood, serological specimens have much less variations than nasopharyngeal viral RNA specimens and can be easily collected. Thirdly, unlike RT-qPCR, serological tests can detect past infection because virus-specific antibodies can persist in the blood for several weeks/months after onset of symptoms, thus being ideal for community surveillance. However, IgM/IgG serological tests too have their limitations, mainly the slow pace of the human antibody response to SARS-CoV-2 [111] which can be host dependent. In the case of SARS-CoV-2, early studies suggest that the majority of patients seroconvert between 7 and 11 days post exposure to the virus, although some patients may develop antibodies sooner. Thereby, antibody testing is not useful in the setting of an acute illness. Antibody tests for SARS-CoV-2 may facilitate (i) contact tracing—RNA-based tests can help here as well; (ii) serologic surveillance at the local, regional, state, and national levels; and (iii) identification of those who have already had the virus and thus may be immune, thus useful to identify individuals who can be source for therapeutic or prophylactic neutralizing antibodies [93]. IgM/IgG is an appropriate test during the chronic phase. Since the exact time of infection is often unknown, both RT-qPCR and IgM/IgG testing have their relevance and importance in COVID-19 diagnosis and management [112].
SARS-CoV-2: Point-of Care Tests (POCT)
Rapid, easy-to-use POCT are now being facilitated outside laboratory settings. These simple test kits are based either on detection of proteins from the COVID-19 virus in respiratory samples (e.g., sputum, throat swab) or detection of human antibodies generated in response to infection in the blood or serum [113]. The WHO applauds the innovative efforts of test developers and rapid respond to needs of the population. However, before these tests can be recommended, they must be validated in the appropriate populations and settings. Inadequate tests may miss patients with active infection or falsely categorize patients as having the disease when they do not, further hampering disease control efforts.
One type of rapid diagnostic test (RDT) detects the presence of viral proteins (antigens) expressed by the COVID-19 virus in a sample from the respiratory tract of a person. If the target antigen is present in sufficient concentrations in the sample, it will bind to specific antibodies fixed to a paper strip enclosed in plastic casing and generate a visually detectable signal, typically within 30 min. The antigen(s) detected are expressed only when the virus is actively replicating; therefore, such tests are best used to identify acute or early infection. The sensitivity of these tests might be expected to vary from 34 to 80% [114]. Another type of test detects the presence of antibodies in the blood of people believed to have been infected with COVID-19 [115]. Antibody detection tests targeting COVID-19 may give false positive results by cross-reacting with other pathogens, including other human CoVs [116, 117].
Research Strategies in the Development of SARS-CoV-2 Vaccines
All vaccines follow the same basic principle—pre-exposing the immune system of a person to either killed or weakened pathogens, or some of the pathogen’s structural parts, which leads the body to mount an immune response [118]. Thus vaccines stimulate the immune system of an individual to prepare it against a future pathogen attack. The development of effective therapeutic and preventive strategies is a research priority. Prophylactic and therapeutic SARS-CoV-2 vaccines are of immense value to deal with the crisis generated by the COVID-19 pandemic [119]. Further, SARS-CoV-2 vaccines will be essential for reducing morbidity and mortality if the virus establishes itself in the population as a seasonal virus.
Vaccines for SARS-CoV-2 are being rapidly developed. These vaccine platforms function by synthesizing viral surface proteins that get attached to the host cells and clamp them into shape. This facilitates easier recognition of antigens by the immune system. Other than these ongoing programs, the Coalition for Epidemic Preparedness Innovation (CEPI) has provided funding to develop COVID-19 vaccines using varied technology [120]. Herein, we summarize approaches and challenges in developing broadly effective SARS-CoV-2 vaccines.
Live-Attenuated Virus Vaccines
These are produced by reducing or eliminating the virulence of a live virus, typically using chemical-driven or site-directed mutagenesis; thus, the virus is capable of productive infection, but the resulting disease is either diminished or eliminated. Live-attenuated vaccines can elicit both innate and adaptive immune responses, and protection can be lifelong. BCG—a live-attenuated vaccine for COVID-19—is in phase 2/3 clinical trial in Australia, the Netherlands, and the USA (Clinical trials Govt.:NCT04327206; NCT04328441) and is being developed by the University of Melbourne and Murdoch Children’s Research Institute, Australia; Radboud University Medical Center, Netherlands; and Faustman Lab at Massachusetts General Hospital, USA. Their production is relatively inexpensive. Codagenix, USA, and the Serum Institute of India are collaborating to make live-attenuated virus vaccines for COVID 19 which they purportedly market-ready by 2022 [66, 121, 122].
Vaccine candidate TMV-083 is an attenuated live virus vaccine using the measles vaccine (MV) as a vector (or vehicle) and expressing a spike protein antigen from the SARS-CoV-2 virus. The spike protein is the “entry key” that allows the virus into cells. The phase 1 trial of this candidate vaccine has started partly at the Cochin-Pasteur Clinical Investigation Center (CIC) in Cochin Hospital (Paris Public Hospital Network (AP-HP)) and partly in Belgium from December 2020. Institut Pasteur announced that the international coalition, CEPI, had allocated 4.3 million euros for the first stages of research and development of this vaccine against SARS-CoV-2 [123].
Inactivated Whole-Virus Vaccine
Inactivated whole-virus comprises the entire disease-causing virion which is inactivated physically (by heat) or chemically. It has several antigenic parts to the host and can induce diverse immunologic responses against the pathogen [124]. The inactivated vaccines being developed for COVID-19 are safest with less adverse reactions [125]. There are several inactivated whole-virus vaccines against SARS-CoV-2 which have got emergency use authorization in China which include inactivated vaccine BBIBP-CorV developed by the Beijing Institute of Biological Products and China National Pharmaceutical Group (Sinopharm); inactivated vaccine developed by the Wuhan Institute of Biological Products and Sinopharm; and inactivated vaccine having formalin with alum adjuvant (CoronaVac) developed by Sinovac Biotech Ltd. [126, 127]. Vaccine candidate, BBV-152, of Bharat Biotech is a whole-virion inactivated SARS-CoV-2 vaccine in phase 1/2 clinical trials [128]. COVAXINTM, India’s indigenous COVID-19 vaccine by Bharat Biotech, is being developed in collaboration with the Indian Council of Medical Research (ICMR) and National Institute of Virology (NIV). Bharat Biotech received Drugs Controller General of India (DCGI) approval for phase 3 clinical trials of COVAXINTM, in 26,000 participants in over 25 centers across India [129].
Viral Vector or Adenoviral Vaccines
These viral gene delivery systems rely on a host viral genome (e.g., adenovirus) that typically lacks the genetic components necessary to produce new virions and that encodes antigenic components of the virus of interest to elicit an immune response. Because viral vector vaccines persist in the host as genetic material, directly infect antigen-presenting cells, and have strong inherent adjuvant activity, they can efficiently induce both innate and B cell- and T cell-mediated immune responses. Adenovirus vectors that express CoV S and nucleocapsid proteins, which are the immune-dominant coronavirus proteins, yield varying results depending on preparation, the route of administration, and the animal model used [130].
A new adenovirus vector vaccine ChAdOx1 nCoV-19 (AZD1222) developed by a group of researchers led by Professor Sarah Gilbert in Jenner Institute, University of Oxford, UK, has completed phase 3 clinical trials. They had entered into a landmark partnership with UK-based global biopharmaceutical giant AstraZeneca and Serum institute of India for further development, large-scale manufacturing, and potential distribution [131]. Two different dosing regimens demonstrated efficacy with one showing a better profile. No hospitalizations or severe cases of COVID-19 in participants treated with AZD1222 have been reported. AstraZeneca will now immediately prepare regulatory submission of the data to authorities around the world that have a framework in place for conditional or early approval. The company will seek an Emergency Use Listing from the World Health Organization for an accelerated pathway to vaccine availability in low-income countries [132]. In the meanwhile, the Serum Institute of India has applied to DCGI for emergency use authorization for its viral vector-based COVID-19 vaccine named Covishield in India [133]. Both Covishield and the ChAdOx1 have the same virus protein but differ in their makeup—the exact nature of the difference has not been publicly disclosed.
Johnson & Johnson is developing an adenovirus vectored vaccine using AdVac®/PER.C6® vaccine platforms. Data from the phase 3 trials of Johnson & Johnson and AstraZeneca’s shots, which are non-replicating viral vector vaccines, are expected by early February 2021. Johnson & Johnson and AstraZeneca are expected to provide 150 million to 200 million doses total of their vaccines in the first quarter of next year if they get the FDA’s nod [134].
Sputnik V is the world’s first registered vaccine for COVID-19 based on a well-studied human adenoviral vector-based platform. It currently ranks among top 10 candidate vaccines approaching the end of clinical trials and the start of mass production on the World Health Organization’s (WHO) list. The ongoing Sputnik V post-registration clinical trial in Russia involves 40,000 volunteers. Clinical trials of Sputnik V have been announced in the UAE, India, Venezuela, and Belarus also. The Sputnik V vaccine’s efficacy is confirmed at 91.4% based on data analysis of the final control point of clinical trials. The Sputnik V vaccine efficacy against severe cases of coronavirus is 100%. The unique substance of the Sputnik V and method of using it has a patent protection in Russia, obtained by Gamaleya National Research Institute of Epidemiology and Microbiology and Acellena Contract Drug Research and Development, Russia [135].
Recombinant Protein-Based Vaccines (Subunit or Molecular Clamp Vaccines)
Subunit vaccines contain pathogen-derived proteins (antigens) with immunogenicity that can elicit the host immune system. Subunit vaccine is safe and easily manufactured by recombinant DNA techniques but requires adjuvant to enhance an immune response. These vaccines work by introducing a fragment of the virus into the body (in the case of COVID-19, the “spike” protein). The idea behind the subunit vaccine is that when the body recognizes the protein, it creates specialized immune cells that block the receptors and effectively shut the door to infection. But the challenge is getting the body to recognize the virus fragment as enough of a threat to create an effective immune response, and injecting the spike protein alone is not enough to do that. “On its own, the SARS-CoV-2 spike protein is unstable. It needs to be ‘locked’ into shape to ensure that the vaccine is inducing the right immune response to the COVID-19 virus if someone is exposed,” Professor Young said! And that is achieved with a molecular clamp [136•].
Additional approaches in the preclinical stage include recombinant protein-based vaccines focused on the S protein. Provoking the immune system without drawing in any infectious viruses is extremely safe. China’s Clover Biopharmaceuticals during preclinical testing of recombinant subunit vaccine detected the antigen-specific neutralizing Abs in the sera of fully recovered patients [137].
Also, GlaxoSmithKline (GSK) is developing a vaccine containing S protein immunogen which in combination with adjuvant (GSK2) produced elevated levels of anti-SARS-CoV IgG Ab response. Sanofi; Canadian biopharmaceutical company, Medicago; and China-based Clover Biopharmaceuticals have joined hands to develop an adjuvanted COVID-19 vaccine candidate. An update was issued in December 2020 on the phase 1/2 clinical trial [138].
DNA-Based Vaccines
DNA vaccines (DVs) consist of DNA that encodes the viral antigenic components; they are directly injected or otherwise inoculated into the vaccine. As in vectored and subunit vaccines, DVs also use spike peptides to elicit high titers of neutralizing antibodies. DVs elicit both the cell-mediated and humoral immune system. DVs induce long-lasting immunity that defends against diseases effectively in the future. DVs are very stable and can be produced within weeks because they do not need culture or fermentation. Of all the vaccines developed for SARS-CoV-2, DVs undoubtedly involve the most novel approach, since no vaccine using this technology has yet been commercialized for humans although this vaccine approach has obtained promising results in tests on animal models [136•]. Interestingly, when DVs are used in combination with inactivated vaccines, the cellular immune response is T1 cell-directed. These offer a precise course to transport antigens to the immune system containing additional sequences of coding molecules to influence the results [139]. This vaccine can encourage activation of T cells via relocating DNA plasmids expressing the SARS-CoV-2 S proteins. Interestingly, live-attenuated vaccines and vaccine combinations (e.g., inactivated virus combined with DV) might hold more promise for the development of successful vaccination strategies for the elderly [139].
Inovio Pharmaceuticals developed a DVs candidate termed INO-4800, which is in phase 1 clinical trial (NCT04336410). Takis/Applied DNA Sciences/Evvivax and Zydus Cadila are developing a DVs candidate for COVID-19 disease which is now in preclinical studies [140, 141].
mRNA-Based Vaccines
mRNA vaccines mimic the natural infection of the virus, but they retain only a short synthetic viral mRNA which encodes only the required antigen [142]. mRNA vaccine is a promising alternative to traditional vaccine approaches due to their safety, potency, quick vaccine development time, and low-cost production. In this approach, RNA that codes for some of the viral proteins is introduced into the body to produce viral proteins, against which the body synthesizes antibodies, thus preparing for future viral attacks. mRNA vaccines strongly induce both cellular and humoral immune responses. It is relatively safe and effective because it is only a transient carrier of message that does not interact with the host genome and it also does not need the whole virus [136•].
Pfizer, one of the world’s largest pharmaceutical companies, based in New York, in collaboration with German biotech company BioNTech developed a COVID-19 vaccine, namely BNT162b2. This vaccine requires two doses taken 21 days apart. The FDA granted emergency approval to Pfizer’s COVID-19 vaccine on December 11, 2020. It is the first vaccine to receive emergency authorization in the USA. Pfizer vaccine is safe and offers strong protection against COVID-19 within 10 days of the first dose, regardless of the recipient’s race, weight, or age. Pfizer has signed a nearly $2-billion contract with the US government to provide 100 million doses for free, though the New York Times reports that only half its supply will go to the USA this year. Canadian health officials have also granted emergency approval to Pfizer’s COVID-19 vaccine [143].
A Massachusetts-based biotech company, Moderna Therapeutics, in collaboration with the National Institutes of Health developed a COVID-19 vaccine, namely mRNA-1273. It relies on injecting snippets of a virus’s mRNA into human cells. It spurs the production of viral proteins that mimic the coronavirus, training the immune system to recognize its presence. This vaccine requires two doses, 4 weeks apart. On December 18, 2020, the FDA granted emergency approval to the mRNA-1273 vaccine [143].
Dr. Mike Ryan, executive director of WHO’s emergencies program rightly said, “Proper trials are vital as there’s only one thing more dangerous than a bad virus and that’s a bad vaccine. We have to be very, very, very careful in developing any product that we’re going to inject into potentially most of the world’s population.”
Conclusion
Our current knowledge of SARS-CoV-2 suggests an intermediate host, very rapid and efficient human-to-human transmission, necessitating robust detection strategies, and speedy development of therapeutic modalities and vaccine development. Prevention, detection, treatment strategies, and our preparedness to deal with human coronavirus outbreaks will decide the evolution of coronaviruses in future. Further, lessons from handling this outbreak will allow us to be better prepared in the future; the viruses will keep coming.
Acknowledgements
We acknowledge all the patients who have been infected with SARS-CoV-2 and all the scientists working on COVID-19 management.
Author Contribution
Not applicable.
Data Availability
Not applicable.
Code Availability
Not applicable.
Declarations
Ethics Approval
Not applicable.
Consent for Publication
All authors have given their consent for publication.
Competing Interests
The authors declare no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Virology
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak– an update on the status. Mil Med Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W, Cao Q, Qin L, Wang X, Cheng Z, Pan A, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): a multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020;S0163-4453:388–393. doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708e20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronavirus (COVID-19) update: FDA issues emergency use authorization for potential COVID-19 treatment. U.S Food and Drug Administration. May 01, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment. Accessed 21 Aug 2020
- 5.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev Res. 2020; 81:537-540. [DOI] [PMC free article] [PubMed]
- 6.Mohamed K, Rodríguez-Román E, Rahmani F, Zhang H, Ivanovska M, Makka SA, et al. Borderless collaboration is needed for COVID-19; a disease that knows no borders. Infect Control Hosp Epidemiol. 2020;41:1245–6. [DOI] [PMC free article] [PubMed]
- 7.••.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Napoli RD. Features, evaluation and treatment coronavirus (COVID-19). StatPearls [internet] Treasure Island (FL): StatPearls Publishing; 2020. [PubMed] [Google Scholar]
- 9.Morrow ES, Roseweir A, Edwards J. The role of gamma delta T lymphocytes in breast cancer: a review. Trans Res. 2019;203:88–96. doi: 10.1016/j.trsl.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Saghazadeh A, Rezaei N. Towards treatment planning of COVID-19: rationale and hypothesis for the use of multiple immunosuppressive agents: anti-antibodies, immunoglobulins, and corticosteroids. Int Immuno Pharmacol. 2020;84:1–6. doi: 10.1016/j.intimp.2020.106560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Mu F, Xie J, Cheng J, Fu Y, Jiang DA. Single ssRNA segment encoding RdRp is sufficient for replication, infection, and transmission of Ourmia-like virus. Front Microbiol. 2020;11:379. doi: 10.3389/fmicb.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.So LKY, Lau ACW, Yam LYC, Cheung TMT, Poon E, Yung RWH, et al. Development of a standard treatment protocol for severe acute respiratory syndrome. Lancet. 2003;361:1615–7. [DOI] [PMC free article] [PubMed]
- 14.Morgenstern B, Michaelis M, Baer PC, Doerr HW, Cinatl J. Ribavirin and interferon-b synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen KY. Coronaviruses: drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Clerk E. New nucleoside analogues for the treatment of haemorrhagic fever virus infections, Minireview. Chem Asian J. 2019;14:3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine respectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020b;30:269–71. [DOI] [PMC free article] [PubMed]
- 18.Glenmark initiates phase 3 clinical trials on antiviral favipiravir for COVID-19 patients in India. Glenmark Pharmaceuticals Ltd. May 26, 2020. https://www.expresspharma.in/latest-updates/glenmark. Accessed 21 Aug 2020
- 19.Yamamoto N, Yang R, Yoshinaka Y, Amari S, Nakano T, Cinatl J, et al. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem Biophys Res Commun. 2004;318:719–25. [DOI] [PMC free article] [PubMed]
- 20.Tchesnokov EP, Feng JY, Porter DP, Guotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheahan TP, Sims AC, Leist SR, Schafer A, Won J, Brown AJ, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–36. [DOI] [PMC free article] [PubMed]
- 23.NIH clinical trial shows remdesivir accelerates recovery from advanced COVID-19. National institute of Allergy and Infectious Diseases. April 29, 2020. https://www.niaid.nih.gov/news-events/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19. Accessed 21 Aug 2020
- 24.Antiviral remdesivir prevents disease progression in monkeys with COVID-19. National institute of Allergy and Infectious Diseases. April 17, 2020. https://www.niaid.nih.gov/news-events/antiviral-remdesivir-prevents-disease-progression-monkeys-covid-19. Accessed 21 Aug 2020
- 25.Savarino A, Trani LD, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 27.Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed]
- 28.Schrezenmeier E, Dorner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 29.Van Roon EN, van den BPM, Jansen TL, Houtman NM, van de LMA, Brouwers JR. An evidence-based assessment of the clinical significance of drug-drug interactions between disease-modifying anti rheumatic drugs and non-rheumatic according to rheumatologists and pharmacists. Clin Ther. 2009;31:1737–1746. doi: 10.1016/j.clinthera.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Parra-Lara LG, Martínez-Arboleda JJ, Rosso F. Azithromycin and SARS-CoV-2 infection: where we are now and where we are going. J Glob Antimicrob Resist. 2020;22:680–684. doi: 10.1016/j.jgar.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sultana J, Cutroneo PM, Crisafulli S, Puglisi G, Caramori G, Trifirò G. Azithromycin in COVID-19 patients: pharmacological mechanism, clinical evidence and prescribing guidelines. Drug Saf. 2020;43:691–698. doi: 10.1007/s40264-020-00976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautret P, Lagier JC, Parola P, Meddeb L, Mailhe M, Doudier B, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;105949:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.González C, Sahagún Prieto AM, Diez L, et al. The pharmacokinetics and interactions of ivermectin in humans-a mini-review. AAPS J. 2008;10:42–46. doi: 10.1208/s12248-007-9000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay MYF, Fraser JE, Chan WKK, Moreland NJ, Rathore AP, Wang C, et al. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor ivermectin. Antivir Res. 2013;99:301–6. [DOI] [PubMed]
- 35.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importing alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calya L, Drucea JD, Cattona MG, Jans DA, Wagstaff KM, et al. The FDA approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivermectin and nitazoxanide combination therapy for COVID-19. U.S National Library of Medicine. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04360356. Accessed 24 Aug 2020
- 38.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 39.Ibrahim IM, Abdelmalek DH, ElshahatME, Elky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020; 80:554-562. [DOI] [PMC free article] [PubMed]
- 40.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bosch BJ, Bartelink W, Rottier PJM. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Kramer-Kuhl A, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87:5502–11. [DOI] [PMC free article] [PubMed]
- 45.Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8:e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchiet F, et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann M, Kleine-Weber H, Pohlmann S. SARS-CoV-2 entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;18:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. [DOI] [PMC free article] [PubMed]
- 49.Diehl WE, Lin AE, Grubaugh ND, Carvalho LM, Kim K, Kyawe PP, et al. Ebola virus glycoprotein with increased infectivity dominated the 2013–2016 epidemic. Cell. 2016;167:1088–98. [DOI] [PMC free article] [PubMed]
- 50.Wang MK, Lim SY, Lee SM, Cunningham JM. Biochemical basis for increased activity of Ebola glycoprotein in the 2013-16 epidemic. Cell Host Microbe. 2017;21:367–375. doi: 10.1016/j.chom.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, He WT, Wang L, Lai A, Ji X, Zhai X, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 2020;26:529–626. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuo L, Hurst KR, Masters PS. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J Virol. 2007;81(5):2249–2262. doi: 10.1128/JVI.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nieto-Torres JL, Dediego ML, Alvarez E, Jiménez-Guardeño JM, Regla-Nava JA, Llorente M, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. [DOI] [PMC free article] [PubMed]
- 54.Gupta MK, Vemula S, Donde R, Gouda G, Behera L, Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. 2020:1–11. 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed]
- 55.Ortego J, Ceriani JE, Patiño C, Plana J, Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368(2):296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–88. [DOI] [PMC free article] [PubMed]
- 58.Bhowmika D, Nandia R, Jagadeesanb R, Kumarc N, Prakashd A, Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect Genet Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coughlin M, Lou G, Martinez O, Masterman SK, Olsen OA, Moksa AA, et al. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using Xeno Mouse. Virology. 2007;361:93–102. [DOI] [PMC free article] [PubMed]
- 60.MmTang XC, Agnihothram SS, Jiao Y, Stanhope J, Graham RL, Peterson EC, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A. 2014;111:E2018–26. [DOI] [PMC free article] [PubMed]
- 61.MmZhang H, Wang G, Li J, Nie Y, Shi X, Lian G, et al. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–45. [DOI] [PMC free article] [PubMed]
- 62.Elshabrawy HA, Coughlin MM, Baker SC, Prabhakar BS. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One. 2012;7:e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyoshi-Akiyama T, Ishida I, Fukushi M, Yamaguchi K, Matsuoka Y, Ishihara T, et al. Fully human monoclonal antibody directed to proteolytic cleavage site in severe acute respiratory syndrome (SARS) coronavirus S protein neutralizes the virus in a rhesus macaque SARS model. J Infect Dis. 2011;203:1574–81. [DOI] [PMC free article] [PubMed]
- 64.Yuan K, Yi L, Chen J, Qu X, Qing T, Rao X, et al. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem Biophys Res Commun. 2004;319:746–52. [DOI] [PMC free article] [PubMed]
- 65.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–3. [DOI] [PMC free article] [PubMed]
- 66.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–5. [DOI] [PMC free article] [PubMed]
- 67.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–2. [DOI] [PMC free article] [PubMed]
- 68.MmZhang H, Baker A. Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Crit Care. 2017;21:305. doi: 10.1186/s13054-017-1882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci. 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 70.Lei C, Fu W, Qian K, Li T, Zhang S, Ding M, et al. Potent neutralization of 2019 novel coronavirus by recombinant ACE2-Ig. BioRxiv. 2020. 10.1101/2020.02.01.929976.
- 71.Gordon DE, Jang MG, Bohaddou M, Xu J, et al. A SARS-CoV-2 human protein-protein interaction map reveals drug targets and potential drug- repurposing. BioRxiv. 2020. 10.1101/2020.03.22.002386.
- 72.Dai W, Zhang B, Su H, Li J, Zhao Y, Xie X, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;80:eabb4489. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rao VK, Carlson EA, Yan SS. Mitochondrial permeability transition pore is a potential drug target for neuro degeneration. Biochim Biophys Acta Mol Basis Dis. 1842;2014:1267–1272. doi: 10.1016/j.bbadis.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong Y, Liao Y, Fang S, Tam JP, Liu DX. Up-regulation of Mcl-1 and Bak by coronavirus infection of human, avian and animal cells modulates apoptosis and viral replication. PLoS One. 2012;07:e30191. doi: 10.1371/journal.pone.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fung TS, Liu DX. Coronavirus infection, ER stress, apoptosis and innate immunity. Front Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mair-Jenkins J, Saavedra-Campo SM, Baillie JK, Cleary P, Khaw F, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020;117(17):9490–6. [DOI] [PMC free article] [PubMed]
- 80.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–9. [DOI] [PMC free article] [PubMed]
- 81.Li L, Zhang W, Hu Y, Tong X, Zheng S, Yang J, et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020;324:460–70. [DOI] [PMC free article] [PubMed]
- 82.Casadevall A, Scharff MD. Return to the past: the case for antibody-based therapies in infectious diseases. Clin Infect Dis. 1995;21(1):150–161. doi: 10.1093/clinids/21.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casadevall A, Scharff MD. Serum therapy revisited: animal models of infection and development of passive antibody therapy. Antimicrob Agents Chemother. 1994;38(8):1695–1702. doi: 10.1128/AAC.38.8.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agarwal A, Mukherjee A, Kumar G, Chatterjee P, Bhatnagar T, Malhotra P. PLACID trial collaborators. BMJ. 2020;371:m3939. doi: 10.1136/bmj.m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leng Z, Zhu R, Hou W, Feng Y, Yang Y, Han Q, et al. Transplantation of ACE2- mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11:216–28. [DOI] [PMC free article] [PubMed]
- 86.Golchin A, Seyedjafari E, Ardeshirylajimi A. Mesenchymal stem cell therapy for COVID-19: present or future. Stem Cell Rev. 2020:1–7. 10.1007/s12015-020-09973-w [DOI] [PMC free article] [PubMed]
- 87.Sarasini A, Percivalle E, Rovida F, Campanini G, Genini E, Torsellini M, et al. Detection and pathogenicity of human metapneumovirus respiratory infection in pediatric Italian patients during a winter--spring season. J Clin Virol. 2006;35:59–68. [DOI] [PMC free article] [PubMed]
- 88.Vemula SV, Zhao J, Liu J, Wang X, Biswas S, Hewlett I. Current approaches for diagnosis of influenza virus infections in humans. Viruses. 2016;8:96. doi: 10.3390/v8040096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goy G, Croxatto A, Posfay-Barbe KM, Gervaix A, Greub G. Development of a real-time PCR for the specific detection of Waddlia chondrophila in clinical samples. Eur J Clin Microbiol Infect Dis. 2009;28:1483–1486. doi: 10.1007/s10096-009-0804-7. [DOI] [PubMed] [Google Scholar]
- 90.Patel R, Fang FC. Diagnostic stewardship: opportunity for a laboratory-infectious diseases partnership. Clin Infect Dis. 2018;67:799–801. doi: 10.1093/cid/ciy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–9. [DOI] [PMC free article] [PubMed]
- 92.Winichakoon P, Chaiwarith R, Liwsrisakun C, Salee P, Goonna A, Limsukon A, et al. Negative nasopharyngeal and oropharyngeal swab does not rule out COVID-19. J Clin Microbiol. 2020;58. 10.1128/JCM.00297-20. [DOI] [PMC free article] [PubMed]
- 93.Patel R, Babady E, Theel ES, Storch GA, Pinsky BA, St George K, et al. Report from the American Society for Microbiology COVID-19. International Summit, 23 March 2020: Value of diagnostic testing for SARS–CoV-2/COVID-19. mBio. 2020;11:e00722–0. [DOI] [PMC free article] [PubMed]
- 94.World Health Organization . Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases: interim guidance. Geneva: WHO; 2020. [Google Scholar]
- 95.CDC 2019-novel coronavirus (2019-nCoV) realtime RT-PCR diagnostic panel; Division of Viral Diseases, U.S. Centers for Disease Control and Prevention: Atlanta, GA, 2020. Catalog # 2019-nCoVEUA-01. CDC-006-00019, Revision: 01 CDC/DDID/NCIRD. Effective: 2/4/2020
- 96.Specific primers and probes for detection 2019 novel coronavirus; China National Institute for Viral Disease Control and Prevention: Beijing, 2020.
- 97.Detection of 2019 Novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR; School of Public Health, Hong Kong University: Hong Kong, 2020. https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn=af1aac73_40
- 98.Naganori N, Shirato K, Katano H, Matsuyama S, Takeda M. Detection of second case of 2019-nCoV infection in Japan. Japan: Department of Virology III, National Institute of Infectious Diseases; 2020. https://www.niid.go.jp/niid/en/2019-ncov-e/9334-ncov-vir3-2.html. Accessed 24 Aug 2020
- 99.Diagnostic detection of novel coronavirus 2019 by real time RT- PCR; Department of Medical Sciences, Ministry of Public Health, Thailand, 2020. Accessed on August 24, 2020.
- 100.Corman VM, Landt O. Detection of 2019-nCoV by real-time RT-PCR. Euro Surveill. 2020;25:23. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Israel develops new Covid pool testing method that can screen nearly 50 samples at once. 22 August, 2020. https://theprint.in/health/israel-develops-new-covid-pool-testing-method-that-can-screen-nearly-50-samples-at-once/487258/. Accessed 2 Sept 2020
- 102.Narayanan KR, Frost I, Heidarzadeh A, Tseng KK, Banerjee S, John J, Laxminarayan R. Pooling RT PCR or NGS samples has the potential to cost-effectively generate estimates of COVID-19 prevalence in resource limited environments. 10.1101/2020.04.03.2005199
- 103.Barrangou R. The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr Opin Immunol. 2015;32:36–41. doi: 10.1016/j.coi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 104.Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 105.CRISPR-CAS9, TALENS and ZFNS - the battle in gene editing. https://www.ptglab.com/news/blog/crispr-cas9-talens-and-zfns-the-battle-in-gene-editing/. Accessed 2 Sept 2020
- 106.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen JS, Ma E, Harrington LB, Costa MD, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–9. [DOI] [PMC free article] [PubMed]
- 108.Chiu C. Cutting-edge infectious disease diagnostics with CRISPR. Cell Host Microbe. 2018;23:702–704. doi: 10.1016/j.chom.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. [DOI] [PMC free article] [PubMed]
- 110.James P, Broughton XD, Guixia Y, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weaver C. Questions about accuracy of coronavirus tests sow 1317 worry. The Wall Street J. 2020. April 2, 2020. https://www.wsj.com/articles/questions-about-accuracy-of-coronavirus-tests-sow-worry-11585836001. Accessed 2 Sept 2020
- 112.Brangel P, Sobarzo A, Parolo C, Miller BS, Howes PD, Gelkop S, et al. A serological point-of-care test for the detection of IgG antibodies against Ebola virus in human survivors. ACS Nano. 2018;12:63–73. [DOI] [PubMed]
- 113.Liu Y, Liu Y, Diao B, Ren F, et al. Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2. 10.1101/2020.03.26.20044883.
- 114.Bruning AHL, Leeflang MMG, Vos JMBW, Spijker R, de Jong MD, Wolthers KC, et al. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin Infect Dis. 2017;65:1026–32. [DOI] [PMC free article] [PubMed]
- 115.Li Z, Yi Y, Luo X, Xion N, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020. https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.25727;92:1518–24. Accessed 2 Sept 2020 [DOI] [PMC free article] [PubMed]
- 116.Wang N, Li SY, Yang XL, Huang HM, Zhang YJ, Guo H, et al. Serological evidence of bat SARS-related coronavirus infection in humans. China Virol Sin. 2018;33:104–7. [DOI] [PMC free article] [PubMed]
- 117.Che X, Qiu L, Liao Z, Wang Y, Wen K, Pan YX, et al. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191:2033–7. [DOI] [PMC free article] [PubMed]
- 118.Klausegger A, Strobl B, Regl G, Kaser A, Luytjes W, Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. 1999;73:3737–3743. doi: 10.1128/JVI.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andrie FE. The future of vaccines, immunisation concepts and practice. Vaccine. 2001;19:2206–2209. doi: 10.1016/S0264-410X(00)00546-6. [DOI] [PubMed] [Google Scholar]
- 120.CEPI to fund three programmes to develop vaccines against the novel coronavirus, nCoV-2019; 2020. https://cepi.net/news_cepi/cepi-to-fund-three-programmes-todevelop-vaccines-against-the-novel-coronavirus-ncov-2019/.
- 121.Codagenix and Serum Institute of India announce commencement of first-in-human trial of COVI-VAC, a single dose, intranasal live attenuated vaccine for COVID-19. Dec 14, 2020. https://www.prnewswire.com/news-releases/codagenix-and-serum-institute-of-india-announce-commencement-of-first-in-human-trial-of-covi-vac-a-single-dose-intranasal-live-attenuated-vaccine-for-covid-19-301191756.html. Accessed 3 Jan 2021
- 122.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.COVID-19: Vaccine against SARS-CoV-2 infection, using the measles vector. Institut Pasteur. https://www.pasteur.fr/en/all-sars-cov-2-covid-19-institut-pasteur/research-projects/covid-19-vaccine-against-sars-cov-2-infection-using-measles-vector. Accessed 3 Jan 2021
- 124.Furuya Y. Return of inactivated whole-virus vaccine for superior efficacy. Immunol Cell Biol. 2012;90:571–578. doi: 10.1038/icb.2011.70. [DOI] [PubMed] [Google Scholar]
- 125.Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2020. 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed]
- 126.WHO. DRAFT landscape of COVID-19 candidate vaccines—20 March 2020. 2020. https://www.who.int/blueprint/prioritydiseases/key-action/novel coronaviruslandscape ncov.pdf?ua=1. Accessed 3 Jan 2021
- 127.Covid: What do we know about China's coronavirus vaccines?. BBC news. 16 December 2020. https://www.bbc.com/news/world-asia-china-55212787. Accessed 3 Jan 2021
- 128.Inactivated Covid vaccine safest with lesser bad reactions: Bharat Biotech. 2020. https://www.business-standard.com/article/current-affairs/inactivated-covid-vaccine-safest-with-lesser-bad-reactions-bharat-biotech-120122100032_1.html. Accessed 3 Jan 2021
- 129.COVAXINTM-India's First indigenous COVID-19 Vaccine. 2020. https://www.bharatbiotech.com/covaxin.html. Accessed 3 Jan 2021
- 130.Chen H, Xiang ZQ, Li Y, Kurupati RK, Jia B, Bian A, et al. Adenovirus-based vaccines: comparison of vectors from three species of Adenoviridae. J Virol. 2010;84:10522–32. [DOI] [PMC free article] [PubMed]
- 131.Van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus. BioRxiv 2020; 10.1101/2020.05.13.093195.
- 132.AZD1222 vaccine met primary efficacy endpoint in preventing COVID-19. AstraZeneca. 23 November 2020. https://www.astrazeneca.com/media-centre/press-releases/2020/azd1222hlr.html. Accessed 3 Jan 2021
- 133.Serum Institute applies for emergency use authorisation for COVID-19 vaccine. 21st December 2020. http://ptinews.com/news/12016707_Serum-Institute-applies-for-emergency-use-authorisation-for-COVID-19-vaccine.html.
- 134.AstraZeneca, J&J COVID-19 shots must succeed to meet U.S. vaccination goal. Fierce Pharma. Dec 10, 2020; https://www.fiercepharma.com/pharma/covid-19-shots-from-astrazeneca-and-j-j-must-succeed-to-meet-us-vaccination-goal-slaoui. Accessed 3 Jan 2021
- 135.Sputnik V: the world’s first registered COVID-19 vaccine - proven human adenoviral vector technology. 2020. https://sputnikvaccine.com/about-vaccine/. Accessed 3 Jan 2021
- 136.•.Belete TM. A review on Promising vaccine development progress for COVID-19 disease. Vacunas. 2020;21:121–128. doi: 10.1016/j.vacun.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Clover Biopharmaceuticals. Clover Biopharmaceuticals vaccines programs. 2020. http://www.cloverbiopharma.com/index.php?m=content&c=index&a=show&catid=11&id=53. Accessed 3 Jan 2021
- 138.Our response to COVID-19. GSK. December 2020. https://www.gsk.com/en-gb/media/resource-centre/our-contribution-to-the-fight-against-2019-ncov/. Accessed 3 Jan 2021
- 139.Rice J, Ottensmeier CH, Stevenson FK. DNA vaccines: precision tools for activating effective immunity against cancer. Nat Rev Cancer. 2008;8:108–120. doi: 10.1038/nrc2326. [DOI] [PubMed] [Google Scholar]
- 140.Inovio accelerates timeline for COVID-19 DNA vaccine INO-4800. INOVIO. 2020. https://www.prnewswire.com/news-releases/inovio-accelerates-timeline-for-covid-19-dna-vaccine-ino-4800-301015031.html. Accessed 3 Jan 2021
- 141.Chan JF, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines-a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Here’s the latest on COVID-19 vaccines. Science Coronavirus Coverage. December 18, 2020. https://www.nationalgeographic.com/science/health-and-human-body/human-diseases/coronavirus-vaccine-tracker-how-they-work-latest-developments-cvd/. Accessed 3 Jan 2021
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.