Abstract
The sudden increase in the COVID-19 epidemic affected by novel coronavirus 2019 has jeopardized public health worldwide. Hence the necessities of a drug or therapeutic agent that heal SARS-CoV-2 infections are essential requirements. The viral genome encodes a large Polyprotein, further processed by the main protease/ 3C-like protease (3CLpro) and papain-like proteases (PLpro) into 16 nonstructural proteins to form a viral replication complex. These essential functions of 3CLpro and PLpro in virus duplication make these proteases a promising target for discovering potential therapeutic candidates and possible treatment for SARS-CoV-2 infection.
This study aimed to screen a unique set of protease inhibitors library against 3CLpro and PLpro of the SARS-CoV-2. A molecular docking study was performed using PyRx to reveal the binding affinity of the selected ligands and molecular dynamic simulations were executed to assess the three-dimensional stability of protein-ligand complexes. The pharmacodynamics parameters of the inhibitors were predicted using admetSAR. The top two ligands (Nafamostat and VR23) based on docking scores were selected for further studies. Selected ligands showed excellent pharmacokinetic properties with proper absorption, bioavailability and minimal toxicity. Due to the emerging and efficiency of remdesivir and dexamethasone in healing COVID-19 patients, ADMET properties of the selected ligands were thus compared with it. MD Simulation studies up to 100 ns revealed the ligands' stability at the target proteins' binding site residues. Therefore, Nafamostat and VR23 may provide potential treatment options against SARS-CoV-2 infections by potentially inhibiting virus duplication though more research is warranted.
Keywords: SARS-CoV-2, 3CLpro, PLpro, Drug, Docking, ADMET, and Simulation
1. Introduction
In December 2019, a new coronavirus caused an outbreak of the pulmonary disease in Wuhan, the capital of Hubei province in China, and has since spread globally [[67], [68], [79]]. The virus has been named SARS-CoV-2 [20], with 96% genome identical to a bat coronavirus and shares 79.6% sequence identity to SARS-CoV [43,67,79]. This pandemic spread worldwide with more than 104.9 million infections and more than 2.27 million deaths till 3rd February 2021 [https://www.worldometers.info/coronavirus/].
The genome size of coronaviruses is ~30,000 nucleotides in length with a 5′-cap structure and a 3′-poly (A) tail and consist of at least six open reading frames (ORFs) [27,13]. The first ORF (ORF 1a/b) is about two-thirds of the genome length, precisely translates two polyproteins, pp1a and pp1ab and are processed by the main protease, also known as the 3C-like protease (3CLpro) and by one or two papain-like proteases (PLpro), into 16 nonstructural proteins (NSPs) and develop into mature proteins which assist in the viral replication [7,26,24]. PLpro facilitates cleavage at the first three polyproteins sites, whereas CLpro facilitates the cleavage at 11 sites [14,29]. The 3CLpro carries out cleavage at the polyproteins' C-terminal while, N-terminal of the polyproteins are cleaved by PLpro [40]. These NSPs engaged in subgenomic RNAs construction that encodes four main structural proteins, namely envelope (E), membrane (M), spike (S), and nucleocapsid (N) proteins and other accessory proteins [56,57].
The ~306 aa long 3CLpro, a key enzyme for coronavirus replication, is also encoded by the polypeptide and responsible for processing the polypeptide into functional proteins [67,77]. The 3CLpro, also known as Nsp5, is the first to get automatically cleaved from polyproteins to produce mature enzymes, and then it further assists in the cleavage of downstream Nsps at 11 sites to release Nsp4–Nsp16 [71]. 3CLpro directly mediates the maturation of Nsps, which is essential in the life cycle of the virus. Studies have shown that 3CLpro of different coronaviruses and all the 3CLpro cleavage sites on polyprotein 1ab are highly conserved in sequences and 3D structures [72,69]. Since there are no known human proteases with a similar cleaving specificity, 3CLpro inhibitors are less likely to be toxic and, together with its functional importance, have made 3CLpro an attractive target for the design of anti-SARS-CoV-2 drugs [2,69,77].
PLpro is responsible for the cleavages of N-terminus of the replicase polyprotein at three conserved cleavage sites to release Nsp1, Nsp2 and Nsp3, essential for correcting virus replication [21,7]. PLpro was also confirmed to be significant to antagonize the host's innate immunity [12,75,37]. As an essential enzyme in the host's coronavirus replication and infection, PLpro has been a popular and valuable target for identifying potential drugs against novel coronavirus 2019 [42].
Molecular docking has become a promising tool for drug discovery and development. By utilizing this tool, the binding interaction of drug-like molecules inside the target protein's binding pockets can be investigated with other factors like identification of hit molecules, optimization of lead compound and virtual screening. Since COVID-19 is a significant outbreak in 215 nations and there are no effective drugs for COVID-19, it is challenging to cure SARS-CoV-2 infection and control the associated pandemic. Therefore, novel drug design and discovery techniques can be utilized to discover potential therapeutic candidates against SARS-CoV-2.
2. Materials and methods
2.1. Protein structure retrieval
The crystal structures for 3CLpro (PDB ID: 6Y2E, Resolution: 1.75 Å) [77] and PLpro (PDB ID:6W9C, Resolution: 2.70 Å) of SARS-CoV-2 were downloaded from the PDB database (https://www.rcsb.org) in PDB format and were utilized for the in silico studies.
2.2. Energy minimization and structure validation
YASARA Energy Minimization Server carried out energy minimization of the crystal structures of 3CLpro and PLpro of the SARS-CoV-2 to obtain an energy-minimized and highly stable protein structure for efficient docking [33]. PROCHECK further validated both the energy minimized crystal structures of SARS-CoV-2 [36].
2.3. Binding site prediction
Binding site residues were anticipated through a literature survey for 3CLpro [[32], [58], [61], [70], [77]] and PLpro [4,66] (Table S1).
2.4. Ligand selection and ligand file preparation
Protease inhibitors, a class of antiviral drugs, are widely used to treat HIV/AIDS and hepatitis C. Inhibition of serine proteases mitigated SARS-CoV pathogenesis in vivo was also reported. Protease inhibitors prevent viral replication by selectively binding to viral proteases and blocking proteolytic cleavage of protein precursors necessary for the production of infectious viral particles [[17], [44], [52], [63]]. The protease inhibitor library, with a unique collection of 227 small molecule inhibitors used for chemical genomics, high-throughput screening (HTS) and high content screening (HCS), was downloaded from Selleckchem.com (https://www.selleckchem.com/) and saved in sdf file. According to our docking protocol, the MDL MOL format was converted to a pdbqt file by Open Babel [49].
2.5. Molecular docking and interaction analysis
Validation of docking protocol is essential in molecular docking to ensure that ligands bind in the correct conformation within the protein's binding site pocket, which is done by validating the size and center of the coordinates of the grid box across the binding pocket [39]. PyRx, a virtual screening software for computational drug discovery, was used to screen the selected ligands (set of protease inhibitors) files against SARS-CoV-2 3CLpro and PLpro [15]. PyRx uses AutoDock 4 and AutoDockVina as docking software implying the Lamarckian Genetic Algorithm and Empirical Free Energy Scoring Function. PyRx was carried out using selected inhibitors' on the energy minimized structure of SARS-CoV-2 3CLpro and PLpro. The macromolecular structure of two proteases (3CLpro & PLpro) and the ligands were prepared and docking was accomplished into the binding site residues inside a grid box with X, Y and Z axis and dimensions adjusted to 16.17 Å × 17.25 Å × 20.14 Å and 49.43 Å × 49.41 Å × 21.06 Å for SARS-CoV-2 3CLpro and PLpro protein respectively. As in PLpro, the catalytic triad Cys111-His272-Asp286 in each of three subunits A, B and C respectively are directed to the center of the protein [66]; the binding pocket for all the three subunits was taken under consideration and was within the validated size and dimensions of the grid box in PyRx docking. The docking protocol was then run at exhaustiveness of 8 and set to only output the lowest energy pose. The interactions between our targeted proteins and the ligands were studied using Ligplot [64], and the pictures were processed and made using Pymol Molecular Visualization Software [38].
2.6. Pharmacokinetics studies
The drug-likeness prediction of the selected ligands was carried out by Lipinski filter (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp), according to which an orally active drug should acquiesce to a minimum of four of the five laid down criteria for drug-likeness, namely: molecular mass, H-bond acceptor, H-bond donor, molar refractive index and LogP [41]. Also, molinspiration (https://molinspiration.com/cgi-bin/properties) was used to calculate the selected inhibitors' molecular properties and bioactivity. The absorption percentage (% AB) was calculated using the formula [[27], [78]]:
| AB% = 109 – (0.345 x TPSA). |
The pharmacodynamics parameters: Absorption, Distribution, Metabolism, Excretion and the Toxicity (ADMET) of the inhibitors were predicted using admetSAR (http://lmmd.ecust.edu.cn:8000/).
2.7. Molecular dynamics (MD) simulation
All-atoms MD simulations were performed on the atomic coordinates of the best-docked complex of SARS-CoV-2 target proteins, i.e., 3CLpro and PLpro using Gromacs v5.1.4 with force field CHARMM27 and water model TIP3P [1] and the ligands parameters were defined from Zoete et al. [80]. The simulation box was defined with buffer distance (10 Å) from the centrally placed protein-ligand complex. The prepared system was solvated with water molecules and neutralized with the addition of 0.15 M counter ions (Na+ and Cl−) [31]. The energy minimization process involves 50,000 steps for each steepest descent, followed by conjugant gradients. PBC condition was defined for x, y and z directions [16] and simulations were performed at physiological temperature; 300 K. SHAKE algorithm was applied to constrain all bonding involved, hydrogen and long-range electrostatic forces treated with PME (Particle mesh Ewald). The system was equilibrated in two steps, NVT and NPT, at 300 K for a period of 500 ps. During the simulation, the Berendsen thermostat [9] and Parrinello-Rahman pressure [51] were used to maintain pressure and temperature. LINC algorithm was used to constrain the bonds and angles [23]. The van der Waals interactions are taken care of by LJ potential with a cutoff of 0.10 nm. Using the NPT ensemble, production runs were performed for the period of 100, with time integration. The energy, velocity and trajectory were updated at the time interval of 10 ps. All production runs were done on CUDA enabled Tesla GPU machine (DELL T640 with V100 GPU) and OS Centos 7 [59,53] and the Gromacs utilities were used for the analyses of obtained MD trajectories.
3. Results and discussions
3.1. Energy minimization and model validation
YASARA [33] was used for energy minimization of the crystal structures for SARS-CoV-2 3CLpro (PDB ID: 6Y2E) and PLpro (PDB ID:6W9C) for low energy and high stability of the protein and was validated using PROCHECK [36]. Ramachandran plot analysis showed that 99.6% residues are in favoured, allowed and generously allowed regions and only 0.4% in the disallowed region for the energy minimized 3CLpro protein. In comparison, 100% residues are in favoured, additionally allowed and generously allowed regions for the PLpro protein (Fig. S1).
3.2. Molecular docking and interaction analysis
The top challenging job in computational chemistry is predicting ligands' actual binding conformation into protein binding sites [6]. After successful docking between the selected ligands (set of protease inhibitors) and the binding site of SARS-CoV-2 3CLpro and PLpro, the ligands' docking scores were noted and the best-ranked poses with the lowest docking score were chosen for interaction studies in detail. The mode of interaction of inhibitor compounds with amino acid residues in protein binding sites results in the compounds' binding affinity and potency [76].
The docking study carried out with the set of protease inhibitors as ligands against 3CLpro and PLpro of SARS-CoV-2 gave a formative revelation of molecular interplay. We find that all ligands interact with 3CLpro and PLpro more or less efficiently. Nafamostat and VR23 [7‑chloro-4-(4-((2,4-dinitrophenyl)sulfonyl) piperazin-1-yl) quinoline] showed the lowest binding energy and high efficiency against both the target protein, i.e., 3CLpro and PLpro of SARS-CoV-2. Nafamostat showed binding energy of −9.0 kcal/mol and −9.2 kcal/mol against 3CLpro and PLpro, respectively, while VR23 possessed binding energy of −9.1 kcal/mol against both 3CLpro and PLpro, respectively (Table 1 ). A recent study suggests that nafamostat mesylate can effectively kill SARS-CoV-2 in vitro by targeting spike protein [74]. Earlier reports also suggested that Nafamostat effectively inhibits MERS-CoV besides its extensive use in Japan to treat patients with a dissemination intravascular coagulation (DIC) and acute pancreatitis [73]. COVID-19 manifested DIC characteristics with enhanced fibrinolysis, and nafamostat was anticipated as a promising solution [5]. Nafamostat mesylate was considered a drug candidate for treating DIC induced by the Ebola virus infection [48]. Nafamostat has been used clinically to treat pancreatitis in Japan [3,28] and was approved by the USA Food and Drug Administration as the generic name, Nafamostat mesylate. VR23 is a small molecule that potentially inhibits trypsin-like proteasomes, chymotrypsin-like proteasomes and caspase-like proteasomes. Structurally, VR23 is a novel proteasome inhibitor with desirable anticancer properties that selectively kills cancer via cyclin E–mediated centrosome amplification without any notable ill effects [55]. The best binding poses for 3CLpro and PLpro with the top 2 ligands are shown in Fig. 1 & 2 , respectively.
Table 1.
Table showing binding energy values and interactions of the top two ligands (protease inhibitors) with the key residues of 3CLpro and PLpro of the SARS-CoV-2 evaluated by PyRx docking.
| Target protein | Inhibitors | Binding energy (kcal/mol) | Key residues interaction | Chain | H-bonds | Bond length (Å) |
|---|---|---|---|---|---|---|
| 3CLpro | Nafamostat | −9.0 | Ser1 | – | N-N4 | 3.04 |
| Thr26 | – | O-N2 | 2.93 | |||
| N-N2 | 3.14 | |||||
| Glu166 | – | OE2-N3 | 2.93 | |||
| VR23 | −9.1 | Ser1 | – | N-O3 | 2.65 | |
| Ser144 | – | OG-O5 | 3.24 | |||
| Glu166 | – | N-O1 | 2.90 | |||
| PLpro | Nafamostat | −9.2 | Trp106 | C | N2-N | 3.21 |
| Tyr273 | B | N5-OH | 2.91 | |||
| Asp286 | C | N1-O | 3.13 | |||
| Ala288 | C | N2-N | 3.10 | |||
| Asp302 | B | N4-OD1 | 3.26 | |||
| VR23 | −9.1 | Trp106 | C | O3-N | 2.80 | |
| Asn267 | A | O5-ND2 | 3.13 | |||
| Tyr273 | A | N5-OH | 2.97 | |||
| Ala288 | C | O3-N | 2.93 |
Fig. 1.
The docking results of (A) Nafamostat (Red) and (B) VR23 (aquamarine) inside binding pocket of the 3CLpro (cartoon and blue) of the SARS- CoV-2. Hydrogen bonded interactions are shown as yellow dotted lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
The docking results of (A) Nafamostat (Red) and (B) VR23 (aquamarine) inside the binding pocket of the PLpro (cartoon and magenta) of the SARS- CoV-2. Hydrogen bonded interactions are shown as yellow dotted lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
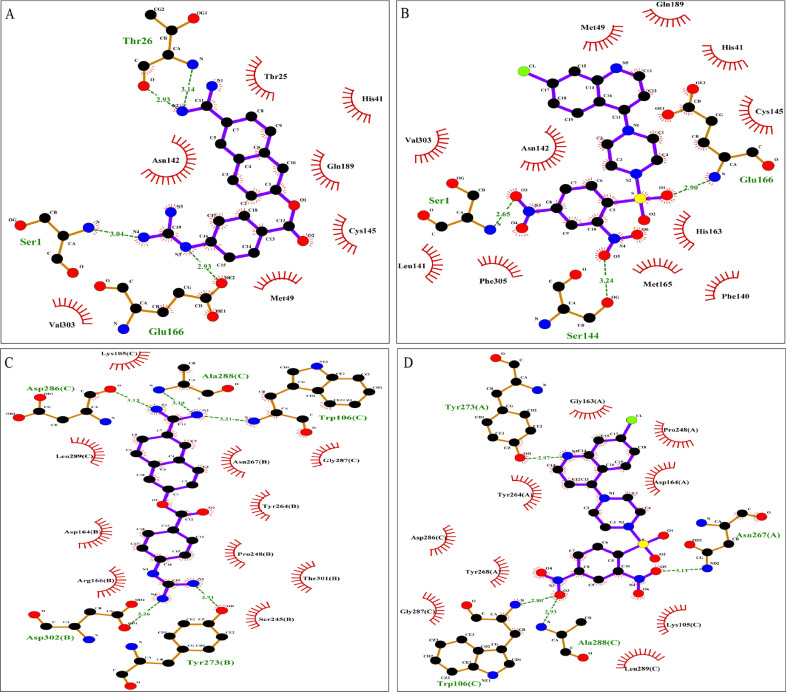
The docking experiment was carried out in PyRx with grid box formation to include all the active site residues for SARS-CoV-2 3CLpro and PLpro. Further bond length, H-bond formation between ligands and target protein were analyzed using Ligplot and interactions were depicted in Fig. 3 A-D. Both the protease inhibitors (ligands) Nafamostat and VR23 did not form any H-bond with the catalytic dyad (Cys145 and His41) of SARS-CoV-2 3CLpro as revealed from the interaction studies, but both Nafamostat and VR23 revealed to form H-bond with the residues Ser1 and Glu166, which provides stability and active conformation of S1 pocket in SARS-CoV-2 3CLpro [67,77]. Apart from these two residues, Nafamostat also forms H-bond with Thr26, while VR23 forms H-bond with Ser144 (Fig. 3 A-B).
Fig. 3.
Diagrammatic sketch illustrating the interactions between (A) Nafamostat and 3CLpro protein. (B) VR23 and 3CLpro protein. (C) PLpro protein and Nafamostat. (D) PLpro protein and VR23 by LigPlot. Ligand is shown in purple and: green dashed lines indicate hydrogen bonds with distance in angstrom (Å), spoked red arcs indicate hydrophobic contacts, atoms are shown in black for carbon, blue for nitrogen, red represents oxygen, green represents fluorine and yellow represents sulfur. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
On the other hand, the 2D interaction of the selected protease inhibitors with SARS-CoV-2 PLpro revealed Nafamostat's interaction and H-bond formation with the residue Asp286 of the catalytic triad Cys111-His272-Asp286 of subunit C apart from other vital interactions with the binding site residues across other subunits (Fig. 3C). VR23 did not form an H-bond with the catalytic triad Cys111-His272-Asp286 of any of the subunits of PLpro. However, VR23 was highly efficient in interacting with other binding site residues such as Trp106 and Ala288 of subunit C and Asn267 and Tyr273 of subunit A respectively, of PLpro (Fig. 3D).
3.3. Pharmacokinetics studies
Lipinski's rule of 5 (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) predicted the selected molecular properties of the ligands from the SMILES imported from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Under Lipinski rule of 5, the calculation for drug-likeness was done with criteria namely: (M.W. <500 Dalton), high lipophilicity expressed as LOGP (LOGP<5), hydrogen bond donors (HBD <5), hydrogen bond acceptors (HBA <10) and molar refractivity should be between 40 and 130 indicated good absorption and permeation across the cell membrane. For an effective drug to be taken orally, it should satisfy at least four criteria out of the 5, thus distinguishing a molecule from a drug or a non-drug [[41]; http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp]. The selected inhibitors Nafamostat and VR23 both satisfied Lipinski's rule of 5 with LOGP value (<5), evaluating them with good permeability across the cell membrane [45] and establishing them as pharmacologically active (Table S2). As evaluated by molinspiration, the molecular property showed excellent results for both the selected ligands (protease inhibitors) to evaluate a single violation for both the inhibitors in Lipinski's rule of 5 (Table 2 ). The AB% (>50%) of a drug is a good sign for its excellent bioavailability, distribution and circulation by oral route [[18], [22]]. Both Nafamostat and VR23 possessed AB% > 50% (Table 2).
Table 2.
Physicochemical proprieties prediction of the selected ligands (Nafamostat and VR23) using Molinspiration software.
| Inhibitors | MW (g/mol) (<500) | MiLogP (≤ 5) | nON (acceptors) (<10) | nOHNH (donors) (<5) | Volume (A3) | TPSA (A2) | AB% (>50%) | Lipinski's violations ≤ 1 |
|---|---|---|---|---|---|---|---|---|
| Nafamostat | 347.38 | 2.16 | 7 | 7 | 306.84 | 140.59 | 60.49 | 1 |
| VR23 | 477.89 | 3.58 | 11 | 0 | 368.86 | 145.16 | 58.91 | 1 |
The selected ligands' bioactivity was checked through molinspiration and calculated the activity against GPCR ligand, ion channel modulator, a kinase inhibitor, nuclear receptor ligand, protease inhibitor and enzyme inhibitor [45]. The bioactivity values were interpreted as: active (bioactivity score > 0), moderately active (bioactivity score: −5.0 – 0.0) and inactive (bioactivity score < −5.0) [62]. The bioactivity score of the selected inhibitors towards GPCR ligand, ion channel modulator, nuclear receptor ligand, kinase, protease and enzyme inhibitions indicated both Nafamostat and VR23 as active protease and enzyme inhibitors. The bioactivity prediction by molinspiration is tabulated in Table 3 .
Table 3.
Bioactivity prediction of the Nafamostat and VR23 through Molinspiration software.
| Inhibitors | GPCR ligand | Ion channel modulator | Kinase Inhibitor | Nuclear receptor ligand | Protease Inhibitor | Enzyme Inhibitor |
|---|---|---|---|---|---|---|
| Nafamostat | 0.28 | 0.14 | −0.03 | −0.16 | 0.57 | 0.19 |
| VR23 | −0.05 | −0.15 | −0.04 | −0.32 | 0.07 | 0.05 |
The pharmacodynamic study for the selected ligands was carried out through admetSAR to understand the drug's action inside a host's body. The ADMET properties of the selected ligands revealed through admetSAR (http://lmmd.ecust.edu.cn:8000/) are presented in Table 4 . The ADMET study in this work focused on the parameters such as solubility (LogS), human intestinal absorption (HIA), Caco-2 permeability, cytochrome substrate/inhibitor, human ether a go-go gene inhibition, AMES toxicity, carcinogens and acute rat toxicity (LD50). The result shows that Nafamostat and VR23 exhibit an ability to cross the blood-brain barrier (BBB), with a probability of 0.92 and 0.68. Besides, an excellent human intestinal absorption for selected ligands and a moderate ability to penetrate human colon adenocarcinoma (Caco-2+), with Caco-2 permeability of 0.53 cm/s and 0.57 cm/s for Nafamostat and VR23, respectively, were predicted (Table 4). These results confirm the high drug absorption (AB%) as predicted by Molinspiration [45] and further supported by the excellent values of HIA of 1.00 and 0.99 for Nafamostat and VR23 respectively in admetSAR (Table 4). LogS values of Nafamostat and VR23 were found to be −4.07 and −4.29, respectively, and were considered moderately soluble (Table 4).
Table 4.
Table indicating pharmacodynamic profile for the selected ligands along with Remdesivir and Dexamethasone as control by admetSAR.
| Inhibitors | Log S (>−4) | Blood Brain Barrier (BBB) | Human Intestinal Absorption (HIA) | Caco2 Permeability (cm/s) | CYP substrate / Inhibitor | Human ether a go-go gene | AMES toxicity | Carcinogenicity | LD50 (Rat Acute Toxicity) (mol/ kg) |
|---|---|---|---|---|---|---|---|---|---|
| Nafamostat | −4.07 | 0.92 | 0.99 | 0.53 | Non-substrate/ Non-inhibitor | weak inhibitor | nontoxic | Non-carcinogen | 2.56 |
| VR23 | −4.29 | 0.68 | 1.00 | 0.57 | Substrate/ Inhibitor | weak inhibitor | nontoxic | Non-carcinogen | 2.52 |
| Remdesivir | −3.47 | 0.74 | 0.88 | −0.14 | Substrate/ Non-inhibitor | weak inhibitor | nontoxic | Non-carcinogen | 2.71 |
| Dexamethasone | −3.70 | 0.97 | 0.99 | 1.08 | Substrate/ Non-inhibitor | weak inhibitor | nontoxic | Non-carcinogen | 2.14 |
In terms of metabolism, Nafamostat was a non-substrate/non-inhibitor of cytochrome P450 (CYP 450), indicating its proper metabolism by CYP450, while VR23 was a substrate for CYP450 3A4 and inhibitor of CYP450 (Table 4).
Toxicity analysis predicted that both the selected protease inhibitors were non-AMES toxic and non-carcinogens. The Nafamostat and VR23 were also weak inhibitors of human ether, a go-go gene, a regulatory potassium channel that leads to long QT syndrome [60,[10], [11]]. Comparing the predicted LD50 doses, a compound with a lower dose is more lethal than the compound having a higher LD50 [47]. From our observation in admetSAR, we found that both the selected protease inhibitors have optimal LD50 doses indicating they are nonlethal and possess excellent pharmacodynamic properties (Table 4).
Due to the emerging and effectiveness of remdesivir and dexamethasone in treating COVID-19 patients in clinical trials [8,65,19,[25], [30]], the ADMET properties of the selected ligands were thus compared with it. All the selected ligands showed immensely satisfying results, with some of the values even better than the control drugs, as shown in Table 4 .
3.4. Molecular dynamics (MD) simulation studies
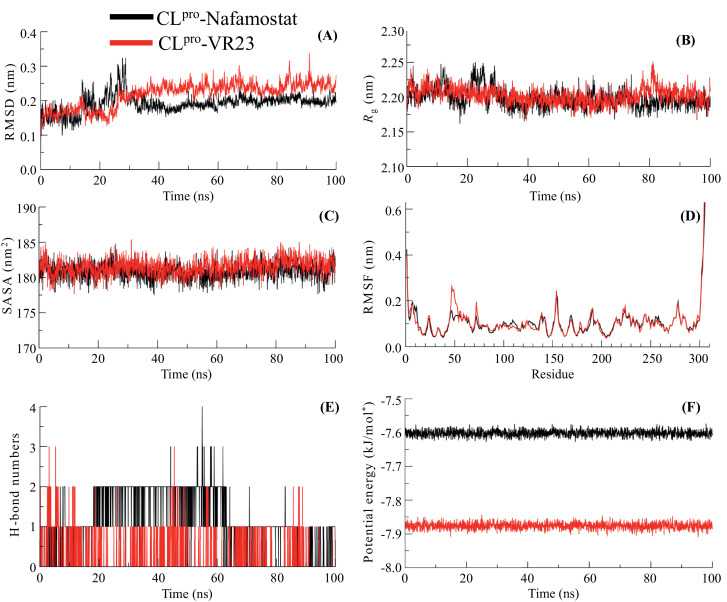
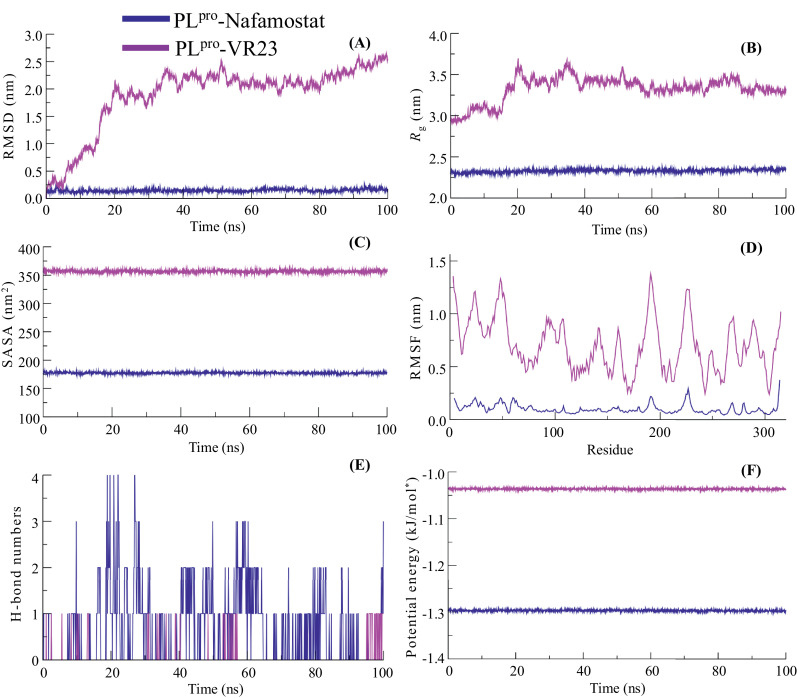
Molecular docking and pharmacokinetic analyses resulted in selecting two possible protease inhibitors, Nafamostat and VR23, against the SARS-CoV-2 protease proteins, 3CLpro and PLpro. In order to examine the molecular stability of the inhibitor's interactions with viral proteases, the best docking poses were selected and all-atoms MD simulations were carried out for the period of 100 ns at the physiological temperature. The changes in the structural features underlying the protease-inhibitors interactions were analyzed through investigating the plots of RMSD, Rg, SASA, RMSF, H-bonds and potential energies, as shown in Fig. 4 and 5 . The obtained backbone Cα-RMSD plot of inhibitors, Nafamostat and VR23 complexed with 3CLpro, during the simulation is depicted in Fig. 4 A. In this Fig. 4A, we can see that the RMSD plot of the 3CLpro-Nafamostat complex reaches the steady equilibrium at ~30 ns. The initial rising in RMSD of ~0.15 nm during 0–25 ns indicates the structural perturbation to well-fit the inhibitor, which is gradually dropped at ~30 ns. The complex structure remains stable with RMSD 0.19±0.02 nm till the end of simulation at 100 ns. The RMSD plot of 3CLpro-VR23 shows that it also attains equilibrium at ~30 ns, after the initial deviation of RMSD ~0.10 ns; however, the structure is observed a stable RMSD value of 0.21±0.03 nm. The RMSD plot of the PLpro-Nafamostat complex suggested relatively stable conformational dynamics of protease-inhibitor, showing a steady equilibrium at RMSD 0.14±0.02 nm can be seen throughout the simulation time (Fig. 5 A). However, the binding of VR23 with PLpro shows the sharp drift of 2.0 nm, optimized at ~40 ns. This equilibrium state is prolonged up to 80 ns and the further increase in RMSD can be seen for the last 20 ns. The result shows that the structure of the PLpro-VR23 complex propagated with an average RMSD 1.89±0.60 nm. These findings indicate a stable global conformational dynamic of Nafamostat with both proteases, 3CLpro and PLpro, and VR23 with 3CLpro. Whereas the binding of VR23 with PLpro underwent significant conformational changes.
Fig. 4.
The 100 ns molecular dynamic simulation results of two protein-ligand complexes (3CLpro- Nafamostat (black) and 3CLpro-VR23 (red)). (A) RMSD values of backbone atoms. (B) Rg of backbone atoms. (C) SASA of the ligand. (D) RMSF values of the chain. (E) Total number of H-bonds between ligands and 3CLpro. (F) Gromacs/ Potential energies values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
The 100 ns molecular dynamic simulation results of two protein-ligand complexes (PLpro- Nafamostat (blue) and PLpro-VR23 (pink)). (A) RMSD values of backbone atoms. (B) Rg of backbone atoms. (C) SASA of the ligand. (D) RMSF values of the chain. (E) Total number of H-bonds between ligands and PLpro.(F) Gromacs/ Potential energies values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Further, protease-inhibitors' conformation stability was determined by the radius of gyration (Rg), which defines the structural compactness of protein. The average Rg values for 3CLpro-Nafamostat, 3CLpro-VR23, PLpro-Nafamostat and PLpro-VR23 complexes were calculated as 2.19, 2.20, 2.32 and 3.32 nm, respectively (Fig. 4 B & 5 B). The Rg plot shows a minor increase in Rg values at 0–10 and 20–25 ns, which gradually settles around 30 ns in the Nafamostat docked structure with 3CLpro (Fig. 4 B). The initial deviation in Rg values may be considered the time taken to optimize Nafamostat's well-fitting at the binding pocket of 3CLpro. After that, no structural drifts were observed in the Rg trajectory of 3CLpro-Nafamostat, suggesting the complex's stability for the remaining period of simulation 30–100 ns. Protease-inhibitor interactions. The Rg plot of PLpro-Nafamostat quickly attains the stable equilibrium and is observed consistently throughout the simulation time (Fig. 5 B). The slight deviation in the Rg plot of 3CLpro-VR23 can be seen during the 0–30 ns, which attained equilibrium at ~30 ns and the structural integrity of the complex is observed stable for the simulation time 30–100 ns. Remarkably, the Rg plot of PLpro-VR23 shows a sharp drift of ~0.75 nm at ~15 ns, which settles gradually around ~40 ns and a stable Rg trajectory is seen up to 100 ns. The substantial jump in Rg values may be due to the significant conformational changes before accommodating the inhibitor at the binding pocket of PLpro. Thus, the significant deviation in Rg values and a slow rate to achieve equilibrium suggested a less stable conformational dynamics of PLpro-VR23 than the binding of VR23 with 3CLpro. Moreover, a better equilibrated and stabilized structure of Nafamostat with both 3CLpro and PLpro, respectively.
The changes in protease-inhibitor complexes' structural features have also been analyzed by computing the solvent-accessible surface area (SASA). The solvent environment around the protein act as a driving force for maintaining the protein folds. SASA is one of the fundamental properties of a protein that maintains the practical orientation of protein and accompanies protein-ligand interactions [50,54]. Thus, it helps to evaluate protein-inhibitor complexes' structural stability under the solvent environment [10]. Fig. 4C shows that the complex structure of 3CLpro-Nafamostat and 3CLpro-VR23 have average SASA values 180.82 ± 1.12 and 189.49 ± 1.14 nm2, respectively. The binding of Nafamostat and VR23 with PLpro has the average SASA values 177.41± 1.15 and 356.70 ± 1.62 nm2, respectively (Fig. 5 C). Thus, comparing the binding of inhibitors with proteases, it is observed that Nafamostat has stable molecular interaction with 3CLpro and PLpro. Eventually, the binding of VR23 with 3CLpro also provides evidence of stable molecular interactions. However, the molecular interaction of VR23 with PLpro indicates the orientational change in the protein surface; thus, we observed a higher value for the accessible area of PLpro-VR23.
To examine protease-inhibitor interactions' dynamic progression, we calculated the root mean square fluctuation (RMSF), which provides a clue about the average positional fluctuations of each amino acid residue [35,46]. Usually, the higher atomistic fluctuations are possessed by the N-and -C terminal residues and loops of protein; however, larger loops exist, interconnecting the stable conformation of α-helices and β-sheets may deviate the RMSF plots [34]. Fig. 4 D shows that the binding of inhibitors with protease 3CLpro complies with a stable molecular interaction and displays almost completely overlapping RMSF plots for 3CLpro-Nafamostat and 3CLpro-VR23. The much higher deviation in the RMSF plot of PLpro-VR23 can be observed compared to PLpro-Nafamostat (Fig. 5 D). The residues of 3CLpro belonging to the stable secondary structure of α-helices and β-sheets display average atomic fluctuations <0.25 nm and no significant perturbation is observed for the region belonging to loops. Indeed, the binding pocket residues (Ser1, Glu166, Thr26 & Ser144) having lower atomic fluctuation, which evident in the stable molecular interaction of inhibitors with 3CLpro. We also find the consistent molecular interaction of Nafamostat with PLpro(Fig. 5 D). However, the RMSF plot of PLpro -VR23 shows the overall higher atomic fluctuations for all residues. Interestingly, the lower atomic fluctuation for residues (Trp106, Ala288, Asn 267 &Tyr273), involved in the interaction with VR23, clearly indicates that ligand remains occupied at the binding pocket during the simulation and higher RMSF is observed due to the conformational shifting of protease, PLpro.
More importantly, we computed the time evolution plot of hydrogen bonds' occupancy (H-bonds) between target proteases and inhibitors. H-bonds play a crucial role in maintaining the shape and stability of protein structure and govern the protein-ligand molecular interactions. Fig. 4 E shows that the maximum propensity of three H-bonds between 3CLpro and VR23. Among them, only one H-bond remains consistent and two appeared and disappeared transiently. The molecular interaction of 3CLpro with Nafamostat displays two H-bonds, which can be seen up to ~65 ns. However, one H-bond is lost during the last ~35 ns of simulation. The H-bond interactions of inhibitors with PLpro is shown in Fig. 5 E. Fig. 5E shows Nafamostat's structure is stabilized at the binding pocket of PLpro through four H-bonds, out of which two are observed consistently during the simulation.
On the other hand, we find the propensity of single H-bond interaction between VR23 and PLpro, which appears intermittently. Thus, H-bond interactions, along with structural parameter analyses described in Fig. 5A-D, provide clear evidence of the loosely bound structure of VR23 with PLpro. Finally, we measured the potential energy of inhibitor bound complexes of 3CLpro and PLpro. Fig. 4 F shows that the structure of 3CLpro-Nafamostat and 3CLpro-VR23 have potential energy values −7.60 × 105 and −7.87 × 105 kJ/mol, respectively. Whereas the structure of PLpro- Nafamostat and PLpro-VR23 shows the potential energy values −1.30 × 105 and −1.03 × 105 kJ/mol (Fig. 5 F). Thus, the obtained higher potential energy values for inhibitors bound complexes of 3CLpro compared to complexes with PLpro suggested the higher molecular affinity of Nafamostat and VR23 towards 3CLpro. This result also indicates Nafamostat's higher molecular affinity with PLpro than VR23, showing approximately 2.7 × 105 kJ/mol higher potential energy.
The MD simulation analyses collectively revealed a higher structural stability of Nafamostat with both proteases, 3CLpro and PLpro. Moreover, VR23 shows preferably stable molecular interaction towards 3CLpro as compared to PLpro. This study provides the structural insights of the molecular interaction of identified inhibitors, Nafamostat and VR23, against the two significant proteases of SARS-CoV-2, 3CLpro and PLpro, which can be explored as a potential lead in the development of anti-CoV drug therapy to control the pandemic of COVID-19.
4. Conclusion
Our study revealed that Nafamostat and VR23 interact with the critical binding site residues of 3CLpro and PLpro of SARS-CoV-2 and probably inhibit the growth of SARS-CoV-2 by targeting 3CLpro and PLpro. Pharmacokinetics properties matching the required criteria for drug-likeness and Nafamostat and VR23 may be considered a potential drug against SARS-CoV-2. Protein-ligand complexes for 3CLpro and PLpro were structurally stable throughout the 100 ns simulation period concerning distance and fluctuation dynamics. However, further experimental studies are needed to check the possible preclinical and clinical efficacy of Nafamostat and VR23 to prevent and treat SARS-CoV-2.
Author contribution statements
DK, RDS, AP and DB carried out the experiment. DB, AP and DK wrote the manuscript. DK, AP and DB contributed to the analysis of the results. DK supervised the project and conceived the original idea.
Declaration of Competing Interest
The authors declare that no conflict of interest exists.
Acknowledgments
The lab is supported by the Science and Engineering Research Board, India (FILE NO. ECR/2015//000155) and the Department of Biotechnology (India) (order no: BT/PR16224/NER/95/176/2015 & BT/PR24504/ NER/95/746/2017) to Dr. Diwakar Kumar. Deep Bhowmik received funding from the SERB grant (FILE NO. ECR/2015//000155).
References
- 1.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 2.Anand K..., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 3.Aoyama T., Ino Y., Ozeki M., Oda M., Sato T., Koshiyama Y., Suzuki S., Fujita M. Pharmacological studies of FUT-175, nafamstatmesilate. I. Inhibition of protease activity in in vitro and in vivo experiments. Jpn. J. Pharmacol. 1984;35(3):203–227. doi: 10.1254/jjp.35.203. [DOI] [PubMed] [Google Scholar]
- 4.Arya R., Das A., Prashar V., Kumar M. Potential inhibitorsagainst papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. ChemRxiv.Preprint. 2020 doi: 10.26434/chemrxiv.11860011.v2. [DOI] [Google Scholar]
- 5.Asakura H., Ogawa H. Potential of heparin and nafamostat combination therapy for COVID-19. Journal of thrombosis and haemostasis: JTH. 2020;18(6):1521–1522. doi: 10.1111/jth.14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azam S.S., Sarfaraz S., Abro A. Comparative modeling and virtual screening for the identification of novel inhibitors for myo-inositol-1-phosphate synthase. Mol. Biol. Rep. 2014;41(8):5039–5052. doi: 10.1007/s11033-014-3370-8. [DOI] [PubMed] [Google Scholar]
- 7.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., … ACTT-1 Study Group Members . The New England journal of medicine; 2020. Remdesivir For the Treatment of Covid-19 - Preliminary Report. NEJMoa2007764. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berendsen H.J.C., Grigera J.R., &Straatsma T.P. The missing term in effective pair potentials. J Phys Chem. 1987;91(24):6269–6271. doi: 10.1021/j100308a038. [DOI] [Google Scholar]
- 10.Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infection, genetics and evolution:journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2020;84 doi: 10.1016/j.meegid.2020.104451. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhowmik D., Jagadeesan R., Rai P., Nandi R., Gugan K., Kumar D. Evaluation of potential drugs against leishmaniasis targeting catalytic subunit of Leishmaniadonovani nuclear DNA primase using ligand based virtual screening, docking and molecular dynamics approaches. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1739557. [DOI] [PubMed] [Google Scholar]
- 12.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X.…Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. 2015. [DOI] [PubMed] [Google Scholar]
- 16.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 17.deLeuw P., Stephan C. Protease inhibitors for the treatment of hepatitis C virus infection. GMS infectious diseases. 2017;5 doi: 10.3205/id000034. Doc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 19.Goldman J.D., Lye D., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., Chen Y.S., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wei X., Gaggar A., Brainard D.M., Towner W.J., …GS-US-540-5773 Investigators Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. The New England journal of medicine. 2020 doi: 10.1056/NEJMoa2015301. 10.1056/NEJMoa2015301Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbalenya A.E., Baker S.C., Baric R.S., et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebert M.F. Impact of Pregnancy on Maternal Pharmacokinetics of Medications. Clinical Pharmacology During Pregnancy. 2013:17–39. doi: 10.1016/b978-0-12-386007-1.00003-9. [DOI] [Google Scholar]
- 23.Hess B., Bekker H., Berendsen H.J.C., &Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. 10.1002/(SICI)1096-987X(199709)18:12%3C1463::AID-JCC4%3E3.0.CO;2-H [Google Scholar]
- 24.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horby P., Lim W.S., Emberson J., et al. 2020. Effect of Dexamethasone in Hospitalized Patients With covid-19: Preliminary Report. medRxiv. 06.22.20137273 [Preprint.] [DOI] [Google Scholar]
- 26.Hsu M.F., Kuo C.J., Chang K.T., Chang H.C., Chou C.C., Ko T.P., Shr H.L., Chang G.G., Wang A.H., Liang P.H. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280(35):31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y., Wu Y., Li Z., Zhu Y., Tien P., Guo D. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79(9):5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwaki M., Ino Y., Motoyoshi A., Ozeki M., Sato T., Kurumi M., Aoyama T. Pharmacological studies of FUT-175, nafamostatmesilate. V. Effects on the pancreatic enzymes and experimental acute pancreatitis in rats. Jpn. J. Pharmacol. 1986;41(2):155–162. doi: 10.1254/jjp.41.155. [DOI] [PubMed] [Google Scholar]
- 29.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson R.M., Vinetz J.M. Dexamethasone in the management of covid -19. BMJ. 2020;370:m2648. doi: 10.1136/bmj.m2648. [DOI] [PubMed] [Google Scholar]
- 31.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 viaintegrated computational approach. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1751298. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 33.Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. Suppl 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar V., Prakash A., Lynn A.M. Alterations in local stability and dynamics of A4V SOD1 in the presence of trifluoroethanol. Biopolymers. 2018;109(3):e23102. doi: 10.1002/bip.23102. [DOI] [PubMed] [Google Scholar]
- 35.Kumar N., et al. Structure-based virtual screening, molecular dynamics simulation and MM-PBSA toward identifying the inhibitors for two-component regulatory system protein NarL of Mycobacterium Tuberculosis. J BiomolStructDyn. 2020;38(11):3396–3410. doi: 10.1080/07391102.2019.1657499. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereo chemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 37.Li S.W., Wang C.Y., Jou Y.J., Huang S.H., Hsiao L.H., Wan L., Lin Y.J., Kung S.H., Lin C.W. SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. Int J Mol Sci. 2016;17(5):678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lill M.A., Danielson M.L. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 2011;25(1):13–19. doi: 10.1007/s10822-010-9395-8. [DOI] [PubMed] [Google Scholar]
- 39.Lim S.V., Rahman M.B., Tejo B.A. Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinformatics. 2011;12(Suppl 13):S24. doi: 10.1186/1471-2105-12-S13-S24. Suppl 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Liu X., Wang X.J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. Journal of genetics and genomics = Yi chuanxuebao. 2020;47(2):119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J.…Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv Z., Chu Y., Wang Y. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV AIDS (Auckl) 2015;7:95–104. doi: 10.2147/HIV.S79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mokhnache K., Madoui S., Khither H., Charef N. Drug-Likeness and Pharmacokinetics of a bis-Phenolic Ligand: evaluations by Computational Methods. Sch J App Med Sci. 2019;7(1):167–173. January. [Google Scholar]
- 46.Mongre R.K., Mishra C.B., Prakash A., Jung S., Lee B.S., Kumari S., Hong J.T., Lee M.S. Novel Carbazole-Piperazine Hybrid Small Molecule Induces Apoptosis by Targeting BCL-2 and Inhibits Tumor Progression in Lung Adenocarcinoma in Vitro and Xenograft Mice Model. Cancers (Basel) 2019;11(9):1245. doi: 10.3390/cancers11091245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nisha C.M., Kumar A., Nair P., Gupta N., Silakari C., Tripathi T., Kumar A. Molecular Docking andInSilicoADMET Study Reveals Acylguanidine 7a as a Potential Inhibitor of β-Secretase. Adv Bioinformatics. 2016;2016:1–6. doi: 10.1155/2016/9258578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishimura, H., & Yamaya, M. (2015). A Synthetic Serine Protease Inhibitor, NafamostatMesilate, Is a Drug Potentially Applicable to the Treatment of Ebola Virus Disease. The Tohoku journal of experimental medicine, 237(1), 45–50. 10.1620/tjem.237.45 [DOI] [PubMed]
- 49.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey P., Meena N.K., Prakash A., Kumar V., Lynn A.M., Faizan Ahmad F. Characterization of heterogeneous intermediate ensembles on the guanidinium chloride-induced unfolding pathway of β-lactoglobulin. Journal of Biomolecular Structure and Dynamics. 2020;38(4):1042–1053. doi: 10.1080/07391102.2019.1593245. [DOI] [PubMed] [Google Scholar]
- 51.Parrinello M., Rahman A. Crystal Structure and Pair Potentials: a Molecular-Dynamics Study. Phys. Rev. Lett. 1980;45(14):1196–1199. doi: 10.1103/physrevlett.45.1196. [DOI] [Google Scholar]
- 52.Patick A.K., Potts K.E. Protease inhibitors as antiviral agents. Clin. Microbiol. Rev. 1998;11(4):614–627. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prakash A., Dixit G., Meena N.K., Singh R., Vishwakarma P., Mishra S., Lynn A.M. Elucidation of stable intermediates in urea-induced unfolding pathway of human carbonic anhydrase IX. J. Biomol. Struct. Dyn. 2018;36(9):2391–2406. doi: 10.1080/07391102.2017.1355847. [DOI] [PubMed] [Google Scholar]
- 54.Prakash A., Luthra P.M. Insilico study of the A2AR–D2R kinetics and interfacial contact surface for heteromerization. Amino Acids. 2012;43:1451–1464. doi: 10.1007/s00726-012-1218-x. [DOI] [PubMed] [Google Scholar]
- 55.Pundir S., Vu H.Y., Solomon V.R., McClure R., Lee H. VR23: a Quinoline-Sulfonyl Hybrid Proteasome Inhibitor That Selectively Kills Cancer via Cyclin E-Mediated Centrosome Amplification. Cancer Res. 2015;75(19):4164–4175. doi: 10.1158/0008-5472.CAN-14-3370. [DOI] [PubMed] [Google Scholar]
- 56.Ramajayam R., Tan K.P., Liang P.H. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans. 2011;39(5):1371–1375. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- 57.Ren Z., Yan L., Zhang N., Guo Y., Yang C., Lou Z., Rao Z. The newly emerged SARS-like coronavirus HCoV-EMC also has an "Achilles' heel": current effective inhibitor targeting a 3C-like protease. Protein Cell. 2013;4(4):248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273(5):1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh R., Meena N.K., Das T., Sharma R.D., Prakash A., Lynn A.M. Delineating the conformational dynamics of intermediate structures on the unfolding pathway of β-lactoglobulin in aqueous urea and dimethyl sulfoxide. J. Biomol. Struct. Dyn. 2019:1–10. doi: 10.1080/07391102.2019.1695669. [DOI] [PubMed] [Google Scholar]
- 60.Sood D., Kumar N., Singh A., Sakharkar M.K., Tomar V., Chandra R. Antibacterial and Pharmacological Evaluation of Fluoroquinolones: a Chemoinformatics Approach. Genomics Inform. 2018;16(3):44–51. doi: 10.5808/GI.2018.16.3.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.UlQamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ungell A.-.L. In Vitro Absorption Studies and Their Relevance to Absorption from the GI Tract. Drug Dev Ind Pharm. 1997;23(9):879–892. doi: 10.3109/03639049709148694. [DOI] [Google Scholar]
- 63.Voshavar C. Protease Inhibitors for the Treatment of HIV/AIDS: recent Advances and Future Challenges. Curr Top Med Chem. 2019;19(18):1571–1598. doi: 10.2174/1568026619666190619115243. [DOI] [PubMed] [Google Scholar]
- 64.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y.…Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Węglarz-Tomczak E., Tomczak J.M., Talma M., Brul S. 2020. Ebselen As a Highly Active Inhibitor of PLproCoV2. bioRxiv. [DOI] [Google Scholar]
- 67.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. ActapharmaceuticaSinica. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Current pharmaceutical design. 2006;12(35):4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay. Antimicrob. Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamoto M., Kiso M., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Takeda M., Kinoshita N., Ohmagari N., Gohda J., Semba K., Matsuda Z., Kawaguchi Y., Kawaoka Y., Inoue J.I. The Anticoagulant Nafamostat Potently Inhibits SARS-CoV-2 S Protein-Mediated Fusion in a Cell Fusion Assay System and Viral Infection In Vitro in a Cell-Type-Dependent Manner. Viruses. 2020;12(6):E629. doi: 10.3390/v12060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan L., Chen Z., Song S., et al. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J Biol Chem. 2015;290(5):3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zafar R., Zubair M., Ali S., Shahid K., Waseem W., Naureen H.…Sadiq A. Zinc metal carboxylates as Potential anti-Alzheimer's candidate: in-vitro anticholinesterase, antioxidant and molecular docking studies. Journal of Biomolecular Structure and Dynamics. 2020:1–15. doi: 10.1080/07391102.2020.1724569. [DOI] [PubMed] [Google Scholar]
- 77.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao Y.H., Abraham M.H., Le J., Hersey A., Luscombe C.N., Beck G., Sherborne B., Cooper I. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002;19(10):1446–1457. doi: 10.1023/a:1020444330011. [DOI] [PubMed] [Google Scholar]
- 79.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S.…Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]