Abstract
In late 2019, a novel Coronavirus emerged in China. Perceiving the modulating factors of cross-species virus transmission is critical to elucidate the nature of virus emergence. Using bioinformatics tools, we analyzed the mapping of the SARS-CoV-2 genome, modeling of protein structure, and analyze the evolutionary origin of SARS-CoV-2, as well as potential recombination events. Phylogenetic tree analysis shows that SARS-CoV-2 has the closest evolutionary relationship with Bat-SL-CoV-2 (RaTG13) at the scale of the complete virus genome, and less similarity to Pangolin-CoV. However, the Receptor Binding Domain (RBD) of SARS-CoV-2 is almost identical to Pangolin-CoV at the aa level, suggesting that spillover transmission probably occurred directly from pangolins, but not bats. Further recombination analysis revealed the pathway for spillover transmission from Bat-SL-CoV-2 and Pangolin-CoV. Here, we provide evidence for recombination event between Bat-SL-CoV-2 and Pangolin-CoV that resulted in the emergence of SARS-CoV-2. Nevertheless, the role of mutations should be noted as another influencing factor in the continuing evolution and resurgence of novel SARS-CoV-2 variants.
Abbreviations: CoV, coronavirus; SARS, severe acute respiratory syndrome; Bat-SL-CoV-2, Bat SARS like Coronavirus 2; RBD, receptor binding domain; MERS, Middle East Respiratory Syndrome; COVID-19, coronavirus disease 2019; hACE2, human angiotensin-converting enzyme 2
Keywords: SARS-CoV-2, Virulence, Phylogenetics, Recombination, Mutation, Pandemic
1. Introduction
Emerging infectious diseases, including Coronaviruses (CoVs), are often the result of a cross-species transmission of viruses from animals to humans. This can happen through several genetic mechanisms including recombination and mutations that give a virus new features and enable the virus to bind and enter into a new host cell with greater efficiency, avoid the immune system, and modifying its virulence (Longdon et al., 2014). An example of this cross species transmission is Severe Acute Respiratory Syndrome (SARS) (China-2002) and Middle East Respiratory Syndrome (MERS) (Saudi Arabia-2012). In the both scenarios, the CoV originated from bats, and amplified in a mammalian host, e.g. Himalayan palm civet (SARS) and dromedary camels (Chan et al., 2020a).
In December 2019, a novel coronavirus was reported in Wuhan, China. The virus was named SARS-CoV-2 and causes Coronavirus disease 2019 (COVID-19) with severe pneumonia symptoms (Guan et al., 2020). In the beginning of 2020, COVID-19 became a national disaster in China, and soon after swept the world. On 11 March 2020, the World Health Organization characterized COVID-19 as a pandemic (Millán-Oñate et al., 2020). In the late 2020, a novel variant of SARS-CoV-2 known as Variant Of Concern (VOC-202012-01) was emerged in England, and very soon became the dominant circulating viral variant in numerous countries around the world (Simmonds, 2020).
The pathogenicity of CoVs differs from each other (Cunha and Opal, 2014). Human CoVs mainly cause mild symptoms (e.g. 229E and OC43), but two Betacoronaviruses cause severe illness and fatality; (i) SARS-CoV infected 8000 humans (774 deaths) (Donnelly et al., 2003), (ii) MERS infected 2494 humans (858 deaths) (Lee et al., 2016). Like all other unknown viruses, very little is known about SARS-CoV-2.
The CoVs have several non-structural proteins (nsps), and structural proteins (sps). The most studied structural protein is the Spike protein (S-protein), which plays a key role in the pathogenicity of SARS-CoV. It is known that SARS-CoV uses human angiotensin-converting enzyme 2 (hACE2) as one of the main receptors (Lai et al., 2020). Mutations in the S-protein of animal SARS-CoVs might change protein structure, especially in RBD, and might make the virus compatible for binding to human cell receptors (Uddin et al., 2020).
Predicting the pathogenicity or transmissibility of a novel agent requires a detailed understanding of multiple factors. A possible tool for forecasting the function of a protein is 3D structure modeling of a protein from its sequences and then uses features of that structure to infer binding domains or other functional characteristics.
Previous studies have demonstrated that bats are the ultimate reservoir hosts for a number of CoVs, including ancestors of SARS-CoV and MERS-CoV (Hu et al., 2015). However, the evolutionary pathways of SARS-CoV-2 elusive. Genetic recombination in different viruses within a species is an important evolutionary process that cause genetic diversity and give new features to an ancestral virus. Studying recombination patterns provide evidence of origin, ecological links, and host range of a recombinant virus (Lam et al., 2013). The occurrence of homologous recombination events has been already detected in coronaviruses (Su et al., 2016). For example, it has been shown that recombination of CoVs in camels caused the emergence of dominant MERS lineage subsequently led to human outbreaks (Zhang et al., 2016).
The objective of a predictive oversight system is forecasting with a high degree of certainty the pathogenic potential of genomes of newly identified pathogens in comparison to the most closely related agent. Using sequence based prediction; we aim to study the mapping of the SARS-CoV-2 genome, modeling of protein architecture, determination of virulence drivers at the genomic level, as well as SARS-CoV-2 evolutionary origin, potential recombination events.
2. Material and methods
2.1. Dataset and gene mapping
A total number of 85 coronavirus sequences covering four genera, in which 35 were full-length sequences of SARS-CoV-2 from clades; L, V, S, G, GR (GISAID nomenclature) available in GenBank was retrieved. Genomic organization analysis, multiple alignments, open reading frames identification, and amino acid (aa) similarity and substitution were performed using Geneious version 11.0.5 (Biomatters Ltd., Auckland, New Zealand) (Shahhosseini et al., 2021).
2.2. 3D modeling of S-protein
The homology model for SARS-CoV-2 S-protein and hACE2 were constructed using Oligomeric modeling, which combined interface conservation, structural clustering, and other template features to provide a quaternary structure quality estimate (QSQE) in the SWISS-MODEL workspace. The secondary structure was extracted from the 3D structure using DSSP program (Joosten et al., 2010; Shahhosseini et al., 2020).
2.3. Phylogenetic tree construction
In order to construct the phylogenetic tree, a Tamura-Nei genetic distances model and Neighbor-Joining (NJ) method were selected with sorted topologies and 70% threshold. Analyses of the sequences were conducted using Geneious software version 11.0.5. To further assess the precise evolutionary origin of SARS-CoV-2, a split network was made by EqualAngel method using the SplitsTree 4.14.8 software (Huson and Bryant, 2006; Shahhosseini et al., 2016).
2.4. Recombination analysis
To analyze the hypothesis whether the SARS-CoV-2 is a result of recombination in parental strains, we used Recombination Detection Program version 4 (RDP4) to detect and characterize recombination events in full-length sequences of SARS-CoV-2 (MN908947). The similarity plots and boot-scanning analyses were performed with Kimura 2-parameter, and window and moving step sizes of 200 and 20 nucleotides respectively. The phylogenetic tree of major and minor parents for recombinant sequence was inferred using UPGMA method in RDP4 software. The number of bootstrap replicates was adjusted on 100 and cutoff percentage on 70% (Chinikar et al., 2016; Lole et al., 1999).
3. Result
3.1. In-depth genome annotation of SARS-CoV-2
The SARS-CoV-2 genome, which encompasses 29,903 nucleotides with 12 open reading frames (ORFs), is compared with genetically relevant Betacoronaviruses in Table 1 . In the SARS-CoV-2 genome, the first two long ORF encompasses two ORFs: ORF1a and ORF1b (21,288 bp) together encodes 7096 aa, which produce non-structural proteins (nsps). The other ORFs encode structural proteins including the S gene (3822 bp) encodes 1273 aa for spike protein (S), the ORF3a gene (825 bp) encodes 275 aa for ORF3a protein, the E gene (225 bp) encodes 75 aa for envelope protein (E), the M gene (666 bp) encodes 222 aa for matrix protein (M), the ORF6 gene (183 bp) encodes 61 aa for ORF6 protein, the ORF7a gene (363 bp) encodes 121 aa for ORF7a protein, the ORF7b gene (129 bp) encodes 43 aa for ORF7b protein, the ORF8 gene (363 bp) encodes 121 aa for ORF8 protein, the N gene (1257 bp) encodes 419 aa for nucleocapsid protein (N), and the ORF10 gene (114 bp) encodes 38 aa for ORF10 protein (Table 1).
Table 1.
Genomic organization, nucleotide and amino acid lengths of each ORF of the SARS-CoV-2 compared with genetically relevant Betacoronaviruses.
| Virus species (nt length)/Gene Bank Acc. No. | Genomic organization/ Open reading frames | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 (29,903 bp)/ MN908947 |
ORF1ab 266–21,555 |
S 21,563–25,384 |
ORF3 25,393–26,220 |
E 26,245–26,472 |
M 26,523–27,191 |
ORF6 27,202–27,387 |
ORF7a 27,394–27,759 |
ORF7b 27,756–27,887 |
ORF8 27,894–28,259 |
N 28,274–29,533 |
ORF10 29,558–29,674 |
|||||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| 21,288 | 7096 | 3822 | 1273 | 825 | 275 | 225 | 75 | 666 | 222 | 183 | 61 | 363 | 121 | 129 | 43 | 363 | 121 | 1257 | 419 | 114 | 38 | |
| Pangolin-CoV (27,213 bp)/ MT084071a |
ORF1ab 266–21,555 |
S 21,635–25,471 |
ORF3 25,479–26,310 |
E 26,334–26,562 |
M 24,001–24,669 |
ORF6 27,291–27,477 |
ORF7a 27,483–27,849 |
ORF7b 27,845–27,977 |
ORF8 27,983–28,348 |
N 28,363–29,628 |
||||||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | aa | ||
| 21,288 | 7096 | 3837 | 1279 | 831 | 277 | 225 | 75 | 666 | 222 | 183 | 61 | 364 | 121 | 129 | 43 | 363 | 121 | 1257 | 419 | |||
| Bat-SL-CoV-2 (29,855 bp)/ MN996532 |
ORF1ab 251–21,537 |
S 21,545–25,354 |
ORF3 25,363–26,190 |
E 26,215–26,441 |
M 26,493–27,158 |
ORF6 27,169–27,354 |
ORF7a 27,360–27,725 |
ORF7b 27,722–27,853 |
ORF8 27,860–28,225 |
N 28,240–29,499 |
||||||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| 21,285 | 7095 | 3807 | 1269 | 825 | 275 | 225 | 75 | 663 | 221 | 183 | 61 | 363 | 121 | 129 | 43 | 363 | 121 | 1257 | 419 | |||
|
Bat-SL -CoV (29,732 bp)/ MG772934 |
ORF1ab 265–13,389 |
S 21,483–25,220 |
ORF3 25,229–26,056 |
E 26,081–26,308 |
M 26,359–27,027 |
ORF7 27,038–27,223 |
ORF8 27,230–27,595 |
ORF10b 27,730–28,095 |
N 28,110–29,369 |
ORF13 28,120–28,413 |
ORF14 28,570–28,782 |
|||||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| 21,210 | 7070 | 3735 | 1245 | 825 | 275 | 225 | 75 | 666 | 222 | 183 | 61 | 363 | 121 | 363 | 121 | 1257 | 419 | 291 | 97 | 210 | 70 | |
| SARS-CoV (29,727 bp)/ AY291315 |
ORF1ab 265–21,485 |
S 21,492–25,259 |
ORF3 25,260–26,092 |
ORF3b 25,689–26,153 |
E 26,117–26,347 |
M 26,398–27,063 |
ORF6 27,074–27,265 |
ORF7 27,273–27,641 |
ORF8 27,779–28,147 |
N 28,120–29,388 |
ORF13 28,130–28,426 |
|||||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| 21,219 | 7073 | 3765 | 1255 | 822 | 274 | 462 | 154 | 228 | 76 | 663 | 221 | 189 | 63 | 366 | 122 | 366 | 122 | 1266 | 422 | 294 | 98 | |
The submitted sequence of Pangolin-CoV (MT084071) to GenBank contains unread regions. In order to fill the gaps, a consensus sequence was generated from Pangolin-CoV metagenome, NCBI BioProject: PRJNA573298.
3.2. Identity of structural proteins
Pairwise identity analysis for ORF1a, ORF1b, S, ORF3, E, M, ORF6, ORF7a, ORF7b, ORF8, N and ORF10 are shown in Table 2 . Given structural proteins, the data showed that SARS-CoV-2 S-protein has highest similarity to Bat-SL-CoV-2 and Pangolin-CoV with 97.4% and 89.7%, respectively. The E-protein of SARS-CoV-2 is 100% identical with Bat-SL-CoV-2, Pangolin-CoV, and Bat-SL-CoV, and is 94.7% identical to SARS-CoV. The identity of SARS-CoV-2 M-protein is the same with Bat-SL-CoV-2, Pangolin-CoV and Bat-SL-CoV with 98.6%, but 89.1% with SARS-CoV. The N-protein of SARS-CoV-2 showed highest identity with Bat-SL-CoV-2 (99%) followed by Pangolin-CoV (97%), Bat-SL-CoV (94.2%), SARS-CoV (90%) (Table 2).
Table 2.
Nucleotide and amino acid sequence identities among the genes of SARS-CoV-2 compared with Pangolin-CoV, Bat-SL-CoV-2, Bat-SL-CoV and SARS-CoV.
| Virus strains | Gene regions (identity %) |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF1a |
ORF1b |
S |
ORF3 |
E |
M |
ORF6 |
ORF7a |
ORF7b |
ORF8 |
N |
ORF10 |
|||||||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| Bat-SL-CoV-2/MN996532 | 96 | 98 | 97.3 | 99.3 | 92.8 | 97.4 | 96.2 | 97.7 | 99.5 | 100 | 95.5 | 98.6 | 98.3 | 100 | 95.5 | 97 | 99.2 | 97.2 | 96.9 | 95 | 96.9 | 99 | 99.1 | 97.3 |
| Pangolin-CoV/MT084071a | 89.3 | 95.8 | 89.6 | 99.2 | 83 | 89.7 | 92.3 | 97 | 99.1 | 100 | 93.3 | 98.6 | 95.5 | 96.4 | 93.3 | 97.4 | 91.4 | 95.2 | 92.1 | 94.2 | 94.6 | 97 | 99.1 | 97.3 |
| Bat-SL-CoV/MG772934 | 90.9 | 95.7 | 86.1 | 95.5 | 74.9 | 79.8 | 88.8 | 91.6 | 98.6 | 100 | 93.4 | 98.6 | 95 | 93.2 | 89.6 | 90 | 95.3 | 92.7 | 88.5 | 94.2 | 91.1 | 94.2 | 100 | 100 |
| SARS-CoV/AY304488 | 76 | 81 | 86.2 | 95.6 | 72.5 | 75.9 | 75.5 | 96.9 | 93.5 | 94.7 | 84.9 | 89.1 | 76.5 | 68.2 | 84.1 | 89 | 86.1 | 83.8 | 40.9 | 16.1 | 88.0 | 90 | 93.1 | 82.4 |
The submitted sequence of Pangolin-CoV (MT084071) to GenBank contains unread regions. In order to fill the gaps, a consensus sequence was generated from Pangolin-CoV metagenome, NCBI BioProject: PRJNA573298.
3.3. Molecular structure of Spike protein
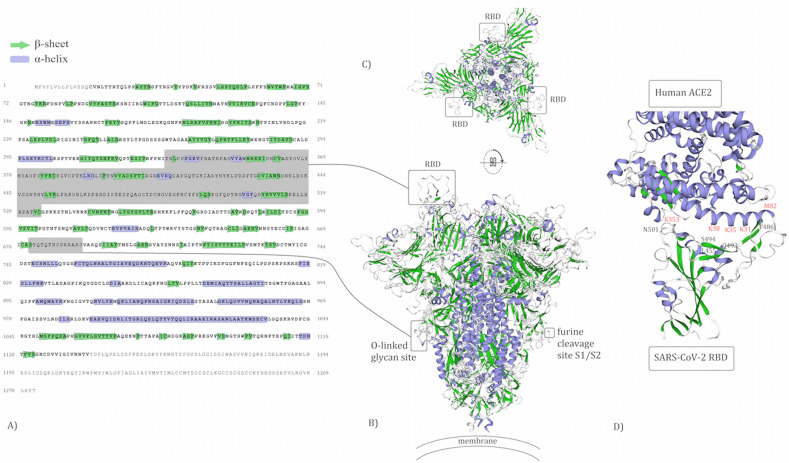
The S-protein of SARS-CoV-2 is composed of three identical units of polypeptide (homotrimer structure). To be able to anchor to host cells, the Spike has a RBD that contains 193 amino acids. Among 193 aa, 51 aa are in contact residues, in which the location of five aa known as key aa directly bind with hACE2, as shown in Fig. 1D. Also, the Spike protein possesses glycosylation sites, which has a key role in the penetration of the outer bilayer leaflet of the host cell membrane to initiate cell entry (Fig. 1).
Fig. 1.
The aa sequence of the spike is colored based on protein structure (A). The predicted 3D structure of the SARS-CoV-2 spike on the surface of membrane protein. The surface of the spike model is colored based on protein structures (B). The representative shows spike from anchor side to host cells (C). The enlarged RBD with key residues binding to hACE2 (D).
3.4. Genetic determinants of virulence in S-proteins
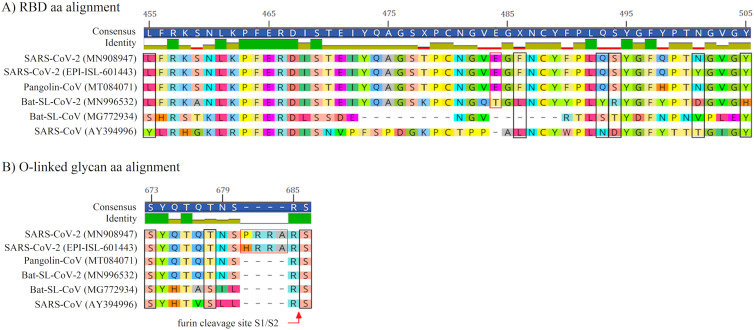
Current sequence prognostication methods might predict the function of an individual protein based on its deduced aa sequences. In order to predict pathogenicity from sequences, we compared 51 aa within the contact residue of RBD in two SARS-CoV-2 variants and SARS-CoV, which showed evolutionary substitutions in 27 aa, in which five aa substitutions (Y455L, L486F, N493Q, D494S, and T501N/Y) occurred in key residues of RBD that are responsible for anchoring with hACE2. Notably, these five aa substitutions in key RBD residues of SARS-CoV-2 are completely identical to the same aa position in Pangolin-CoV (except at position 501 in the new variant of SARS-CoV-2 ‘VOC-202012-01’), but not Bat-SL-CoV-2. In addition, SARS-CoV-2 and Pangolin-CoV has a Glutamic acid insertion at position 484, which is unique and not seen in others compared aa sequences. Among key residues of RBD, only position 505 is strictly conserved in all compared aa sequences, except in Bat-SL-CoV-2 (Fig. 2A).
Fig. 2.
The amino acids alignment in RBD (A) and O-linked glycan (B) of SARS-CoV-2 (MN908947), SARS-CoV-2 VOC-202012-01 (GISAID ID: EPI-ISL-601443), Pangolin-CoV (MT084071), Bat-SL-CoV-2 (MN996532), Bat-SL-CoV (MG772934), and SARS-CoV (AY394996) are shown respectively. Key residues in RBD are marked with black boxes (A). Three residues in O-linked glycan are marked with black boxes. The amino acid insertion in SARS-CoV-2 is marked with red box. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In O-linked glycan residues, three aa are critical for O-linked glycosylation attachment in SARS-CoV. Similarly in SARS-CoV-2, at two positions (S673 and S686), no aa substation was observed among aa alignments, but at position 678, Threonine in SARS-CoV2, Bat-SL-CoV-2 and Pangolin-CoV is substituted with Serine in Bat-SL-CoV and SARS-CoV. Notably, SARS-CoV-2 has four extra aa (P/HRRA) in O-linked residues, which is a unique feature in compared to other Sarbecoviruses. In addition, we observed that the acquisition of the polybasic cleavage site (RRAR) is restricted to SARS-CoV-2 at the junction (between aa position 685 and 686) of two subunits of the spike (Fig. 2B).
3.5. Phylogenetic tree construction based on full genes
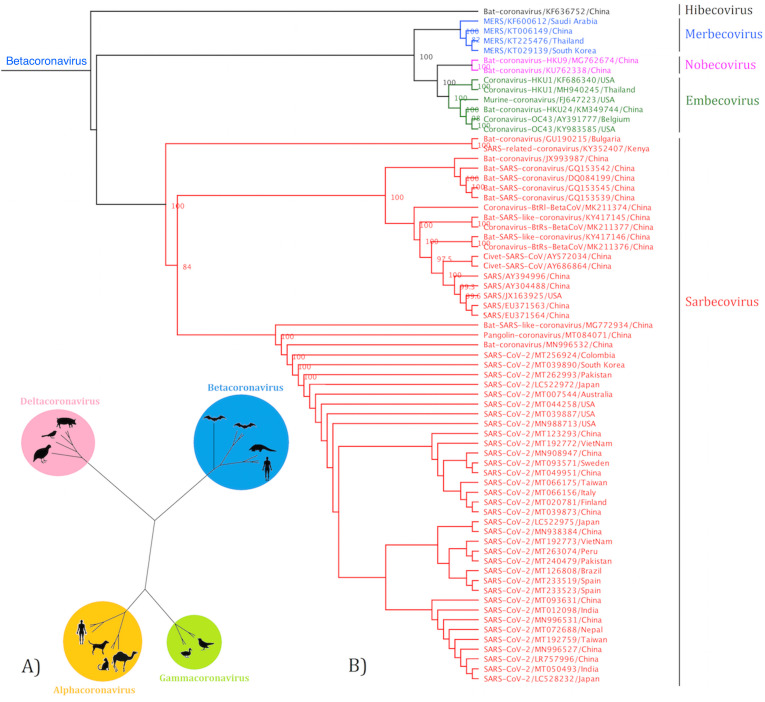
The construction of cladogram revealed the grouping of coronaviruses into four distinct groups (Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus) (Fig. 3A). Further analysis showed that Betacoronaviruses have five distinct clades of viruses classified as Sarbecovirus, Merbecovirus, Embecovirus, Nobecovirus, and Hibecovirus, in which the SARS-CoV-2 fell within Sarbecovirus (Fig. 3B).
Fig. 3.
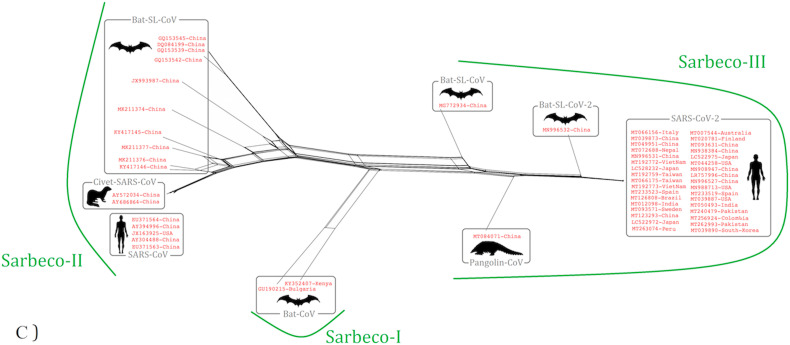
The cladogram of coronaviruses (A) and the phylogenetic NJ tree of Betacoronaviruses (B) were constructed from aligned sequences using tools implemented within the Geneious software. The bootstrap values and number of bootstrap replications were greater than 70% and 1000, respectively. The neighbor-net network based on the full-length sequence of Sarbecoviruses was constructed using the Split tree software (C).
Split tree networking illustrated that the Sarbecovirus clade split into three subclades: Sarbeco-I isolates from bat species in Kenya and Bulgaria, Sarbeco-II divided to three host-specific groups including SARS-CoV isolates from humans, CoV isolates from civets and SARS-like CoV isolates from bats, whilst Sarbeco-III consist (i) SARS-CoV-2 isolated from humans, (ii) SARS-like CoVs isolated from bats, and Pangolin-CoV. All SARS-CoV-2 isolates tightly clustered together with more than 99% similarity (Fig. 3C).
3.6. Evidence for recombination
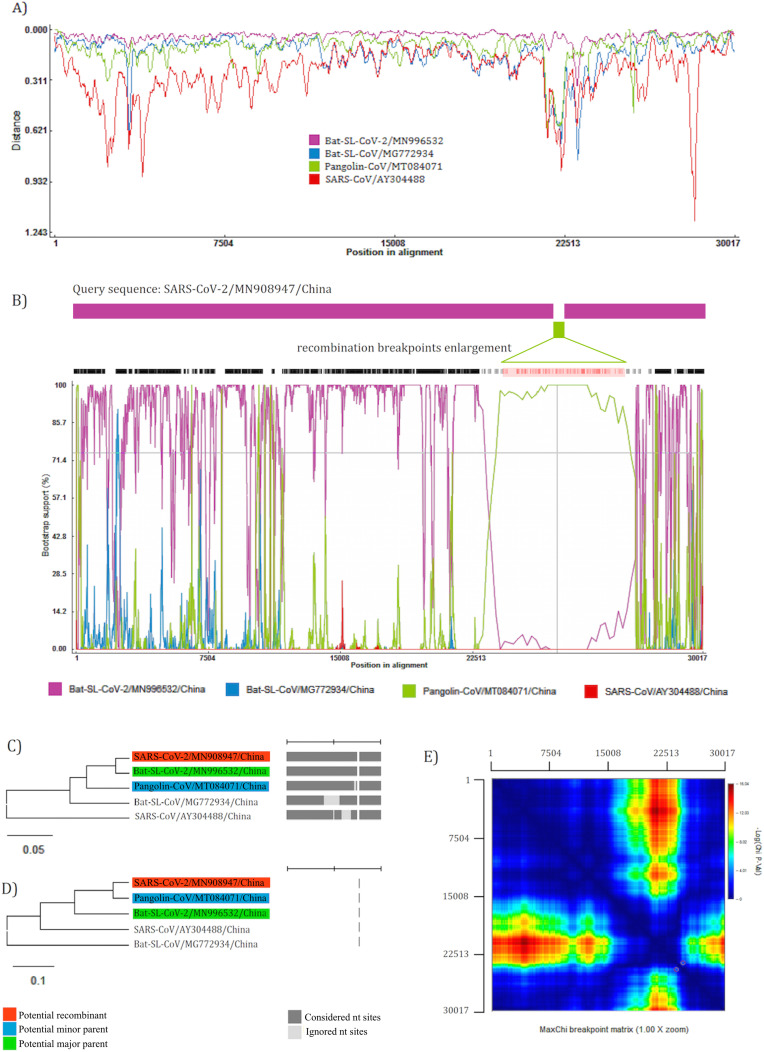
Similarity plotting analysis between SARS-CoV-2 and reference sequences showed recombination event in SARS-CoV-2 (Fig. 4 -A). The recombinant sequence (SARS-CoV-2) is shown as query, while sequences from the major (Bat-SL-CoV-2) and minor parents (Pangolin-CoV) are in purple and green, respectively. Further analysis with Bootscanning indicates recombination breakpoints beginning at nt 22,849 (99% CI: nt 22,711- nt 22,889) and ending at nt 23,093 (99% CI: nt 23,044- nt 23,145). Bat-SL-CoV-2 is the major parent with 98.4% similarity and Pangolin-CoV is the minor parent with 88.2% similarity for recombinant SARS-CoV-2. MaxChi matrix shows the statistically optimal positions of breakpoint pairs. The phylogenetic tree of alternative parents showed close similarity of SARS-CoV-2 (recombinant virus) with Bat-SL-CoV-2 (major parent) at the scale of the complete virus genome, except recombination point that shows highest evolutionary similarity to Pangolin-CoV (minor parent) (Fig. 4 B, C, D, E).
Fig. 4.
Recombination analysis by RDP4 of SARS-CoV-2 as query sequence and close strains in Sarbecovirus are shown with Similarity plotting on a window size of 200 nucleotides and moving in steps of 20 nt along the alignment (A). Schematic sequence displays of recombinant region in green. The Bootscan analysis shows the most probable positions of breakpoint pairs, which are enlarged (B). Phylogenetically plausible alternative parents trees inferred in different genomic regions of recombinant sequence based on the major parent (C) and minor parent (D). MaxChi breakpoint matrix. Colors represent chi-squared values for breakpoint; dark red peaks indicate the most probable positions of breakpoint pair (E). The number of bootstrap replicates was adjusted on 100 and cutoff percentage on 70%. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
SARS-CoV-2 is a novel coronavirus and pathogenicity mechanisms are not yet clear. Given the evolution of pathogenicity in viruses, we used computational genomics to solve the controversy on origin and pathogenicity of SARS-CoV-2. Comparison of aa at S-protein showed that SARS-CoV-2 has the highest similarity to Pangolin-CoV than SARS-CoV. In addition, the critical residues binding to hACE2 are 100% identical in SARS-CoV-2 and Pangolin-CoV. This finding suggests that natural selection might happen in pangolin and gave the virus new features for recognition of hACE2. Our sequence-homology data is in accordance with a previous report discussing the low similarity between SARS-CoV S-protein and SARS-CoV-2 S-protein (Yuan et al., 2020). However, the conserved regions in RBD in two SARS-CoV and SARS-CoV-2 is sufficient to expect that SARS-CoV-2 can bind to hACE2 (Ralph et al., 2020).
The 3D modeling of S-protein showed the location of O-linked domain and RBD and its correspondent binding site at hACE2, implying the key role of S-protein in binding to host cells. Importantly, the overall homology of S-protein in SARS-CoV-2 and SARS-CoV are similar, suggesting use of the same cell entry receptor (Zhou et al., 2020). It is shown that at position 493, Gln is compatible for binding to salt bridge between Lys31 and Glu35 (hot spot 31) in hACE2. Also, the Leu455 and Phe486 make a favorable interaction with hot spot 31. The Asn501and Ser494 has favorable interactions with salt bridge between Lys353 and Asp38 (hot spot 353), but interactions are weaker than binding between the counterpart aa in SARS-CoV with hACE2 (Wan et al., 2020). In addition, a Glu484 insertion mutation changes the length of the loop, consequently altering aa positions in the RBD. The new variant of SARS-CoV-2 from England (VOC-202012-01) is defined with two deletions at positions 69–70 and 144. It is demonstrated that deletions (H69 and V70) in the unit S1 of S-protein is likely induce a conformational alteration comparing to other SARS-CoV-2 variants (Xie et al., 2021). The changes in homology in combination with other mutations in the RBD is highly likely enhancing the transmissibility of SARS-CoV-2 (Lauring and Hodcroft, 2021).
Both SARS-CoV-2 and SARS-CoV has 23 conserved aa at contact residue for binding to hACE2, supporting the idea that hACE2 is also the receptor of SARS-CoV-2. In addition, an in-vitro study to explore a treatment for COVID-19 showed that the production of human recombinant ACE2 can inhibit SARS-CoV-2 infection in early stages, supporting the idea that hACE2 is a receptor for SARS-CoV-2 (Monteil et al., 2020). To explore why SARS-CoV-2 has rapid human-to-human transmission, the binding affinity of SARS-CoV-2 RBD to hACE2 should be studied. The aa substitution in key residues of RBD of SARS-CoV-2 in comparison to SARS-CoV created stronger binding to hACE2, which is around 10–20 fold higher (Wrapp et al., 2020). Hence, the high binding affinity between SARS-CoV-2 RBD and hACE2 makes stronger interaction, which increase chances of virus attachment to host cell for virus entry, and promoting high infectivity. Moreover, it is assumed that decreased virulence of SARS-CoV-2 than SARS or MERS results in a longer time for tissue damage and onset of infection symptoms. It is estimated that the median incubation time for COVID-19 is 5 days (between 1 day to 14 days or longer) (Li et al., 2020), but apparently healthy patients are capable of shedding virus (Lauer et al., 2020). The combination of a long incubation time combined with virus shedding is another factor that contributed in the rapid spread of SARS-CoV-2 causing 2019–21 pandemic. In addition, among 13 non-synonymous mutations identified in SARS-CoV-2 VOC-202012-01, seven mutations (N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) are in the S-gene (Rambaut et al., 2020). Of note, mutation N501Y, where amino acid Asparagine has been replaced with Tyrosine, in the key contact residue of RBD is demonstrated to increase infectivity and virulence in a mouse model (Gu et al., 2020).
The O-linked glycan domain in cleavage site of SARS-CoV-2 is identical to the SARS-CoV at positions Ser673 and Ser686, but position 678 in SARS-CoV-2 is Threonine while the same position in SARS-CoV is Serine. Linkage between Serine or Threonine in S-protein with a glycan has a critical role in viral entry into host cell (Bagdonaite and Wandall, 2018). In addition, we detected a unique aa (Proline) insertion (PRRA) at position 681 in O-linked glycan domain. However, new variant of SARS-CoV-2 (VOC-202012-01) has one non-synonymous mutation (P681H) at this domain. Hence, any changes at this site may play a role in altering pathogenicity capability of SARS-CoV-2. Moreover, it has been shown that introduction of aa into a furin cleavage site at the junction of S1 and S2 in SARS-CoV can mediate membrane fusion and virus infectivity (Andersen et al., 2020; Belouzard et al., 2009). Furthermore, the insertion of a polybasic cleavage site (RRAR) at the junction between unit S1 and S2 of spike can facilitate furin recognition and consequently allowing an efficient cleavage by host cell proteases, which play an important role in viral fusion into host cell membrane (Nao et al., 2017), which might be responsible for the efficient spread of SARS-CoV–2 (Coutard et al., 2020). Similarly, it has been shown that acquisition of a polybasic cleavage site is one of the necessary elements in altering a low-pathogenic avian influenza to a highly pathogenic strain (Stech et al., 2009).
The phylogenetic tree shows that SARS-CoV-2 has the closest evolutionary relationship with Bat-SL-CoV-2 at the scale of the complete virus genome, and less similarity to Pangolin-CoV. In this context, it can be assumed that the recombination between Bat- and Pangolin-CoVs happened deep in an ancestry of the bat lineage. However, the RBD of Bat-SL-CoV-2 showed low aa identity (78%) to SARS-CoV-2, suggesting that SARS-CoV-2 possibly was not transmitted directly from bats to humans. In contrast, the RBD of Pangolin-CoV is almost identical to SARS-CoV-2 (98%) at the aa level, proposing that mutations in Pangolin gave new features to CoV which enable the virus to bind to hACE2. This finding suggests the Pangolin as the intermediate host for SARS-CoV-2 (Li et al., 2020b). In addition, the role of adaptation pressures should be considered as an influencing factor that may have shaped RBD and mutations induced by host system apparently shaped SARS-CoV-2 genome including the critical amino acids within the RBD (Matyášek and Kovařík, 2020; Simmonds, 2020).
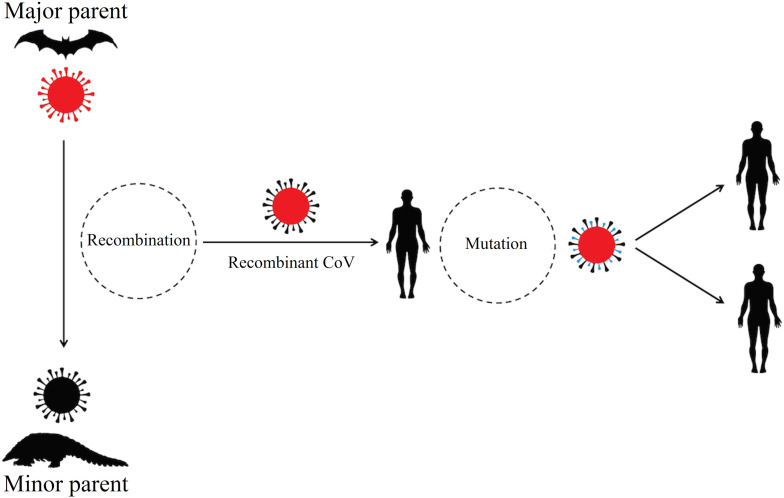
To clarify this contrast that SARS-CoV-2 has highest similarity to Bat-SL-CoV-2 when comparing the complete virus genome, but bats cannot be the direct origin of SARS-CoV-2, we investigated recombination events. Together Bootscanning curves and comparison of phylogenetic tree topology of major and minor parental viruses revealed recombination event between Bat-SL-CoV-2 and Pangolin-CoV with position of breakpoint pairs at RBD. Due to recombination event, the key residues of RBD from Pangolin-CoV integrated to Bat-SL-CoV-2, then zoonotic transmission occurred by a recombinant virus with capability to bind to hACE2. Also, we propose several mutations in the recombinant CoV leading to the emergence of different variants of SARS-CoV-2 in the human community with new features for rapid transmission and severity (Fig. 5 ).
Fig. 5.
The proposed cycle for the emergence and resurgence of SARS-CoV-2 variants causing 2019–21 pandemic.
The recombination and phylogenetic analysis support the reliability of our model for SARS-CoV-2 origin and spillover transmission and further suggests that the previously proposed origins of SARS-CoV-2 which identify each bat or pangolin solely may not be accurate. For example, we observed several bias in the previous studies looking for the origin of SARS-CoV-2. The first bias was observed in those articles that identify bat as animal origin for SARS-CoV-2. Such studies argued that SARS-CoV-2 is very likely to be originated from Bat-SL-CoV-2 (RaTG13). However, they ignored Pangolin-CoV in the genomic analysis (Stech et al., 2009). Such approaches lead to this claim to exclude the role of recombination in emergence of SARS-CoV-2 (Stech et al., 2009).
The second bias was observed in articles that compared SARS-CoV-2 with Bat-SL-CoVs (MG772933 and MG772934), and ignored more genetically relevant Bat-SL-CoV-2 and Pangolin-CoV in analysis (Stech et al., 2009). Hence, they predicted that Bat-SL-CoV is the most probable source of SARS-CoV-2. Concurrently, authors could detect the occurrence of recombination events in the S-gene, but due to bias in dataset gathering they suggested recombination is more likely occurring in bat coronaviruses and is not the reason for emergence of SARS-CoV-2 (Chan et al., 2020a; Lv et al., 2020; Zhou et al., 2020). Whereas, another study suggested bat might be the original host of SARS-CoV-2 and an unknown animal might present an intermediate host (Paraskevis et al., 2020). Similarly, another study suggesting SARS-CoV-2 might be a recombinant virus between the Bat-SL-CoV and an origin-unknown CoV (Chan et al., 2020b; Malik et al., 2020; Xiong et al., 2020).
Since the initial stage of COVID-19 pandemic in early 2020, several SARS-CoV-2 variants have emerged in human host due to mutations (Volz et al., 2020). In the SARS-CoV-2 VOC, mutations in the S-gene, particularly N501Y in RBD, were postulated as a causative factor that gave the virus new features for rapid transmission. Consequently, the S-gene and its RBD domain are rapidly and frequently evolving in human hosts (Simmonds, 2020). Collectively, although recombination played a role in the emergence of SARS-CoV-2, convergence has a critical role in the recurrent emergence of novel SARS-CoV-2 variants in different phases of COVID-19 pandemic (Korber et al., 2020).
5. Conclusions
Here, we provide evidence for recombination event between Bat-SL-CoV-2 and Pangolin-CoV that resulted in the emergence of SARS-CoV-2. Our findings are in accordance with previous findings that viral recombination between different CoVs within animal populations may lead to the emergence of novel zoonotic CoVs that are lethal to humans (Wu et al., 2020).
CRediT authorship contribution statement
NSH, SC designed the study. NSH analyzed data. NSH, SC, GW, GK wrote the manuscript. NSH, GW revised the manuscript.
Declaration of competing interest
No conflict of interests to declare.
Acknowledgements
Not applicable.
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28(7):443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences. Proc. Natl. Acad. Sci. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Kok K.-H., To, K.K.-W, Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinikar S., Shah-Hosseini N., Bouzari S., Shokrgozar M.A., Mostafavi E., Jalali T., Khakifirouz S., Groschup M.H., Niedrig M. Assessment of recombination in the S-segment genome of Crimean-Congo hemorrhagic fever virus in Iran. J. Arthropod. Borne Dis. 2016;10(1):12–23. [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C.B., Opal S.M. Middle East respiratory syndrome (MERS) a new zoonotic viral pneumonia. Virulence. 2014;5(6):650–654. doi: 10.4161/viru.32077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C.A., Ghani A.C., Leung G.M., Hedley A.J., Fraser C., Riley S., Abu-Raddad L.J., Ho L.-M., Thach T.-Q., Chau P. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361(9371):1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Ge X., Wang L.-F., Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;12(1):221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Joosten R.P., Te Beek T.A., Krieger E., Hekkelman M.L., Hooft R.W., Schneider R., Sander C., Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2010;39(suppl_1):D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;105924 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Chong Y.L., Shi M., Hon C.-C., Li J., Martin D.P., Tang J.W.-T., Mok C.-K., Shih S.-R., Yip C.-W. Systematic phylogenetic analysis of influenza a virus reveals many novel mosaic genome segments. Infect. Genet. Evol. 2013;18:367–378. doi: 10.1016/j.meegid.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020 doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? Journal of the American Medical Association. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Lee J., Chowell G., Jung E. A dynamic compartmental model for the Middle East respiratory syndrome outbreak in the Republic of Korea: a retrospective analysis on control interventions and superspreading events. J. Theor. Biol. 2016;408:118–126. doi: 10.1016/j.jtbi.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Giorgi E.E., Marichannegowda M.H., Foley B., Xiao C., Kong X.-P., Chen Y., Gnanakaran S., Korber B., Gao F. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 2020 doi: 10.1126/sciadv.abb9153. (eabb9153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B., Brockhurst M.A., Russell C.A., Welch J.J., Jiggins F.M. The evolution and genetics of virus host shifts. PLoS Pathog. 2014;10(11) doi: 10.1371/journal.ppat.1004395.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Li G., Chen J., Liang X., Li Y. Comparative genomic analysis revealed specific mutation pattern between human coronavirus SARS-CoV-2 and bat-SARSr-CoV RaTG13. BioRxiv. 2020 doi: 10.1101/2020.02.27.969006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M., Tiwari R., Chaicumpa W. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40(1):68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyášek R., Kovařík A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased towards C> U transitions, indicating rapid evolution in their hosts. Genes. 2020;11(7):761. doi: 10.21203/rs.3.rs-21377/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán-Oñate J., Rodriguez-Morales A.J., Camacho-Moreno G., Mendoza-Ramírez H., Rodríguez-Sabogal I.A., Álvarez-Moreno C. A new emerging zoonotic virus of concern: the 2019 novel coronavirus (COVID-19) Infectio. 2020;24(3) doi: 10.22354/in.v24i3.848. [DOI] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., del Pozo C.H., Prosper F. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nao N., Yamagishi J., Miyamoto H., Igarashi M., Manzoor R., Ohnuma A., Tsuda Y., Furuyama W., Shigeno A., Kajihara M. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. MBio. 2017;8(1) doi: 10.1128/mBio.02298-16. e02298-02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph R., Lew J., Zeng T., Francis M., Xue B., Roux M., Ostadgavahi A.T., Rubino S., Dawe N.J., Al-Ahdal M.N. 2019-nCoV (Wuhan virus), a novel coronavirus: human-to-human transmission, travel-related cases, and vaccine readiness. J Infect Dev Ctries. 2020;14(01):3–17. doi: 10.3855/jidc.12425. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Genom. Epidemiol. 2020:1–5. [Google Scholar]
- Shahhosseini N., Chinikar S., Nowotny N., Fooks A.R., Schmidt-Chanasit J. Genetic analysis of imported dengue virus strains by Iranian travelers. Asian Pac. J. Trop. Dis. 2016;6(11):850–853. doi: 10.1016/S2222-1808(16)61144-1. [DOI] [Google Scholar]
- Shahhosseini N., Moosa-Kazemi S.H., Sedaghat M.M., Wong G., Chinikar S., Nowotny N., Hajivand Z., Mokhayeri H., Kayedi M.H. Autochthonous transmission of West Nile virus by a new vector in Iran, vector-host interaction modeling, and virulence gene determinants. Viruses. 2020;12:144. doi: 10.3390/v12121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahhosseini N., Frederick C., Letourneau-Montminy M-P., Marie-Odile B-B., Kobinger G.P., Wong G. Computational genomics of Torque teno sus virus and Porcine circovirus in swine samples from Canada. Research in Veterinary Science. 2021;134:171–180. doi: 10.1016/j.rvsc.2020.12.010. [DOI] [PubMed] [Google Scholar]
- Simmonds P. Rampant C→ U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short-and long-term evolutionary trajectories. Msphere. 2020;5(3) doi: 10.1101/2020.05.01.072330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stech O., Veits J., Weber S., Deckers D., Schröer D., Vahlenkamp T.W., Breithaupt A., Teifke J., Mettenleiter T.C., Stech J. Acquisition of a polybasic hemagglutinin cleavage site by a low-pathogenic avian influenza virus is not sufficient for immediate transformation into a highly pathogenic strain. J. Virol. 2009;83(11):5864–5868. doi: 10.1128/JVI.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M., Mustafa F., Rizvi T.A., Loney T., Suwaidi H.A., Al-Marzouqi A.H.H., Eldin A.K., Alsabeeha N., Adrian T.E., Stefanini C. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12(5):526. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2020 doi: 10.1101/2020.07.31.20166082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1101/2020.02.11.944462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021 doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Xiong C., Jiang L., Chen Y., Jiang Q. Evolution and variation of 2019-novel coronavirus. Biorxiv. 2020 doi: 10.1101/2020.01.30.926477. [DOI] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T., Lv H., Mok C.K., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Shen L., Gu X. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci. Rep. 2016;6(1):1–10. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]