Abstract
Background
The immunological factors involved in protection against the disease caused by SARS-CoV-2 are insufficiently defined and understood. However, previous knowledge pertaining to the related SARS virus and other human coronaviruses may prove useful. Population-based serosurveys measuring anti-SARS-CoV-2 antibodies may provide a pattern for estimating infection degrees and observing the development of the epidemic. In this study, we aimed to investigate the persistence of antibody against the SARS-CoV-2 in recovered patients in Al Madinah region of Saudi Arabia.
Materials and methods
A total of 150 recovered COVID-19 patients participated in this study. All the patients tested positive for the presence of SARS-CoV-2 RNA, using qualitative RT-PCR. An ELISA was used to measure anti-Spike (S) IgG antibodies in serum samples and screen for their persistence at various time points post-infection.
Results
The patients were categorized as asymptomatic (27.3%), mild (28%) and moderate (44.7%) according to the disease severity. Amongst them, 35.3% were females (n = 53) and 64.7% were males (n = 97). Significant anti-S IgG antibody levels were observed among the different groups, with the patients in moderate group exhibiting the highest levels followed by the mild group; while the lowest levels were detected among the asymptomatic. There was a significant positive correlation between the patients’ age and anti-S IgG antibody concentrations (Pearson r = 0.45; p < 0.001).
Conclusion
Our findings provide a solid evidence to support the use of an anti-S IgG ELISA as a diagnostic tool to indicate SARS-CoV-2 infection. IgG seropositivity was sustained in recovered patients up to a hundred days' post-infection, the latest time point for antibody measurement in our study. Ours is the first report in Saudi Arabia to investigate the durability of humoral immune response in recovered COVID-19 patients.
Keywords: SARS-CoV-2, Antibody, Convalescents, Persistence, Humoral immunity
1. Introduction
In late 2019, cases of pneumonia from an unfamiliar source were first described in China’s Hubei province (Chen et al., 2020). Later, a worldwide occurrence of severe acute respiratory syndrome (SARS) was confirmed to be caused by a new coronavirus, thereafter named SARS-CoV-2, the causative agent of the illness known as COVID-19 (Cordes and Heim, 2020). The World Health Organization (WHO) designated this novel illness a “Public Health Emergency of International Concern” on January 30th, 2020; shortly thereafter, it was declared a “Global Pandemic” (Cucinotta and Vanelli, 2020).
The immunological parameters mediating protective immunity against SARS-CoV-2 are not yet fully understood. Nonetheless, experience gained from treating SARS-related coronavirus infections may give an insight into probable treatment modalities and vaccines for COVID-19 (Barbareschi et al., 2020).
Antibodies play a vital role in neutralizing viruses and defending the host against re-infection. The spike (S) protein of SARS coronavirus 2 (SARS-CoV-2) attaches to the angiotensin-converting enzyme 2 (ACE2) on host cells to initiate viral entry. Therefore, ACE2 receptor is an important target for scientists seeking therapeutic candidates to prevent the virus from causing infection (Chan et al., 2020).
Understanding antibodies’ permanency within the bloodstream is vital for determining seroprevalence in people. Moreover, this would inform choices regarding obtaining plasma from recovered individuals for therapeutic purpose, in addition to designing vaccines against SARS-CoV-2. Recently, it has been reported in a longitudinal analysis of immune memory subsequent to COVID-19 that the infection elicited memory lymphocytes that exhibited functional hallmarks of antiviral immunity. It presented characteristics linked with potent antiviral functions; (IgG) antibodies, neutralizing plasma, in addition to memory B and T cells that persisted for a minimum of three months (Rodda et al., 2021). In another study, anti-spike/nucleocapsid IgG-antibody titers were detected that stabilized at comparatively higher levels over the six months surveillance period. (Wu et al., 2020). Moreover, they showed positive rates for binding as well as higher neutralizing SARS-CoV-2-specific antibody percentages.
Till date, there are no reports on the durability of antibody response in convalescent patients following COVID-19 in Saudi Arabia. In this study, we aimed to investigate the persistence of IgG antibody over a period of time following recovery from COVID-19 infection in Al Madinah region of Saudi Arabia. This would help to ascertain whether there is a detectable level of antibodies against SARS-CoV-2 that would consequently induce protection upon subsequent exposure to the virus.
2. Materials and methods
2.1. Patients samples and controls
A total of 150 recovered COVID-19 patients were enrolled in the study. The participants were asked to fill a consent form prior to conducting the study. Archived serum samples (n = 22) collected one-year before the pandemic were used as negative control as well as to check the specificity of the ELISA. Medical records were obtained and data collected for non-hospitalized patients and asymptomatic contacts with laboratory-confirmed COVID-19, as reported to the Ministry of Health (MOH) in the Madinah region between March 25th, 2020, and July 1st, 2020. The study was approved by the Research Ethics Committee of the General Directorate of Health Affairs in Al Madinah (IRB number: 446).
2.2. RT-PCR test for SARS-CoV-2
Nasopharyngeal (NP) swabs were collected with sterile synthetic tip flocked swabs (BD, USA). The swabs were left in place for a few seconds to absorb the secretions. Swabs were placed immediately into sterile tubes containing 2–3 ml of viral transport media. A full MOH protocol is available online: https://www.moh.gov.sa/en/CCC/healthp/regulations/Documents/Novel%20Corona%20Virus%20Infection%20Guidelines.pdf (accessed on 12–01-2021).
Extraction of nucleic acid was done using Roche Magna Pure LC (RNA Viral Isolation Kit, USA). Briefly, 200 µL of each sample was added to 96-well MagNA pure LC plate. Reaction reagents were then loaded and checked before running the samples according to the manufacturer’s instructions for nucleic acid extraction in the specimen. A full protocol is available online: https://www.altona-diagnostics.com/files/public/Content%20Homepage/-%2002%20RealStar/INS%20-%20RUO%20-%20EN/RealStar%20SARS-CoV-2%20RT-PCR%20Kit%201.0_WEB_RUO_EN-S02.pdf (accessed on 12–01-2021).
2.3. The one step RT-PCR real time amplification
After the viral RNA extraction was performed, the RT-PCR reaction mixture was prepared by using Altona diagnostics detection kit RealStar® SARS-Cov-2 RT-PCR kit 1.0 targeting specific RNA SARS-Cov-2 Envelope gene (E gene) and SARS-Cov-2 spike protein (S gene) based on one step RT-PCR real time method. A full protocol is available online: (https://www.altonadiagnostics.com/files/public/Content%20Homepage/-%2002%20RealStar/INS%20-%20RUO%20-%20EN/RealStar%20SARS-CoV-2%20RT-PCR%20Kit%201.0_WEB_RUO_EN-S02.pdf).(accessed on 12–01-2021).
All the tubes containing the one step RT-PCR mixture was sealed and carefully transferred to Real Time LC 480 (Roche, USA) and subjected to the following cycling program: a single cycle of 55 0C for 20 min, single cycle of 95 0C for 2 min, 45 cycles of 95 0C for 15 s, 45 cycles of 55 0C for 45 s and 45 cycles of 72 0C for 15 s. This procedure was carried out in all the steps to prevent any contamination. A full protocol is available online: https://diagnostics.roche.com/global/en/products/params/cobas-sars-cov-2-test.html (accessed on 12–01-2021).
2.4. Enzyme-linked immunosorbent assay (ELISA)
The assay was performed using the SARS-CoV-2 Spike (S) polyhistidine tag at the C-terminus of recombinant protein for plating and subsequent detection of anti-S IgG antibodies. (W. H. Mahallawi, 2020). In brief, 96-well ELISA plates (Costar; Corning, Corning, NY, USA) were coated with 100 µL/well of SARS-CoV-2 recombinant S protein (Sino Biological, Beijing, China) at 2 µg/mL in phosphate-buffered saline (PBS; pH 7.2), concealed with an adhesive seal and left overnight at 4 °C. Following five times washing of the plates with washing buffer (PBS containing 0.05% Tween-20; Sigma-Aldrich, St. Louis, MO, USA), the plates were blocked with 150 µL/well blocking buffer (PBS containing 0.05% heat-inactivated fetal bovine serum (FBS); Sigma-Aldrich) for one hour at room temperature. Serum samples (diluted 1:100 using blocking buffer) were added at 100 µL/well. The plates were incubated for 30 min at room temperature and washed again five times. A specific alkaline phosphatase-conjugated goat anti-human IgG secondary antibody (1:1,000 in blocking buffer, Sigma-Aldrich) was then added at 100 µL/well, and the plates were left at room temperature for 30 min before being washed another five times. All the washing steps were performed using an automated microplate washer (Elx50; Bio Tek, Winooski, VT, USA). Lastly, 100 µL/well of p-nitrophenyl phosphate substrate (p-NPP, Sigma-Aldrich), was added. The plates were retained in the dark, till yellow color established. To end the reaction after 30 min, 100 µL of the stopping solution (1.2 N sodium hydroxide, Reagecon, UK), was added to all the wells. The optical density (OD) at 405 nm was measured using a microplate reader (ELX800; BioTek).
2.5. Statistical analysis
All the calculations and statistical analysis were performed using GraphPad Prism statistical software (version 9, USA). Differences between participants’ categories optical densities (in unit of ODs) were shown as median. Data were expressed for IgG antibody concentrations in ODs as median. Differences between groups were analyzed using Student’s t test. Correlation was observed using Pearson r. A p < 0.05 was considered significant for all tests.
3. Results
3.1. Categorization of patients samples
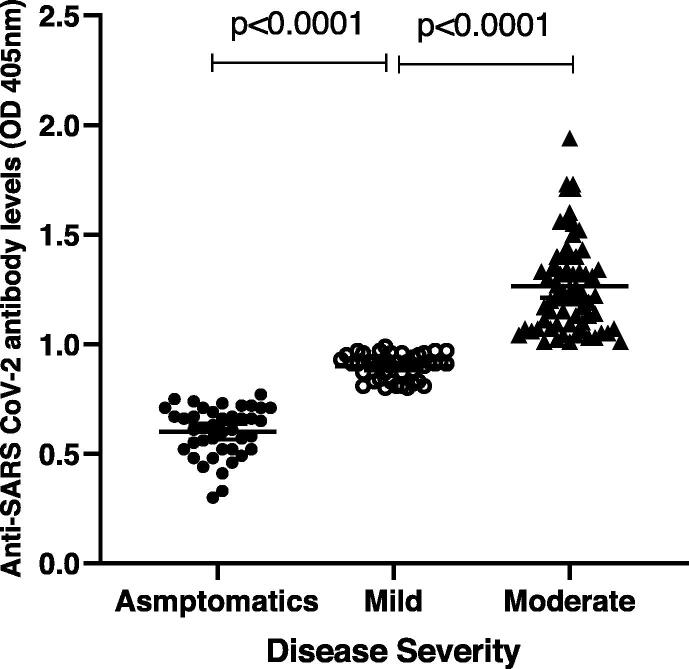
The participants were divided into three categories according to disease severity: 27.3% were asymptomatic (OD median = 0.62, n = 41), 28% were mild (OD median = 0.91, n = 42), and 44.7% were moderate (OD median = 1.22, n = 67). Interestingly, we found significant differences among the groups (p < 0.0001) (Fig. 1). Patients categorizations were selected based on MOH guidelines (https://www.moh.gov.sa/Ministry/MediaCenter/Publications/Documents/MOH therapeutic-protocol-for-COVID-19.pdf).
Fig. 1.
Participants’ categories according to the disease severity: 27.3% asymptomatic (OD median = 0.62; n = 41), 28% mild (OD median = 0.91; n = 42), and 44.7% moderate (OD median = 1.22; n = 67; p < 0.0001).
3.2. Measurement of anti-S IgG antibody of SARS-CoV-2 using ELISA
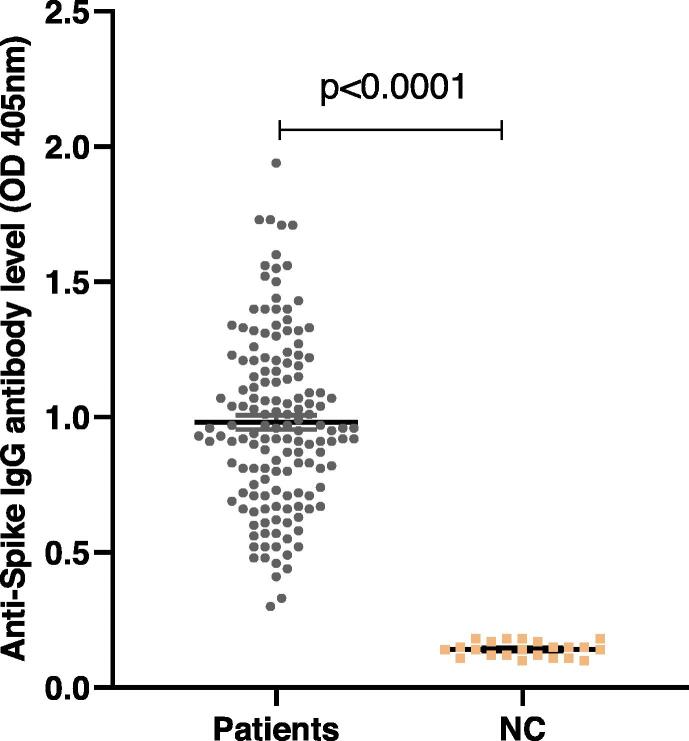
The study population consisted of 150 recovered COVID-19 patients (mean age 30.79 ± 8.95 years; range 16–62); 35.3% were female (n = 53) and 64.7% were male (n = 97). ELISA was used to measure concentrations (in units of optical density) of specific anti-S IgG antibodies against SARS-CoV-2 among the participants. Fig. 2 shows significant (p < 0.0001) anti-SARS-CoV-2 IgG antibody concentrations (in units of ODs) in patients’ serum samples (OD values 0.32–1.94; mean 0.98 ± 0.32; n = 150) when compared to negative controls’ stored serum samples obtained from one year prior to the pandemic (OD values 0.10–0.18; mean ± SD = 0.14 ± 0.02; n = 22). All the patients' samples showed detectable antibody against the S protein of the virus.
Fig. 2.
Significant anti-SARS-CoV-2 IgG antibody concentrations (in units of OD) (p < 0.0001; n = 150) in patients’ serum samples were observed when compared to stored serum samples from one-year pre-pandemic, used as negative controls, NC (OD median of patients’ sera = 0.96; n = 150; OD median of NC = 0.14; n = 22).
3.3. Correlation between antibody concentrations and patients’ age and gender
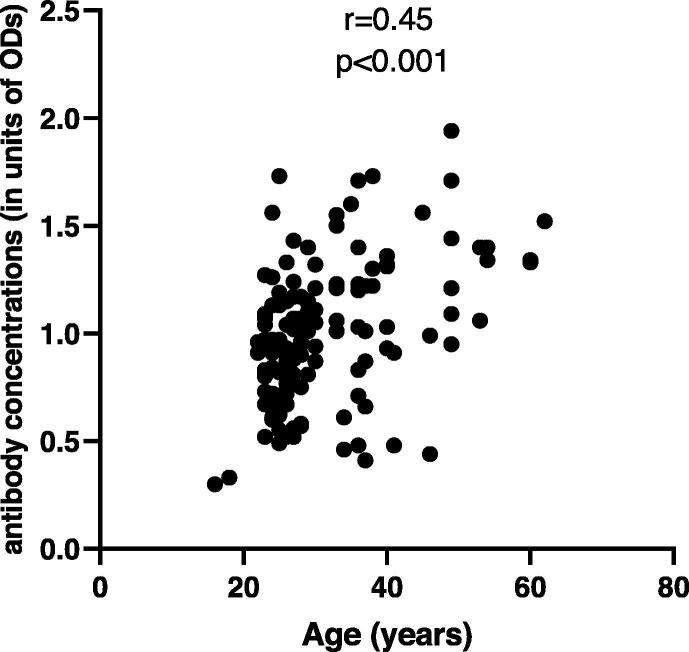
We found a significant positive correlation between patients’ age and anti-SARS-CoV-2 IgG antibody concentrations (Pearson r = 0.45; p < 0.001 (95% CI = 0.32 to 0.57); n = 150) (Fig. 3). This indicates that the humoral immune response to the virus was greater in older participants than in younger participants; and could be considered as an age-dependent immunity.
Fig. 3.
Positive correlation between age and anti-SARS-CoV-2-S IgG antibody concentrations (in units of ODs) (Pearson r = 0.45; p < 0.001; n = 150).
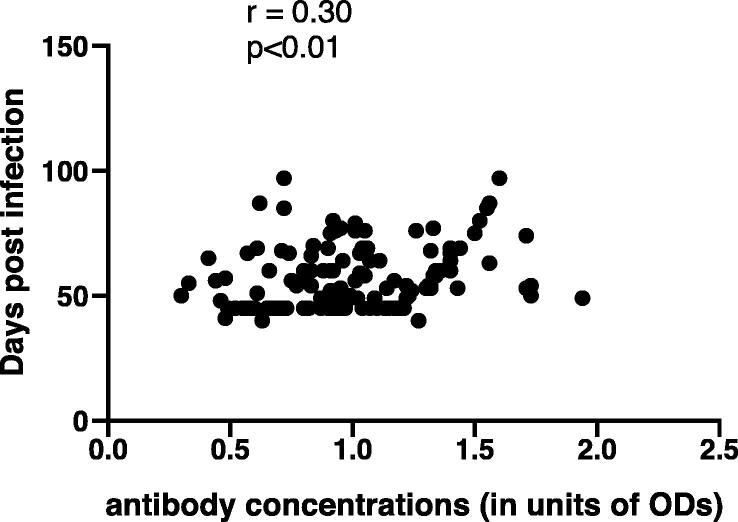
We also stratified the quantities of specific anti-S IgG antibody against the time post-infection of all the participants. Interestingly, we found a significant positive correlation between anti-S IgG levels and time post onset of illness (Pearson r = 0.30; p < 0.01n = 150), Fig. 4. No correlation was observed between antibody concentrations and the patients’ gender (data not shown).
Fig. 4.
Positive correlation was observed when antibody concentrations (in units of ODs) were stratified against the days post infection. (Pearson r = 0.3, p < 0.01, n = 150).
4. Discussion
The current occurrence and fast blowout of SARS-CoV-2 pose an abundant warning of a pandemic outbreak of COVID-19 globally. Diagnostic techniques are the frontline tactic for detecting this infection. Presently, SARS-CoV-2 can be identified by RT-PCR, nonetheless insufficient acquisition of reagents as well as machines, the necessity for advancement of laboratory services with limiting biosafety levels in addition to technical complexity, and lastly the high degree of false-negative consequences initiated mostly by an unstandardized acquisition of respiratory samplings have led to low efficacy of in-time recognition of the disease.
There is scarcity of data on the serologic analysis using antibodies targeting SARS-CoV-2. Currently, the antibody responses against SARS-CoV-2 remain poorly defined and the clinical value of serological testing is unclear. It has been shown that the seroconversion, as well as the antibody intensities, develop quickly throughout the first two weeks and that the increasing seropositive amount extends up to 50% on the 11th day and up to 100% on the 39th day (Zhao et al., 2020) Serological analysis is supportive for the identification of prospective patients with copies of the virus undetectable by RT–PCR and for the recognition of asymptomatic infections (Long, Liu, et al., 2020)
We screened the antibody response to SARS-CoV-2 in 150 recovered COVID-19 patients. All the participants recruited in this study were confirmed by RT-PCR to have infection by the virus. Clinical records were extracted from the Regional Laboratory to obtain the date of the positive sample. All samples were obtained at different time points since the onset of the infection.
Several studies have reported that IgG antibodies to SARS-CoV-2 Spike were detectable in the blood of more than 90% of patients within 10–11 days of symptom onset. (Amanat et al., 2020, Long et al., 2020a) It remains unclear whether these IgG antibodies specific for SARS-CoV-2 antigens persist (Iyer et al., 2020) or decline(Long, Tang, et al., 2020) over time.
In our study, the time interval varied from 16 to a hundred days' post-infection (mean ± SD = 54 ± 13.99). Our results confirm that the IgG antibody response to SARS-CoV-2 is detected and persists for more than three months. This finding is the first in Saudi Arabia regarding the durability of the humoral immune response among recovered COVID-19 patients; it aligns with a study that similarly confirmed the persistence of the IgG response to the SARS-CoV-2 Spike protein. (Iyer et al., 2020). Having supported this finding, we also observed the tendency of the humoral immune response to maintain continuous antibody production over a long period. Furthermore, we studied the correlation between antibody concentrations and time elapsed since the patients were tested positive for the virus. A positive correlation was observed when we associated the anti-SARS-CoV-2 IgG antibody with the days-post infection.
It is promising that the specific antiviral IgG antibody generated by natural infection could still be detected at such a long period of time following recovery. Almost all of the participants showed a detectable and measurable antibody response. This is due to the healthy and functional humoral immune response, which would be on the front lines combatting future viral attacks. Therefore, this result provides hope regarding the development of vaccines against the virus. A vaccine is urgently needed to prevent COVID-19 and the attending complications and deaths occurring due to the community transmission of SARS-CoV-2 (Heaton, 2020).
To lessen the effects of the virus on public health, the economy and people, a vaccine is urgently necessary. It is crucial to ensure the immunogenicity and safety of any vaccine before it is licensed. The most recent vaccine candidate produced by Oxford University, called ChAdOx1 nCoV-19, is now in the final phase of clinical trials and has yielded optimistic results (Folegatti et al., 2020) An mRNA vaccine named mRNA-1273, is agreed to be used by the U.S. FDA for emergency use authorization after the final phase assessment produced by Vaccine Research Center (VRC) at the National Institutes of Health and Moderna. (https://www.nature.com/articles/d41587-020–00022-y) accessed on 01–12-2020.
Serological analysis is useful for the identification of suspected patients with undetectable viral RNA by RT–PCR results and for the recognition of asymptomatic infections. Interestingly, our results indicate that specific anti-Spike IgG antibody levels increase with disease severity. We found that the IgG titers in the moderate category were greater than those among the asymptomatic patients. This result is consistent with a study that assessed the seroconversion of SARS-CoV-2 patients (Long, Liu, et al., 2020) Moreover, we also observed much higher antibody concentrations among hospitalized patients than among recovered patients (unpublished data). This correlates with disease severity, as shown in a study conducted by Jiang, who found that greater quantities of anti-Spike IgG were associated with worse clinical readouts and more elderly patients (Jiang et al., 2020)
It is commonly acknowledged that antibodies are vital for neutralizing virus particles and conferring protection to a host exposed to viral re-infection. Anti-Spike antibody capacities are essential, as this protein harbors the receptor binding domain (RBD). The RBD eases the entrance of SARS-CoV-2 to human cells by attaching to the host receptor, ACE2 (Letko et al., 2020). Thus, neutralizing antibodies have been revealed to block the RBD. Whether the specific IgG antibody targeting SARS-CoV-2 perseveres or declines remains uncertain; this gap in our understanding persists. At odds with our findings, waning humoral immunity in patients with SARS-CoV-2 infection has also been reported. Therefore, this raises the possibility that naturally developed humoral immunity counter to SARS-CoV-2 might not be long-lasting (Choe et al., 2021, Shioda et al., 2020).
More studies are needed to investigate whether this long-term IgG antibody is functional or not. Moreover, the persistence of this immunity has not been fully studied due to the recent onset of the pandemic.
We must specify that, while our assay recognizes individuals who have been exposed to SARS-CoV-2 infection, it does not offer evidence about whether recovered patients are protected from re-infection. It is important to investigate the best immunological links of protection against SARS-CoV-2 in humans (Iyer et al., 2020)
In numerous human trial observations of common cold coronavirus infections, the existence of pre-existing neutralizing antibodies has been linked to protection, alongside the development of symptomatic infection and lower viral shedding (Huang et al., 2020). A recent study using various assays for detection of antibodies reactive with the SARS-CoV-2 S glycoprotein suggests that pre-existing humoral immunity to the novel coronavirus is present in uninfected and unexposed humans. Moreover, this study indicates that antibodies were primarily of the IgG class and targeted the S2 subunit (Ng et al., 2020)
Interestingly, we found that antibody concentrations did not decline over the time. A positive correlation was observed when we associated the anti-SARS-CoV-2 IgG antibody with the days post infection. Our result is in an agreement with a recent study that showed the same trend when they compared the kinetics of IgG and IgM against the virus, using two different assays (Liu et al., 2020)
A recent study showed protection from reinfection by the novel SARS-CoV-2 in a recovered patient who was in close contact with his family and had been confirmed to be positive for the virus (W. Mahallawi, 2020). Additionally, when rhesus macaques were vaccinated against SARS-CoV-2, they were protected from re-infection when challenged. They also developed neutralizing antibodies targeting the S protein and were found to be carrying a strong associate of protective immunity (Chandrashekar et al., 2020)
Our study has a few limitations. Firstly, we did not have sufficient number of samples at prolonged time periods. Secondly, because of the narrow obtainability of the biosafety level 3 (BSL3) laboratory, we could not assure whether the detected antibodies of serum samples were functional or not by performing virus neutralization assays.
In conclusion, our data indicated sustained humoral immunity in recovered patients who suffered from symptomatic COVID-19, suggestive of durable immunity. Hence our findings support the feasibility of development of vaccines against SARS CoV-2 with induction of protective immunity.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Waleed Mahallawi, Email: wmahallawi@gmail.com.
Mohammad Alzahrani, Email: mohammada.z@iu.edu.sa.
Ziab Alahmadey, Email: zal-ahmadey@moh.gov.sa.
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020 doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbareschi M., Facchetti F., Fraggetta F., Sapino A. What are the priorities of pathologists' activities during COVID-19 emergency? Pathologica. 2020 doi: 10.32074/1591-951X-15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020 doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Barouch D.H. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020 doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe P.G., Kang C.K., Suh H.J., Jung J., Song K.H., Bang J.H., Oh M.D. Waning Antibody Responses in Asymptomatic and Symptomatic SARS-CoV-2 Infection. Emerg. Infect. Dis. 2021;27(1) doi: 10.3201/eid2701.203515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes A.K., Heim A. Rapid random access detection of the novel SARS-coronavirus-2 (SARS-CoV-2, previously 2019-nCoV) using an open access protocol for the Panther Fusion. J. Clin. Virol. 2020;125 doi: 10.1016/j.jcv.2020.104305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Oxford C.V.T.G. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton P.M. The Covid-19 Vaccine-Development Multiverse. N Engl. J. Med. 2020 doi: 10.1056/NEJMe2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. T., Garcia-Carreras, B., Hitchings, M. D. T., Yang, B., Katzelnick, L. C., Rattigan, S. M., . . . Cummings, D. A. T. (2020). A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. doi:10.1101/2020.04.14.20065771 [DOI] [PMC free article] [PubMed]
- Iyer, A. S., Jones, F. K., Nodoushani, A., Kelly, M., Becker, M., Slater, D., . . . Charles, R. C. (2020). Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. doi:10.1101/2020.07.18.20155374
- Jiang H.W., Li Y., Zhang H.N., Wang W., Yang X., Qi H., Tao S.C. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11(1):3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Zheng S. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Mahallawi W. Case Report: A Recovered SARS CoV-2 Patient Protected From Reinfection. Front. Med. 2020;7(649) doi: 10.3389/fmed.2020.564264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahallawi W.H. A serological assay to detect human SARS-CoV-2 antibodies. J. Taibah Univ. Med. Sciences. 2020 doi: 10.1016/j.jtumed.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K. W., Faulkner, N., Cornish, G. H., Rosa, A., Harvey, R., Hussain, S., . . . Kassiotis, G. (2020). Pre-existing and <em>de novo</em> humoral immunity to SARS-CoV-2 in humans. bioRxiv, 2020.2005.2014.095414. doi:10.1101/2020.05.14.095414
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Pepper M. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell. 2021;184(1):169–183.e117. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda K., Lau M.S., Kraay A.N., Nelson K.N., Siegler A.J., Sullivan P.S., Lopman B.A. Estimating the cumulative incidence of SARS-CoV-2 infection and the infection fatality ratio in light of waning antibodies. medRxiv. 2020 doi: 10.1101/2020.11.13.20231266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Liang, B., Chen, C., Wang, H., Fang, Y., Shen, S., . . . Zheng, X. (2020). SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. medRxiv, 2020.2007.2021.20159178. doi:10.1101/2020.07.21.20159178 [DOI] [PMC free article] [PubMed]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]