Abstract
Background and Objectives:
Intestinal pathotypes of Escherichia coli belong to the companion animals may poses potential risk to public health following zoonotic transmission. Therefore, this study was proposed to determine the virulence genes associated to diarrheagenic E. coli strains isolated from healthy pet dogs and their owners in the southeast of Iran, Kerman province.
Materials and Methods:
Totally 168 E. coli isolates were collected from 49 healthy household dogs and their owners. Seventy isolates were obtained from non-pet owners as control group. Presence or absence of the virulence genes including eae, stx1, stx2, st1, lt1, ipaH, cnf1 and cnf2 were screened by conventional polymerase chain reaction (PCR) and dissemination pattern of the genes were studied among the various hosts.
Results:
PCR examinations showed that the most frequent virulence gene was ipaH (6.1%) in dogs followed by eae in dog owners (6.1%) and in controls (8.6%). The most frequent pathotypes in dogs, their owners and controls were EIEC (6.1%), EHEC (4.08%) and EPEC (8.5%), respectively. In one of studied houses, both of dog and its owner harbored E. coli strains with same virulence profile (stx1/eae) and pathotype (EHEC).
Conclusion:
These results collectively indicate that healthy household dogs probably are the mild reservoir of potential virulent E. coli strains with possible active transmission to their contact owner. However, even non-pet owners seemed to be a notable source of intestinal pathotypes, especially EPEC, for their environment. Transmission of E. coli pathotypes may occurs by direct contact with the reservoirs or ingestion of contaminated food. These pathotypes are potentially virulent and creates public health hazards. Further studies are needed for better understanding of dissemination mechanisms of E. coli pathotypes among humans and their pets.
Keywords: Escherichia coli, Virulence, Dog, Zoonotic enteropathogens
INTRODUCTION
Zoonotic enteropathogens comprise diverse range of microorganisms that could be transmitted to humans by consumption of meat or dairy products, by direct contact with companion and farm animals (or their feces) or by consumption of food or water contaminated with animal feces. In the United States, researchers estimated that 14% of enteric infections with 7 groups of zoonotic enteropathogens were attributable to direct contact with animals (1). Dogs have a significant situation in public health because they are among the most common household animals and may be the reservoir of many bacterial agents (2). Close contact between household dogs and their owners may involuntarily represent the transmission risk of zoonotic entropathogens for humans.
Escherichia coli strains are routinely isolated from feces of human and warm-blooded animal species such as farmed and companion animals (3). Therefore these hosts may be the potential source of pathogenic E. coli strains which often can express the virulence factors for colonization and invasion. Intra-intestinal pathogenic strains of E. coli are classified into the six main pathotypes including Shiga toxin-producing E. coli (STEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) as described below (4).
Enterotoxigenic E. coli have isolated from human, lambs, calves and dogs, causing profuse diarrhea in neonatal animals and cholera-like disease in humans. ETECs can be colonized through the factors such as F5, F6, F17 and F41 and produce enterotoxins including LTI, LTII, STa and STb. Most of ETECs stains isolated from diarrhoeic dogs produce the STa (5).
Shiga toxin-producing E. coli pathotype have reported from human patients with hemolytic uremic syndrome (HUS) and hemorrhagic colitis (HC), healthy ruminants, canine and wildlife (6). Generally, STECs carry either or both of stx1 and stx2 which are responsible for production of Shiga toxins. Some STEC strains may cause the A/E (attaching and effacing) lesions because of LEE (locus of enterocyte effacement) in their genome, suggesting a subset of STEC pathotype called enterohemorrhagic E. coli (EHEC). The strains which cause A/E lesions without Shiga toxin production are referred to enteropathogenic E. coli (EPEC) pathotype. This pathotype is the most identified bacterial agent of persistent diarrhea in children (7).
Enteroaggregative E. coli (EAEC) adhere to intestinal mucosa using AAF (aggregative adherence fimbriae), creates a thick biofilm and secret enterotoxins and cytotoxins (8). Enteroinvasive E. coli (EIEC) invade to the epithelial cells of colon and spread via cell-to-cell immigration (9). Diffusely adherent E. coli (DAEC) produce F1845, binds to decay-accelerating factor (DAF) on the small intestine enterocytes and interfere with microvillia growth (10). Necrotoxigenic E. coli (NTEC) which produce cytotoxic necrotizing factor (CNF) induce the intestinal and extra-intestinal infections in both humans and animals (11). The most common diseases caused by EAEC, EIEC and DAEC are travelers diarrhea (in adults), shigellosis (in all age groups of human) and persistent watery diarrhea (in children), respectively (12).
Vega-Manriquez et al. (2020) recently reported several E. coli pathotypes (ETEC, STEC and EHEC) from healthy dogs, in contrast with diarrheic animals that showed only EPEC (13). Therefore zoonotic diarrheagenic pathotypes of E. coli usually have no effect on healthy dogs. These findings present the high risk of zoonotic transmission of E. coli pathotypes through environmental contamination by asymptomatic dogs. To the best of our knowledge, several studies have been conducted on the enteric and extra-intestinal E. coli strains of pet dogs and their owners. In order to innovation, in this study we aimed to detect diarrheagenic pathotypes of E. coli in household dogs and their owners in compare with non–pet owner (control) group. Results of this project can be useful for awareness of the epidemiological situation of healthy household dogs as shedders of virulent E. coli strains. These data can help to assign preventive and control strategies targeted at transmission pathways.
MATERIALS AND METHODS
Fecal sampling.
Totally, 168 rectal swabs were obtained from pet dogs (n=49), their owners (n=49) and the persons without pet (n=70) as controls. Informed consent was obtained from pet owners and control group (Ethical code: IR.KMU.REC.1397.03). Information about each dog including their name, address, age, sex, breed, breeding type, health history, medical history and antibiotic consumption were collected (Table 1). All sampled animals and humans were apparently healthy. Over the past month, not all humans have had any infections or antibiotics. The non-pet owners were selected from the students who did not keep the pet and did not deal with the animal at work. There was no similarity in association with age and gender between pet owners and non-pet owners groups and selection was completely random; we just considered the facts that these people did not own pet and the sex and age factors in people was not considered during sampling. The sampling process was performed in 2015 to 2017. Finally, the swabs were placed in Amies medium (Merck, Germany) and referred to veterinary microbiology laboratory within 12 hours.
Table 1.
No. of E. coli isolates from dogs in different gender, age, breed and diet groups.
| Variable | Different groups in each variable | No. of E. coli isolates in each group (%) | 95% confidence interval (CI) |
|---|---|---|---|
| Gender | Female | 33 (67.3%) | 54.2%–80.4% |
| Male | 16 (32.7%) | 19.5%–45.7% | |
| Age (years) | <1 | 10 (20.4%) | 9.1%–31.6% |
| 1–2 | 9 (18.3%) | 7.5%–29.2% | |
| 2–3 | 10 (20.4%) | 9.1%–31.6% | |
| 3–4 | 7 (14.2%) | 4.4%–24% | |
| 4–5 | 8 (16.3%) | 5.9%–26.6% | |
| 5–6 | 1 (2.1%) | 0%–10.8% | |
| 6–7 | 1 (2.1%) | 0%–10.8% | |
| 7–8 | 3 (6.2%) | 0%–12.8% | |
| Breed | Terrier | 15 (30.6%) | 2.02%–12.12% |
| German Shepherd | 13 (26.5%) | 14.1%–38.8% | |
| Shih Tzu | 5 (10.3%) | 0%–34.99% | |
| Boxer | 4 (8.1%) | 0%–15.8% | |
| Poodle | 4 (8.1%) | 0%–15.8% | |
| Dobermann | 2 (4.1%) | 0%–9.6% | |
| Spitz | 2 (4.1%) | 0%–9.6% | |
| Yorkshire Terrier | 2 (4.1%) | 0%–9.6% | |
| Mixed-breed dogs | 2 (4.1%) | 0%–9.6% | |
| Diet | Homemade raw food | 7 (14.3%) | 4.4%–24% |
| Homemade cooked food | 33 (67.3%) | 54.2%–80.4% | |
| Commercial cooked food | 9 (18.4%) | 7.5%–29.2% | |
| Antibiotic therapy | Yes | 4 (8.1%) | 0%–15.8% |
| over the past month | No | 45 (91.9%) | 84.1%–99.5% |
| Total | All E. coli isolates | 49 isolates | 2.24%–9.07% |
E. coli isolation.
Two suspected E. coli colonies were selected for biochemical confirmation and standard isolation procedure was done (14); for confirmation, each suspected colony was examined by IMViC (Indole, Methyl red, Voges-Proskauer and Citrate) and TSI (triple sugar iron agar) tests. Confirmed E. coli isolates (Indole-positive, Methyl red-positive, Voges-Proskauer-negative, Citrate-negative and acid/acid in TSI) were saved for next steps; the confirmed isolates were cultured in Luria-Bertani broth (Merck, Germany) and incubated at 37°C, overnight. Then sterile glycerol was added to achieve 25% concentration, and they were vortexed and immediately saved at –80ºC for next steps (15).
Molecular detection of pathotypes.
One confirmed isolates was randomly selected from each sample for molecular detection of pathotypes. DNA extraction was done by boiling method. In brief, a single colony from overnight and pure culture of each sample on LB agar was selected and suspended in 350 µl distilled and sterile water and boiled for 10 min in 98–100ºC. Then, the bacterial suspensions were centrifuged at 10,000 × g for 5 min, and the supernatants (approximately 200 µl) were used as DNA templates (16, 17). Then, eight virulence genes including eae, stx1, stx2, st1, lt1, ipaH, cnf1 and cnf2 were screened by conventional polymerase chain reaction (PCR) for pathotyping. The primers of eae, stx1 and stx2 were used in a triplex PCR to detect EPECs (eae-positives), STECs (stx1 and/or stx2-positives) and EHECs (positives for eae and one or both of stx1 and stx2 genes). The primers of st1 and lt1 were employed in a duplex PCR to detect ETECs (Table 2) (18, 19). The virulence genes ipaH (EIECs), cnf1 and cnf2 (NTECs) were detected via simplex PCR technique (Table 2) (19, 20). The referernce strains including 28°C (cnf1+), 1404 (cnf2+), ETEC H10407 (st+ and lt+), Sakai (stx1+, stx2+ and eae+) and 85b (ipaH+) were utilized as the positive controls. PCR products were visualized in an electrophoresis process (85V for 45 min) on an 1.5% agarose gel, staining via FluoroVue (SMOBio, Taiwan) and imaging by a GelDoc 1000 (Vilber Lourmat, France).
Table 2.
Primer sequences for screening of pathotypes in PCR
| Pathotype | Gene | Sequence (5′–3′) | PCR condition | Product size (bp) | Reference |
|---|---|---|---|---|---|
| STEC/EPEC/EHEC | stx1 | AGAGCGATGTTACGGTTTG TTGCCCCCAGAGTGGATG |
30 cycles: 94°C (30 s), 50°C (30 s), 72°C (30 s) | 388 | (18) |
| stx2 | TGGGTTTTTCTTCGGTATC GACATTCTGGTTGACTCTCTT |
30 cycles: 94°C (30 s), 50°C (30 s), 72°C (30 s) | 807 | ||
| eae | AGGCTTCGTCACAGTTG CCATCGTCACCAGAGGA |
30 cycles: 94°C (30 s), 50°C (30 s), 72°C (30 s) | 507 | ||
| ETEC | st1 | ATTTTTMTTTCTGTATTRTCTT CACCCGGTACARGCAGGATT |
40 cycles: 95°C (45 s), 50°C (1 min), 72°C (1 min) | 190 | (19) |
| lt1 | GGCGACAGATTATACCGTGC CGGTCTCTATATTCCCTGTT |
40 cycles: 95ºC (45 s), 50°C (1 min), 72°C (1 min) | 450 | ||
| EIEC | ipaH | GTTCCTTGACCGCCTTTCCGATACCGTC GCCGGTCAGCCACCCTCTGAGAGTAC |
40 cycles: 95°C (45 s), 50°C (1 min), 72°C (1 min) | 600 | (19) |
| NTEC | cnf1 | GGGGGAAGTACAGAAGAATTA TTGCCGTCCACTCTCACCAGT |
30 cycles: 94°C (1 min), 54°C (30 s), 72°C (1 min) | 1111 | (20) |
| cnf2 | TATCATACGGCAGGAGGAAGCACC GTCACAATAGACAATAATTTTCCG |
30 cycles: 94°C (1 min), 55°C (1 min), 72°C (1 min) | 1240 |
Statistical analysis.
All data were entered into the Excel (Microsoft 2016) and SPSS (SPSS 24; IBM) programs as binomial information (presence or absence of virulence genes in each isolate) to calculate the prevalence percentages as descriptive statistical analysis of the data. Finally, confidence intervals and significant differences between statistical rates were calculated via Chi-squared test (95% confidence level and P value <0.05).
RESULTS
One hundred and sixty eight E. coli isolates were obtained from same number of various hosts. Of them 18 (5 healthy dogs, 4 dog owners and 9 controls) possessed at least one of the examined virulence genes (Tables 3–5 and Fig. 1). eae was the most common virulence gene which were found in 10 isolates including one healthy dog, three owners and six controls (Table 3). The eae gene was present alone in six isolates. It co-existed with stx1 in three isolates and cnf1 in one isolate (Table 5). The genes ipaH and stx1 were just observed in dogs (n=4) and their owners (n=3) whereas lt1, stx2 and cnf1 genes were found only in three controls (Table 3). ST encoding gene was identified just in one dog isolate. Four isolates possessed multiple-virulence-gene pattern including stx1/eae (n=3) and eae/cnf1 (n=1).
Table 3.
Prevalence of virulence genes among E. coli isolates from healthy household dogs, their owners and controls
| Virulence gene | Healthy dogs | Owners | Controls | Total |
|---|---|---|---|---|
| eae | 1/49 (2.04%) | 3/49 (6.1%) | 6/70 (8.5%) | 10/168 (5.9%) |
| ipaH | 3/49 (6.1%) | 1/49 (2.04%) | - | 4/168 (2.3%) |
| stx1 | 1/49 (2.04%) | 2/49 (4.08%) | - | 3/168 (1.7%) |
| lt1 | - | - | 2/70 (2.8%) | 2/168 (1.1%) |
| stx2 | - | - | 1/70 (1.4%) | 1/168 (0.5%) |
| st1 | 1/49 (2.04%) | - | - | 1/168 (0.5%) |
| cnf1 | - | - | 1/70 (1.4%) | 1/168 (0.5%) |
Table 5.
Comparative assessment of dog-owner pair samples and controls which at least one of them is positive for virulence genes
| Order | Sample code | Host | Virulence gene profile | Pathotype |
|---|---|---|---|---|
| 1 | 15 D | Dog | stx1/eae | EHEC |
| 15 O | Owner | stx1/eae | EHEC | |
| 2 | 16 D | Dog | st1 | ETEC |
| 16 O | Owner | - | - | |
| 3 | 31 D | Dog | - | - |
| 31 O | Owner | eae | EPEC | |
| 4 | 36 D | Dog | ipaH | EIEC |
| 36 O | Owner | - | - | |
| 5 | 37 D | Dog | ipaH | EIEC |
| 37 O | Owner | stx1/eae | EHEC | |
| 6 | 8 D | Dog | - | - |
| 8 O | Owner | ipaH | EIEC | |
| 7 | 14 D | Dog | ipaH | EIEC |
| 14 O | Owner | - | - | |
| 8 | 44 C | Control | eae/cnf1 | EPEC |
| 9 | 10 C | Control | eae | EPEC |
| 10 | 43 C | Control | eae | EPEC |
| 11 | 45 C | Control | eae | EPEC |
| 12 | 63 C | Control | eae | EPEC |
| 13 | 55 C | Control | eae | EPEC |
| 14 | 66 C | Control | stx2 | STEC |
| 15 | 18 C | Control | lt1 | ETEC |
| 16 | 21 C | Control | lt1 | ETEC |
D, Dog; C, control; O, owner; STEC, Shiga toxin-producing E. coli; EPEC, enteropathogenic E. coli; EHEC, enterohemorrhagic E. coli; ETEC, enterotoxigenic E. coli; EIEC, enteroinvasive E. coli; NTEC, necrotoxigenic E. coli
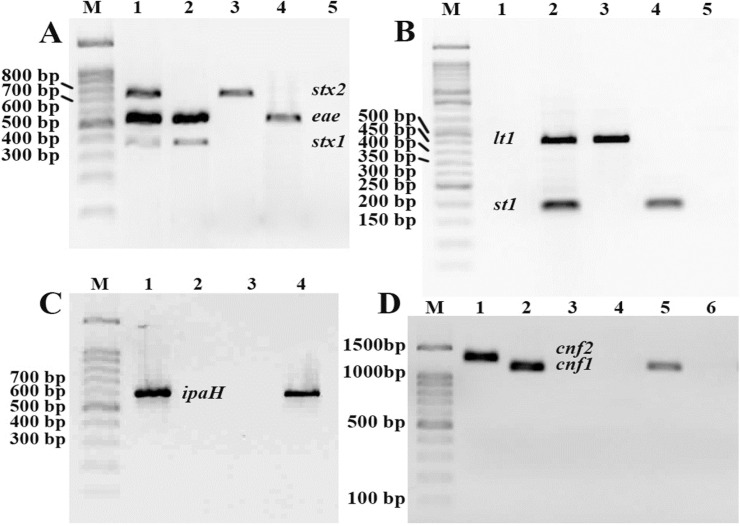
Fig. 1.
PCR products on electrophoresed agarose gel (1.5%) for screening of virulence genes among E. coli isolates. A, M, marker (100 bp); lane 1, stx1/stx2/eae (positive-control); lane 2, stx1/eae; lane 3, stx2; lane 4, eae; lane 5, negative-control. B, M, marker (50 bp); lane 1, negative-control; lane 2, st1/lt1 (positive-control); lane 3, lt1; lane 4, st1; lane 5, negative-sample. C, M, marker (100 bp); lane 1, ipaH (positive-control); lane 2, negative-control; lane 3, negative-sample; lane 4, ipaH. D, M, marker (100 bp); lane 1, cnf2 (positive-control); lane 2, cnf1 (positive-control); lane 3–4, negative-sample; lane 5, cnf1; lane 6, negative-control.
Based on virulence gene profiles, five pathotypes including EPEC (4.1%), EIEC (2.3%), EHEC (1.7%), ETEC (1.7%) and STEC (0.5%) were detected among all isolates. EIEC in healthy household dogs (6.1%), EHEC in their owners (4.08%) and EPEC in controls (8.5%) were the most frequent pathotypes in each host (Table 4). In the present work, virulence genes were identified in seven pet dogs, which three of them were found to carry at least one of the examined virulence genes that were not detected in their owners. The reverse situation occurred in two household dogs that only family members were positive for virulence genes while their pet dog was negative. In one household, the same virulence gene pattern stx1/eae was detected from dog and its owner. This pattern is related to the important pathotype EHEC. There was no significant and notable similarity among age and gender groups in association with virulence and antimicrobial resistance factors.
Table 4.
Prevalence of pathotypes among E. coli isolates from healthy household dogs, their owners and controls
| Pathotype | Healthy dogs | Owners | Controls | Total |
|---|---|---|---|---|
| EPEC | - | 1/49 (2.04%) | 6/70 (8.5%) | 7/168 (4.1%) |
| EIEC | 3/49 (6.1%) | 1/49 (2.04%) | - | 4/168 (2.3%) |
| EHEC | 1/49 (2.04%) | 2/49 (4.08%) | - | 3/168 (1.7%) |
| ETEC | 1/49 (2.04%) | - | 2/70 (2.8%) | 3/168 (1.7%) |
| STEC | - | - | 1/70 (1.4%) | 1/168 (0.5%) |
STEC, Shiga toxin-producing E. coli; EPEC, enteropathogenic E. coli; EHEC, enterohemorrhagic E. coli; ETEC, enterotoxigenic E. coli; EIEC, enteroinvasive E. coli; NTEC, necrotoxigenic E. coli
DISCUSSION
E. coli have been implicated in many clinical cases of canine diarrhea however it is not the primary cause of enteritis and isolation of virulent diarrheagenic pathotype were severally reported from healthy dogs (21). According to our findings in healthy household dogs, only EHEC (stx1 and eae-positive strains), ETEC (st1-positive strains) and EIEC (ipaH-positive strains) were found, while in Brazil, only EPEC (eae-positive strains) and EAEC (aggR-positive strains) were detected (2). The frequency of stx1, eae, stx2 and cnf1 in Iranian diarrheic dogs at 2016 were recorded as 64.3%, 50%, 35.7% and 7.1% respectively which is significantly high in compare with our results from healthy animals (22). Differences in frequency in various studies can be due to several reasons. For example, differences in sample size, sample type (urine or feces), sampling method (stool or swab), sample transfer (with or without transfer medium), isolation method (such as biochemical techniques), number of selected colonies, detected genes, etc. may cause variation in prevalence of E. coli pathotypes in this study compared to previous study in Iran and other countries.
Shiga toxin-encoding genes are more widely studied in companion dogs and a high prevalence among diarrheic dogs were reported in Canada (23), which is supported by Paula and Marin in Brazil during 2008 to 2009 (24, 25). These results are contrary to other reports from Iranian studies such as Koochakzadeh et al. (2014), Zahraei et al. (2011) and the present work that showed the low frequency of stx-positive strains in dogs including 18.9%, 4% and 2.3% respectively (26, 27). Results of the present study is in agreement with our previous research which was done on healthy dogs in Kerman (28), suggesting that apparently healthy pet dogs could be considered as the mild asymptomatic carriers of diarrheagenic pathotypes of E. coli.
Prevalence and dissemination of virulence genes among dogs and their environment is related to different variables such as nutrition type and raising place. Indoor animals contaminate their floor and the outdoor pets contaminate the soil via feces excretion and shedding of pathogenic or commensal E. coli strains in the environment (2). Thus, the reservoir role of household dogs may be associated to dominant practices of dog-keeping which is varying in different countries and regions (29). In our study, all dogs have been fed by cooked or commercial foods. This type of nutrition could reduce the prevalence of food-born virulent E. coli strains in household dogs which is noted in the present study.
In our work, prevalence of virulence genes in pet owners was similar to pet dogs, and different with controls group. It illustrates the differences between dissemination pattern of virulence genes between pet owner and non-pet owner group. One pet owner and his dog completely showed the same virulence gene profile. Derakhshandeh et al. (2018) in the Iran, Harada et al. (2012) in the Japan and Stenske et al. (2009) in the United States, reported that canine feces are a significant source of pathogenic E. coli strains for urinary tract infections for their owners (30–32). Based to our data pet dogs may be considered as a mild source of pathogenic and a major reservoir of commensal E. coli strains. Commensal microorganisms of healthy companion animals usually are not pathogeic for human, except for immunocompromised or peoples with impaired immune system such as very young, pregnant, and elderly ones (33). Dog owners and control group in the present study were all aged between 25 to 45 years old and they were apparently healthy.
Based to our results, stx2, lt1 and cnf1 genes were just detected in controls group, not in dogs and their owners. Moreover, eae was found considerably in controls in compare with other groups. Furthermore, three pathotypes including EPEC, ETEC and STEC were detected in control group which is in agreement with findings in north and north-west provinces of Iran (34). Alizade et al. (2019) presented a comprehensive review about the prevalence of diarrheagenic E. coli strains in Iranian peoples during 27 years (35). The average frequency of the ETEC (16%), EAEC (11%), EPEC (11%), STEC (9%), DAEC (6%) and EIEC (4%) were less than 20% that is in agreement with low prevalence of the pathotypes in humans in our study. These findings show that infections in the Iranian population have low frequency and the prevalence of the pathotypes is diverse in different regions of Iran.
In human, there is relationship between the virulent genes and potential signs and symptoms; stx1 and stx2 genes encode Shiga toxin 1 and Shiga toxin 2 which they depurinate rRNA, inhibit protein synthesis and induce apoptosis in digestive and urinary systems. So, a wild range of lesions (from non-bloody diarrhea to severe hemorrhagic colitis, bloody diarrhea and lethal hemolytic uremic syndrome) could be observed in STEC infections. eae gene encodes an important protein named Intimin which induces adhesion of the EPECs to and effacement of enterocytes and finally induces Th1 response. These lesions lead to various signs and symptoms in EPEC infections including diarrhea which may be accompanied with mucus, rarely blood, abdominal pain, fever, myalgia, vomiting and nausea. st1 and lt1 genes encode heat-stable and heat-labile enterotoxins in ETEC pathotype. These toxins cause mild watery diarrhea (without the appearance of gross blood or mucus in the feces) to severe life-threatening cholera-like infection by activation of guanylate cyclase and increasing of intracellular calcium resulting in ion secretion in gut lumen. ipaH gene encode an E3 ubiquitin ligase which downregulates host inflammatory response in EIEC infections with signs and symptoms associated with dysentery, fever, severe abdominal cramps, tenesmus, and diarrhea containing watery feces, mucus and traces of blood. cnf1 and cnf2 genes encode cytotoxic necrotizing factors in NTECs which cause necrosis in various tissues. These factors are produced by NTECs in pyelonephritis and may also be involved in kidney invasion (4, 36–38). In our study there was no finding about the role of screened virulence genes with the potential symptoms in human, and all human hosts were apparently healthy. This finding could be due to the fact that either the number of strains with the virulence gene did not reach the infectious dose or the gene encoding the virulence factors was silent.
CONCLUSION
In conclusion, healthy household dogs in the southeast of Iran is not the primary, major and main reservoir of the E. coli pathotypes and could be considered as a mild and secondary source of pathogenic E. coli strains for environment and their owners. Dogs may have received the patotypes via the food and water contaminated with ruminant feces. Due to the fact that the frequency of E. coli pathotypes in dogs is very low, so the rate of shedding of these pathotypes to the environment is also very low compared to ruminants. Even non-pet owners seemed to be a significant source of EPECs. Transmission of E. coli pathotypes may occurred by direct contact with reservoirs or ingestion of contaminated food. These pathotypes are potentially virulent and creates public health hazards. Further studies are needed for better understanding of dissemination mechanisms of E. coli pathotypes among humans and their pets.
ACKNOWLEDGEMENTS
The authors would like to thank Shahid Bahonar University of Kerman and Research Center of Tropical and Infectious Diseases (Kerman University of medical sciences) for their financial support.
REFERENCES
- 1.Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, et al. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin Infect Dis 2012;54 Suppl 5:S472–479. [DOI] [PubMed] [Google Scholar]
- 2.Damborg P, Nielsen SS, Guardabassi L. Escherichia coli shedding patterns in humans and dogs: insights into within-household transmission of phylotypes associated with urinary tract infections. Epidemiol Infect 2009;137:1457–1464. [DOI] [PubMed] [Google Scholar]
- 3.Corzo-Ariyama HA, García-Heredia A, Heredia N, García S, León J, Jaykus L, et al. Phylogroups, pathotypes, biofilm formation and antimicrobial resistance of Escherichia coli isolates in farms and packing facilities of tomato, jalapeño pepper and cantaloupe from Northern Mexico. Int J Food Microbiol 2019;290:96–104. [DOI] [PubMed] [Google Scholar]
- 4.Donnenberg M. (2013). Escherichia coli: pathotypes and principles of pathogenesis. Academic Press. [Google Scholar]
- 5.Dubreuil JD, Isaacson RE, Schifferli DM. Animal enterotoxigenic Escherichia coli. EcoSal Plus 2016;7:10.1128/ecosalplus.ESP-0006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persad AK, Lejeune JT. (2015). Animal reservoirs of Shiga toxin-producing Escherichia coli. In: Enterohemorrhagic Escherichia coli and other Shiga toxin-producing E. coli. American Society of Microbiology. p:231–244. [Google Scholar]
- 7.Pearson JS, Giogha C, Wong Fok Lung T, Hartland EL. The genetics of enteropathogenic Escherichia coli virulence. Annu Rev Genet 2016;50:493–513. [DOI] [PubMed] [Google Scholar]
- 8.Jensen BH, Olsen KEP, Struve C, Krogfelt KA, Petersen AM. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin Microbiol Rev 2014;27:614–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Servin AL. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin Microbiol Rev 2014;27:823–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 2010;8: 26–38. [DOI] [PubMed] [Google Scholar]
- 11.Mainil J. Escherichia coli virulence factors. Vet Immunol Immunopathol 2013;152:2–12. [DOI] [PubMed] [Google Scholar]
- 12.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 2013;26:822–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega-Manriquez XD, Ubiarco-López A, Verdugo-Rodríguez A, Hernández-Chiñas U, Navarro-Ocaña A, Ahumada-Cota RE, et al. Pet dogs potential transmitters of pathogenic Escherichia coli with resistance to antimicrobials. Arch Microbiol 2020;202:1173–1179. [DOI] [PubMed] [Google Scholar]
- 14.Markey B, Leonard F, Archambault M, Cullinane A, Maguire D. (2013). Clinical veterinary microbiology e-book. Elsevier Health Sciences. [Google Scholar]
- 15.Alizade H, Jajarmi M, Aflatoonian MR, Kalantar-Neyestanaki D, Shoja S, Ghanbarpour R. Comparative prevalence of bla gene with virulence genes and serotypes in Klebsiella pneumoniae. Jundishapur J Microbiol 2018; 11 (4); e61285. [Google Scholar]
- 16.Dashti AA, Jadaon MM, Abdulsamad AM, Dashti HM. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J 2009;41:117–122. [Google Scholar]
- 17.Ghanbarpour R, Aflatoonian MR, Askari A, Abiri Z, Naderi Z, Bagheri M, et al. Domestic and game pigeons as reservoirs for Escherichia coli harboring antimicrobial resistance genes. J Glob Antimicrob Resist 2020;22:571–577. [DOI] [PubMed] [Google Scholar]
- 18.China B, Pirson V, Mainil J. Typing of bovine attaching and effacing Escherichia coli by multiplex in vitro amplification of virulence-associated genes. Appl Environ Microbiol 1996;62:3462–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aranda KRS, Fagundes-Neto U, Scaletsky ICA. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol 2004;42:5849–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tóth I, Hérault F, Beutin L, Oswald E. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J Clin Microbiol 2003;41:4285–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puño-Sarmiento J, Medeiros L, Chiconi C, Martins F, Pelayo J, Rocha S, et al. Detection of diarrheagenic Escherichia coli strains isolated from dogs and cats in Brazil. Vet Microbiol 2013;166:676–680. [DOI] [PubMed] [Google Scholar]
- 22.Torkan S, Bahadoranian MA, Khamesipourc F, Anyanwu MU. Detection of virulence and antimicrobial resistance genes in Escherichia coli isolates from diarrhoiec dogs in Iran. Arch Med Vet 2016;48:181–190. [Google Scholar]
- 23.Hammermueller J, Kruth S, Prescott J, Gyles C. Detection of toxin genes in Escherichia coli isolated from normal dogs and dogs with diarrhea. Can J Vet Res 1995;59:265–270. [PMC free article] [PubMed] [Google Scholar]
- 24.Paula CJS de, Marin JM. Occurrence of non-O157 Shiga toxin-producing Escherichia coli in dogs with diarrhea. Cienc Rural 2008;38:1682–1686. [Google Scholar]
- 25.Paula CJS de, Marin JM. Multidrug-resistant Shiga toxin-producing Escherichia coli in dogs with diarrhea. Arq Bras Med Vet Zootec 2009;61:511–514. [Google Scholar]
- 26.Koochakzadeh A, Salehi TZ, Fasaei BN, Badouei MA. Detection of verotoxin (Shiga-like toxin)-producing and eae harboring Escherichia coli in some wild captive and domestic Equidae and Canidae. Arch Razi Inst 2014;69:157–163. [Google Scholar]
- 27.Salehi TZ, Badouei MA, Gohari IM. Molecular detection and antibacterial susceptibility of enteropathogenic Escherichia coli (EPEC) and shigatoxigenic Escherichia coli (STEC) strains isolated from healthy and diarrhoeic dogs. Comp Clin Path 2011;20:585–589. [Google Scholar]
- 28.Ghanbarpour R, Akhtardanesh B, Afsahi E, Sookhtanloo S. Molecular characterization of Escherichia coli pathotypes from diarrheic and healthy dogs. Online J Vet Res 2010;14:316–324. [Google Scholar]
- 29.Younis K, Baddour M, Ibrahim MS. Detection of diarrheagenic Escherichia coli in pet animals and its antibiotic resistance in Alexandria governorate. AJVS 2015;45: 113–118. [Google Scholar]
- 30.Derakhshandeh A, Eraghi V, Boroojeni AM, Niaki MA, Zare S, Naziri Z. Virulence factors, antibiotic resistance genes and genetic relatedness of commensal Escherichia coli isolates from dogs and their owners. Microb Pathog 2018;116:241–245. [DOI] [PubMed] [Google Scholar]
- 31.Harada K, Niina A, Nakai Y, Kataoka Y, Takahashi T. Prevalence of antimicrobial resistance in relation to virulence genes and phylogenetic origins among urogenital Escherichia coli isolates from dogs and cats in Japan. Am J Vet Res 2012;73:409–417. [DOI] [PubMed] [Google Scholar]
- 32.Stenske KA, Bemis DA, Gillespie BE, D’Souza DH, Oliver SP, Draughon FA, et al. Comparison of clonal relatedness and antimicrobial susceptibility of fecal Escherichia coli from healthy dogs and their owners. Am J Vet Res 2009;70:1108–1116. [DOI] [PubMed] [Google Scholar]
- 33.O’Neil J. Zoonotic infections from common household pets. J Nurse Pract 2018;14:363–370. [Google Scholar]
- 34.Miri ST, Dashti A, Mostaan S, Kazemi F, Bouzari S. Identification of different Escherichia coli pathotypes in north and north-west provinces of Iran. Iran J Microbiol 2017;9:33–37. [PMC free article] [PubMed] [Google Scholar]
- 35.Alizade H, Teshnizi SH, Azad M, Shojae S, Gouklani H, Davoodian P, et al. An overview of diarrheagenic Escherichia coli in Iran: A systematic review and meta-analysis. J Res Med Sci 2019;24:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bien J, Sokolova O, Bozko P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol 2012;2012: 681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol 2004;2:123–140. [DOI] [PubMed] [Google Scholar]
- 38.Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev 1998;11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]