Abstract
Background and Objectives:
Foodborne pathogens are among the serious problems all around the world and thus a novel and natural strategy to control and to inhibit such pathogens is highly demanded nowadays. The aim of this study was to isolate a specific bacteriophage of Escherichia coli O157:H7 from sewage in Fars province, Iran to determine its morphological and antimicrobial activities.
Materials and Methods:
In order to isolate the bacteriophage of E. coli O157:H7, 10 samples of slaughterhouse wastewaters were used. Double-Layer Agar method was employed to isolate the bacteriophage. To identify the fine structure of the bacteriophage, electron microscope was employed. Host range and antibacterial activity of the phage was also investigated, in vitro.
Results:
The morphological and biological characteristics of a virulent Siphoviridae phage, PI, are reported. It was found that infection of E. coli O157:H7 strains with this specific bacteriophage produce clear plaques. In the one-step growth analysis, it was confirmed that the phage has been characterized with a very short rise period (around 15 min), an average burst size of 193 PFU/cell, high infectivity and potent lytic action. The bacteriolytic activity of PI was also investigated, in vitro. It was also clarified that at the MOI of 100, 10 and 1, the phage rapidly lysed the bacterial cells within 0.5 or 2 h.
Conclusion:
These results indicate that the phage PI is a newly discovered phage against E. coli O157:H7 in Iran which may be recommended to use as bio-control purposes.
Keywords: Escherichia coli O157:H7, Bacteriophage, Siphoviridae
INTRODUCTION
Since 1983, when Escherichia coli O157:H7 was identified, it has been considered as an important foodborne pathogen (1). The low infective dose of this pathogen in humans is about 10 to 100 cells. E. coli O157:H7 is one of the major causes of gastroenteritis that may cause severe symptoms, such as hemorrhagic colitis or the hemolytic uremic syndrome, thrombotic thrombocytopenic purpura and also acute renal failure in children. Infection in children and elderly persons can be fatal (2, 3).
A variety of preservatives have been currently developed to control the pathogens in food. Although antibiotics are the most effective method of controlling bacterial pathogens, one of the disadvantages is that these cause antibiotic-resistant strains. The Food and Drug Administration (FDA) in the USA has not allowed the use of antibiotics and other chemical antimicrobial agents to control bacteria in food, (4). This is important in foods that are eaten raw and/or unprocessed, because the thermal processes will reduce the quality of this type of food. Today, the use of safe and effective natural control methods to reduce foodborne pathogens in foods is an urgent need, such as bacteriophages (5).
Bacteriophages are the virallytic organisms that infect and lyse only specific bacterial cells (6, 7). Phages are feasible, natural and non-toxic and have no effect on human, animal and plant cells (8). Phages were discovered about 100 years ago and recently antibacterial potential of phages to control of pathogenic bacteria is taken into consideration, especially for antibiotics resistant bacteria (9, 10). High availability, high level of specificity and rapid replication and propagation in the presence of their own specific host, are benefits of phages (8, 11). They are naturally present in the gastrointestinal and environmental ecosystem and can be isolated from various sources (5, 12).
The use of bacteriophages to control and improve food safety has increased, one of the natural ways to reduce the pathogenic bacteria in the food supply chain (13). Increase the consumer pressure to ensure food safety and reduce the use of chemical disinfectants and detergents that are harmful to nature, are the most important reasons to use bacteriophages. In food safety, bacteriophages can be used directly to food and food-related surfaces, on harvested and processed foods (14).
Bacteriophages are effective in reducing the levels of target pathogen in many foods without change in their organoleptic properties and high level of specificity does not affect the commensal and desirable bacteria used in the targeted food (15, 16). In general, there is very little resistance to bacteria compared to chemical agents. They can also be easily separated from the environment and can be produced in high volumes and low cost (17).
The present study was aimed to isolate specific bacteriophage of E. coli O157:H7 from sewage in Fars province, Iran to determine its morphological and antimicrobial activities.
MATERIALS AND METHODS
Host bacterial cultures and growth conditions.
E. coli O157:H7 (ATCC 43895) (provided by Department of Food Hygiene and Public Health, School of Veterinary Medicine, Shiraz University, Shiraz, Iran), was used as host bacteria. Twenty-five bacterial strains (Table 1) were tested for phage host range. These strains included a selection of E. coli strains that have O157 and non-O157 serotypes and a variety of bacteria belong to different genera. Additionally, Nalidixic acid resistant E. coli O157:H7 was examined. Before the experiments, overnight culture of each bacterial strain was prepared in the brain heart infusion broth (BHI # 110493, Merck, Germany) and incubated at 37°C for 18 h. Soft BHI agar used in the plaque assay was prepared with BHI broth supplemented with 0.7% agar.
Table 1.
Bacterial strains used for the host range spectrum of the bacteriophage PI (109 PFU/mL)
| Bacterial strain | Sources | Spot test | Efficiency of plating (EOP) |
|---|---|---|---|
| E. coli O157:H7* | ATCC 43895 | + | High |
| E. coli O157:H7 | NCTC 12900 | + | High |
| E. coli O157:H7 | Wild type | + | High |
| E. coli O157:H7 (nalidixic acid resistance) | ATCC35218 | + | High |
| E. coli O126:K71 | PTCC 1276 | _ | _ |
| E. coli | PTCC1270 | _ | _ |
| E. coli | PTCC1399 | _ | _ |
| E. coli | PTCC1395 | _ | _ |
| E. coli | PTCC 1338 | _ | _ |
| E. coli O6 | ATCC 25922 | _ | _ |
| E. coli | PTCC1551 | + | Low |
| E. coli | PTCC1533 | + | High |
| E. coli O55:K59 | PTCC1269 | _ | _ |
| E. coli | ATCC 8739 | + | High |
| E. coli | ATCC 47013 | _ | _ |
| Salmonella Typhimurium | ATCC 14028 | _ | _ |
| Yersinia enterocolitica | PTCC 1786 | _ | _ |
| Listeria monocytogenes | PTCC1297 | _ | _ |
| Staphylococcus aureus | PTCC 1337 | _ | _ |
| Staphylococcus aureus | ATCC 6538 | _ | _ |
| Bacillus subtilis | PTCC 1023 | _ | _ |
| Bacillus cereus | ATCC 14579 | _ | _ |
| Enterococcus | Wild type | _ | _ |
| Micrococcus | Wild type | _ | _ |
| Proteus | Wild type | _ | _ |
| Citrobacter | Wild type | _ | _ |
EOP was determined using double agar overlay method.
represents host bacterium; – = no activity; + = phage active; EOP was considered as ‘high’ when the difference in ratio between the host vs. test bacterium was 50 and 50 was considered as ‘low.
Phage isolation.
For isolation of E. coli O157:H7-specific phages, 10 environmental samples such as sewage were collected from two different slaughterhouse areas (Shiraz and Marvdasht) in Fars, Iran. The collected samples were transferred to the laboratory beside the ice and examined immediately.
At first, the sewage samples were centrifuged (6,000 ×g for 15 minutes) and then the supernatant was filtered using a syringe filter (pore size 0.22 µm). Twenty ml of the filtrate were mixed with 2 ml of overnight bacterial culture of E. coli O157:H7 and 3 ml of double BHI broth and incubated at 37°C for 24 h (enrichment method). After incubation, chloroform (50 µl / ml) was added and mixed vigorously. Bacterial debris was centrifuged (6,000 ×g for 15 min) and the supernatant was filtered through a 0.22 µm pore size syringe filter and stored at 4°C. The filtrate was then plated on BHI agar (1.5% w/v agar, # 113825, Merck, Germany) through the double layer agar method to detect the phage against E. coli O157: H7 (18). Briefly, 3 ml of molten soft agar BHI mixed with 300 μl of the host strain and then suspension was overlaid on top of a plate containing BHI agar. When the over layer agar was solidified, 5–10 μl of the sample containing phage were spotted on it. After incubation at 37°C for 24 h, all plates were checked for clear zone formation.
Purification of bacteriophages.
Single plaques were picked with a sterile glass Pasteur pipette and re-suspended in 100 μl of sterile SM buffer (50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 8 mM MgSO4.7H2O and 0.01 % (w/v) gelatine), and put in the refrigerator for two hours. Then, 50 μl / ml of chloroform was added and centrifuged at 9000 ×g, 4°C for 20 min. The supernatant was serially diluted and 100 μl of each dilution and 300 μl of the host strain were mixed with 3 ml molten BHI soft agar and poured onto BHI agar plate. Once the top agar solidified, plates incubated at 37°C. After 24 h, plates were observed for a clear zone and checked the plaque morphology. To ensure the purity of the isolated phage, each plaque was re-isolated three times. The phage was stored at –20°C in the BHI broth containing 30% (v/v) glycerol for further characterization.
Phage enumeration.
Bacteriophages titration was enumerated using the double layer assay as described by Adams (1959). Phage lysate was serially diluted using SM buffer. Then, 10 μl of filtrate and 300 μl of the host strain were mixed with 3 ml molten BHI soft agar and poured onto BHI agar plate. Once the top agar solidified, plates incubated at 37°C for 24 h. Double layer assay was conducted for each dilution in triplicates. After incubation, the dilution that formed 30 to 200 plaques were selected and counted the number of plaques. The data were used to calculate the titration of phage lysate (PFU / ml) using the standard formula (18).
Preparation of phage stoke.
To prepare large amounts of purified phage, 1 ml of fresh E. coli O157: H7 culture and 100 μl of purified phage were added to 100 ml of BHI broth and incubated at 37°C for 24 h. After adding chloroform (50 μl / ml) (for bacterial cell lysis), the solution was centrifuged (9,000 ×g for 13 minutes) and the supernatant was filtered (0.22 μm). Then, the number of phages was counted in the stoke using double layer method.
Transmission electron microscopy.
Purified and concentrated phage with polyethylene glycol-8000 was used for transmission electron microscopy (TEM) at Plant Virology Research Center of Shiraz University, Iran. Identification and classification were done according to the size, tail structure, and head structure. The phage was negative stained by 2% uranyl acetate on a mesh copper grid with carbon-coated formvar film and visualized under Leo/Philips 906E TEM.
Bacteriophage host range and efficiency of plating (EOP).
To determine host range of isolated E. coli O157: H7 phages, various types of Gram-negative and Gram-positive bacterial species were used. Table 1 shows the bacterial strains used for the determination of phage susceptibility. 300 μl of the overnight culture of each bacterial strain (test bacteria) was added to the BHI soft agar and then poured onto the agar base plate. Then, 10 μl of dilutions of phages (host bacteria) (10–1–10–6) were placed on it. After incubation at 37°C for 24 h, the plates were examined for plaque formation (19). To determine the efficacy of each phage against different strains of E. coli O157: H7, the EOP test was performed (Phage titer of the host bacteria/phage titer on target bacteria). A solution of 1 to 10 bacteriophages (100 μl) was mixed with 300 μl of the test bacteria and 3 ml of soft BHI medium was added and cultured on the BHI agar medium as a double-layer method. After 24 h, phage titer of the test bacteria was enumerated. Experiments were performed in triplicates. When the difference in ratio between the host vs. test bacterium was < 50, the EOP was considered as “high”, other values were recorded as “low” (20).
Degree of adsorption.
When the host bacterium reaches to mid-exponential phase, the phages added at the MOI of five. After 5 minutes incubation at 37°C, the culture put on ice, 1% chloroform added and then centrifuged at 8000 ×g, 4°C for 10 minutes. Then supernatant was filtered and the PFU was measured. The degree of adsorption was calculated as the number of phages adsorbed in five minutes by subtracting the residual phage concentration from the concentration at the beginning of the experiment (21).
One-step growth curve.
To determine the latent period and burst size, when the E. coli O157:H7 was grown at 37°C and OD at 600 nm reached to 0.1 (~108 CFU ml–1), 50 ml of the culture was harvested. Phage was added at a MOI of 0.01 and allowed to adsorb for 5 min at room temperature. The mixture was centrifuged (8,000 ×g for 5 min) to remove the excess phages, and the supernatant was discarded. The pellets were re-suspended in the 50 ml of fresh BHI broth and incubated at 37°C with shaking at 200 rpm min–1. Samples were collected every 10 min during a 2 h incubation and were immediately centrifuged at 8,500 ×g for 1 min and supernatant was 10-fold serially diluted and plated onto BHI agar to determine the phage titration. Based on the number of PFU per ml, one step growth curve was drawn (7, 22, 23). The burst size was calculated by this formula: (titer after burst - titer at T0)/(phage added - titer at T0) (24). All experiments were done in triplicate.
Bacterial challenge test.
To determine the in vitro phage bacteriolytic activity, E. coli O157:H7 was inoculated into BHI broth for overnight at 37°C, then 1% of this culture was inoculated into 50 ml of fresh BHI broth and incubated at 37°C with shaking at 200 rpm until the OD 600 reached 0.6. Soon afterward, phage was added at the MOI of 0.01, 0.1, 1.0, 10 and 100, and 200 μl of each one was transferred into 96-well microplate in triplicate. Bacterial growth was monitored by turbidity measurements every 60-min interval for 24 h using microplate reader (Biotek, USA) at the OD 600 nm. A culture containing only bacteria (phage-free cultures) and a culture containing only phage (cell-free cultures) were used as controls (25).
RESULTS
Phage isolation and purification.
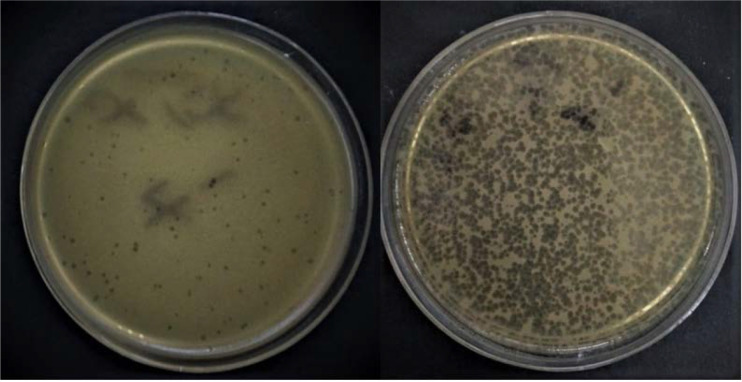
From the 10 slaughterhouse sewage samples tested, one phage, specific to E. coli O157:H7, was isolated from a sewage sample collected from a pond located in the Shiraz slaughterhouse using the double-layer agar assay technique. The isolated phage formed medium-sized (ca. 1–5 mm in diameter) clear plaques on the lawn of its host bacteria (Fig. 1). High titer of phage suspension (3 × 109 PFU/ml) was obtained.
Fig. 1.
Morphology of lytic plaques. Large plaque and medium plaque without halo in titer of 107 PFU/ml
Bacteriophage morphology.
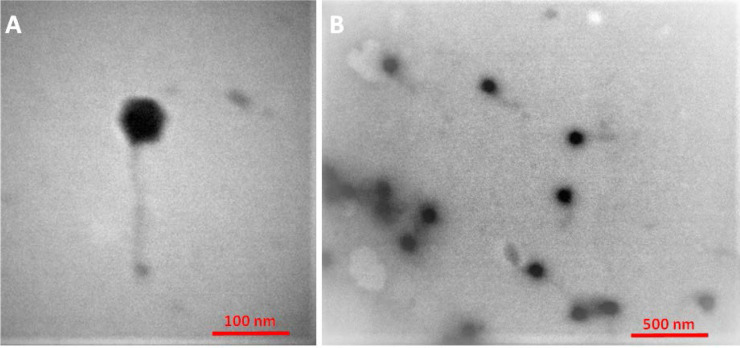
The electron micro-graphs showed that phage PI had a hexagonal head (approximately 57 nm in diameter) with an extremely thin, long flexible and non-contractile tail (ca. 157 ± 5 nm in length and 11 ± 1 nm in width) and no visible collar or terminal knobs (Fig. 2). They were identified as the Siphoviridae family phage of the Caudovirales order.
Fig. 2.
Transmission electron microscopy images of phage PI belonging to the family of Siphoviridae.
Analysis of phage host range and efficiency of plating (EOP).
Host range of the isolated phage was determined against three E. coli O157:H7 and other bacterial strains. The isolated phage formed clear spots on the lawn of 3 (100%) E. coli O157:H7 isolates and 3 (13.6%) of other E. coli strains and bacterial isolates. It is suggesting that this phage has high host specificity to E. coli O157:H7. EOP results indicate that the PI phage formed phage lysis plates in the presence of all the positive target strains (Table 1).
Degree of adsorption.
Phage adsorption assays showed that nearly 99.8% of the phage particles adsorbed to the host bacterial cells after 5 min.
Burst size and latent period.
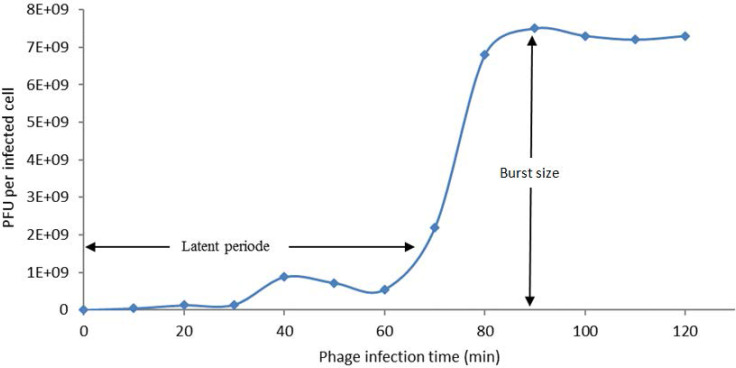
One-step growth curve analysis for phages PI was conducted with E. coli O157:H7 ATCC 43895 in the BHI broth at 37°C (Fig. 3). A latent period of 70 min and a burst size of 193 PFU / host cell were calculated for phage PI from the triphasic curve.
Fig. 3.
One-step growth curve of phage PI performed with BHI broth at 37°C.
Kill curve.
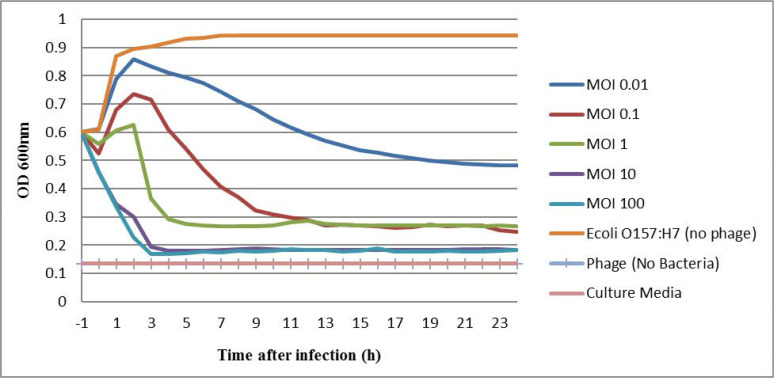
The host lysis activity of phage PI was determined in an in vitro culture condition, using the bacterial challenge test. Phage PI was added at the MOI of 100, 10, 1, 0.1 and 0.01 to the culture of E. coli O157:H7 ATCC 43895 at an OD 600 of 0.6, the host strain was significantly decreased in the MOI of 100, 10 in 2.5 h. But in the MOI of 1, 0.1 and 0.01, the reduction time of bacterial cells were longer at these concentrations (Fig. 4).
Fig. 4.
The effect of different multiplicity of infection (MOI) of phage PI on the growth of the E. coli O157:H7 (ATCC 43895) at 37°C (n = 3). Line orange: host bacteria (positive control), no phage added. Line navy blue (MOI 0.01), Line red: (MOI 0.1), Line green: (MOI 1), Line purple (MOI 10), Line blue (MOI 100), Line pale blue and Line pink (negative control), phage and culture media, no bacteria added.
DISCUSSION
E. coli O157:H7, as a food-borne pathogen is one of the problems in food safety. Using different antibiotics to control this pathogen has led to antibiotic-resistant bacteria. Among the different methods of controlling pathogens, bacteriophages have considered as a novel control strategy and have been studied as therapeutic and bio-control agents for their specific lytic activity (22, 26). Efficacy of several isolated phages that infecting and control E. coli O157:H7 strains have been evaluated before (27). High host specificity and very narrow host range for bacterial inhibition are typical features of phages (8, 11, 13, 17, 19, 22, 28). Here, we have isolated phage PI from slaughterhouse sewage samples, which specifically inhibited E. coli O157:H7. Numerous investigations have reported the presence of lytic specific phages for E. coli O157:H7 in sewage samples that supported our findings and confirm that wastewater samples are richest sources for lytic bacteriophage isolation (17, 19, 29, 30). Clear plaques formed by PI, with sizes ranging from 2 to 5 mm in diameter. Depending on growth conditions plaque size may vary, but typical virulent phages produce clear plaques, whereas turbid plaques form by lysogenize phages (31), This is confirmatory to the idea that phage PI is among the virulent phage. Based on guidelines of the International Committee on Taxonomy of Viruses (32), phage PI is a member of Siphoviridae family in the order Caudovirales. Reported over 95% of phages belong to the Caudovirales. Recently isolated phages for reduction of E. coli O157:H7 belong to the Siphoviridae family are vB_EcoS_FFH_1 and vB_EcoS_ FFH_3 (33) and the phage that isolated by Adibi et al. (17).
Phage PI showed a very specific host range against all strains of E. coli O157:H7 and could only infect three non-O157 E. coli strains (Table 1). Specific receptors on the bacterial are required for phage infection. Fimbriae, pili, outer membrane proteins, H antigen (flagella) and O antigen of lipopolysaccharide (LPS) are the common receptors on E. coli (34). Based on the data it can be concluded that the phage is not specified to the O157 strains, however, the phage was also able to destroy other strains harboring the O antigen. It seems that these antigens may not be seriously involved in the host binding and recognition mechanisms. Different strains and serotypes can be shared in cell wall receptors (35). As a result, phage PI has revealed a wide inhibitory spectrum and was effective against six tested strains. Earlier researches have also reported that a broad host range phages are able to lyse EHEC (36–38). When the lytic ability of phage PI against nonpathogenic E. coli was examined, it was about 13.6% of the strain collection. Viscardi et al. have also reported that half of their phages represented a 13.9% lytic ability against the nonpathogenic strains, which is partly similar to our data, but the collection of nonpathogenic E. coli strains used was different (37). In our study, EOP method was used to determine the lethalness of the PI on the specific strains. It was found that the isolated phage was effective on 83% of the strains identified as host. The specific host strain is one of the factors that affect plating efficiency which emphasizing the significance of examining the relative EOP on a susceptible hosts (39). Similar results were found in the investigation by Viazis et al. in which, all phages were extremely effective against EHEC O157 strains (38). The high host specificity of phage PI makes it as a bio-control agent to protect food from foodborne bacteria (E. coli O157:H7) without affecting probiotic or beneficial bacteria in food.
The burst size of phage PI was calculated based on the final concentration of the phage PI and the concentration of the E. coli O157:H7 cell that was infected by the phage. The average burst size of phage PI was calculated about 193 PFU per bacterial cell. Analysis of phage PI one-step growth showed a moderately short rise period and a large burst size that revealed high lytic activity and strong propagation of the phage and thus suggesting the PI as a good candidate for the bio-control purposes against E. coli O157:H7. It developed rapidly in the host cell and was completely lysed the host culture within 20 to 25 min with a rise period of 15 min. Generally, most Siphoviridae bacteriophages showed short latent periods and for many Myoviridae and Siphoviridae phages, typical burst size was ranged between 50–100 PFU per cell (22, 40). Therefore to develop bio-control methods against E. coli O157:H7 as antimicrobial agent in the food chain, these characteristics of isolated phages could be applied, because the burst size is reasonably related to the phage propagation (41).
In this study, challenge test in vitro against E. coli O157:H7 was performed by using phage PI in five different MOI up to 24 h (Fig. 4). This phage significantly decreased E. coli O157:H7 in culture media at MOI 100, 10 and 1 rather than the control. The E. coli O157:H7 growth inhibition of phage phiLLS was pretty much similar to the PI phage that we have identified here (7). Also, no re-growth of bacterial cells was observed up to 24 h. The growth of bacterial cells after this time was likely due to the presence of phage resistant host or bacterial insensitive mutants (7, 42). PI lytic ability was significantly reduced at MOI of 0.01 because the concentration of phage was very low and many bacterial cells were not infected and the division continued. The short length of this host growth inhibition activity of phage PI in this test recommends that phage PI may be a suitable antibacterial candidate against E. coli O157:H7.
In conclusion, we have isolated a lytic phage, PI, against E. coli O157:H7. This phage is characterized by a wide host spectrum and belongs to the Siphoviridae family. Furthermore, this phage showed a short rise period and burst size of 193 PFU/infected cell and high lytic ability to inhibit the growth of E. coli O157:H7 cell. Based on all these characteristics, phage PI is recommended as a suitable bio-control agent in food safety. However, further in vivo experiments are needed to ensure the safety of this phage.
ACKNOWLEDGEMENTS
The work was kindly supported by School of Veterinary Medicine, Shiraz University. Authors wish to thank all staffs at the Department of Food Hygiene and Public Health especially Mrs Younesian and Miss Aghazi.
REFERENCES
- 1.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 1983; 308:681–685. [DOI] [PubMed] [Google Scholar]
- 2.Todd W, Dundas S. The management of VTEC O157 infection. Int J Food Microbiol 2001; 66:103–110. [DOI] [PubMed] [Google Scholar]
- 3.Sperandio V, Nguyen Y. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol 2012; 2:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Ku HJ, Lee DH, Kim YT, Shin H, Ryu S, et al. Characterization and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157: H7 and Shigella flexneri: potential as a biocontrol agent in food. PLoS One 2016; 11(12):e0168985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sillankorva SM, Oliveira H, Azeredo J. Bacteriophages and their role in food safety. Int J Microbiol 2012; 2012:863945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul Toxicol Pharmacol 2005; 43:301–312. [DOI] [PubMed] [Google Scholar]
- 7.Amarillas L, Rubí-Rangel L, Chaidez C, González-Robles A, Lightbourn-Rojas L, León-Félix J. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol 2017; 8:1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Breidt F. Escherichia coli O157: H7 bacteriophage Φ241 isolated from an industrial cucumber fermentation at high acidity and salinity. Front Microbiol 2015; 6:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Twort FW. An investigation on the nature of ultra-microscopic viruses. Acta Kravsi 1961;2:37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haq IU, Chaudhry WN, Akhtar MN, Andleeb S, Qadri I. Bacteriophages and their implications on future biotechnology: a review. Virol J 2012; 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safwat Mohamed D, Farouk Ahmed E, Mohamed Mahmoud A, Abd El-Baky RM, John J. Isolation and evaluation of cocktail phages for the control of multi-drug-resistant Escherichia coli serotype O104: H4 and E. coli O157: H7 isolates causing diarrhea. FEMS Microbiol Lett 2018; 365(2): 10.1093/femsle/fnx275. [DOI] [PubMed] [Google Scholar]
- 12.Premarathne J, Thung T, New C, Huat J, Basri DF, Rukayadi Y, et al. Distribution of bacteriophages in food and environment samples. Int Food Res J 2017; 24:888–896. [Google Scholar]
- 13.Callaway TR, Edrington TS, Brabban AD, Anderson RC, Rossman ML, Engler MJ, et al. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157: H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog Dis 2008; 5:183–191. [DOI] [PubMed] [Google Scholar]
- 14.Sulakvelidze A. Using lytic bacteriophages to eliminate or significantly reduce contamination of food by foodborne bacterial pathogens. J Sci Food Agric 2013; 93:3137–3146. [DOI] [PubMed] [Google Scholar]
- 15.Hagens S, Loessner MJ. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 2007; 76:513–519. [DOI] [PubMed] [Google Scholar]
- 16.Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 2008; 62:1–11. [DOI] [PubMed] [Google Scholar]
- 17.Adibi M, Mobasher N, Ghasemi Y, Mohkam M, Mobasher MA. Isolation, purification and identification of E. coli O157 phage for medical purposes. Trends Pharmacol Sci 2017; 3:43–48. [Google Scholar]
- 18.Oliveira A, Sillankorva S, Quinta R, Henriques A, Sereno R, Azeredo J. Isolation and characterization of bacteriophages for avian pathogenic E. coli strains. J Appl Microbiol 2009; 106: 1919–1927. [DOI] [PubMed] [Google Scholar]
- 19.Yıldırım Z, Sakin T, Çoban F. Isolation of anti-Escherichia coli O157: H7 bacteriophages and determination of their host ranges. TURJAF 2018; 6:1200–1208. [Google Scholar]
- 20.Manohar P, Tamhankar AJ, Lundborg CS, Ramesh N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS One 2018; 13(10):e0206278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirzaei MK, Nilsson AS. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One 2015; 10(3):e0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park M, Lee J-H, Shin H, Kim M, Choi J, Kang D-H, et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl Environ Microbiol 2012; 78:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HS, Choi S, Shin H, Lee J-H, Choi SH. Vibrio vulnificus bacteriophage SSP002 as a possible biocontrol agent. Appl Environ Microbiol 2014; 80:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casey E, Mahony J, Neve H, Noben J-P, Dal Bello F, van Sinderen D. Novel phage group infecting Lactobacillus delbrueckii subsp. lactis, as revealed by genomic and proteomic analysis of bacteriophage Ldl1. Appl Environ Microbiol 2015; 81:1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J-H, Bai J, Shin H, Kim Y, Park B, Heu S, et al. A novel bacteriophage targeting Cronobacter sakazakii is a potential biocontrol agent in foods. Appl Environ Microbiol 2015; 82:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisien A, Allain B, Zhang J, Mandeville R, Lan C. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J Appl Microbiol 2008; 104:1–13. [DOI] [PubMed] [Google Scholar]
- 27.Peng Q, Yuan Y. Characterization of a newly isolated phage infecting pathogenic Escherichia coli and analysis of its mosaic structural genes. Sci Rep 2018; 8:8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyman P, Abedon ST. (2010). Bacteriophage host range and bacterial resistance, in Advances in Applied Microbiology. Elsevier; pp: 217–248. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y-D, Park J-H. Characterization and application of phages isolated from sewage for reduction of Escherichia coli O157: H7 IN biofilm. LWT-Food Sci Technol 2015; 60:571–577. [Google Scholar]
- 30.Topka G, Bloch S, Nejman-Faleńczyk B, Gąsior T, Jurczak-Kurek A, Necel A, et al. Characterization of bacteriophage vB-EcoS-95, isolated from urban sewage and revealing extremely rapid lytic development. Front Microbiol 2019; 9:3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abedon ST, Yin J. (2009). Bacteriophage plaques: theory and analysis, in Bacteriophages. Springer; pp: 161–174. [DOI] [PubMed] [Google Scholar]
- 32.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (2005). Virus taxonomy: VIIIth report of the International Committee on Taxonomy of Viruses. Academic Press. [Google Scholar]
- 33.Hong Y, Pan Y, Ebner P. Development of bacteriophage treatments to reduce E. coli O157: H7 contamination of beef products and produce. J Anim Sci 2014; 92:1366–1377. [DOI] [PubMed] [Google Scholar]
- 34.Wilson SGS, Parker M, Collier LH. (1990). Topley and Wilson’s Principles of Bacteriology, Virology and Immunology. Edward Arnold. [Google Scholar]
- 35.Drulis-Kawa Z, Majkowska-Skrobek1 G, Maciejewska B. Bacteriophages and phage-derived proteins – application approaches. Curr Med Chem 2015; 22: 1757–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raya RR, Varey P, Oot RA, Dyen MR, Callaway TR, Edrington TS, et al. Isolation and characterization of a new T-even bacteriophage, CEV1, and determination of its potential to reduce Escherichia coli O157: H7 levels in sheep. Appl Environ Microbiol 2006; 72:6405–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viscardi M, Perugini AG, Auriemma C, Capuano F, Morabito S, Kim K-P, et al. Isolation and characterisation of two novel coliphages with high potential to control antibiotic-resistant pathogenic Escherichia coli (EHEC and EPEC). Int J Antimicrob Agents 2008; 31:152–157. [DOI] [PubMed] [Google Scholar]
- 38.Viazis S, Akhtar M, Feirtag J, Brabban A, Diez-Gonzalez F. Isolation and characterization of lytic bacteriophages against enterohaemorrhagic Escherichia coli. J Appl Microbiol 2011; 110:1323–1331. [DOI] [PubMed] [Google Scholar]
- 39.Kutter E. (2009). Phage host range and efficiency of plating, in Bacteriophages. Springer; pp: 141–149. [DOI] [PubMed] [Google Scholar]
- 40.Litt PK, Jaroni D. Isolation and physiomorphological characterization of Escherichia coli O157: H7-infecting bacteriophages recovered from beef cattle operations. Int J Microbiol 2017; 2017:7013236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallet R, Kannoly S, Wang N. Effects of bacterio-phage traits on plaque formation. BMC Microbiol 2011; 11:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khakhum N, Yordpratum U, Wongratanacheewin R. Bacteriophages and their medical applications. Srinagarind Med J 2010; 25:47–53. [Google Scholar]