Abstract
Background: Rheumatoid arthritis (RA) is related to several pivotal susceptibility genes, including cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and costimulatory molecule (CD80/CD86) genes. Although the connection between polymorphisms of CTLA-4 and CD86 genes in different populations of RA have been studied extensively, the results are controversial.
Objective: To clarify the correlation in the Chinese Han population between CTLA-4, CD80/86, and CD28 gene polymorphisms, and RA susceptibility.
Methods: A case-control study (574 RA patients and 804 controls) was conducted to determine the correlation between CTLA-4 rs231775 and rs16840252 gene polymorphisms, CD86 rs17281995 gene polymorphisms, and the risk of RA for the Chinese Han population. Furthermore, an additional meta-analysis, including three single nucleotide polymorphisms (SNPs) (CTLA-4 rs231775, CTLA-4 rs3087243, and CTLA-4 rs5742909) from 32 citations, including 43 studies, 24,703 cases and 23,825 controls was performed to elucidate the relationship between known SNPs in the CTLA-4 genes and RA for more robust conclusions.
Results: The results showed that CTLA-4 rs231775 gene polymorphism decreased the RA risk (GA vs. AA, OR = 0.77, P = 0.025), whereas CTLA-4 rs16840252 and CD86 rs17281995 gene polymorphisms were not related to RA susceptibility. Stratification analyses by RF, ACPA, CRP, ESR, DAS28, and functional class identified significant associations for CTLA-4 rs231775 and rs16840252 gene polymorphisms in the RF-positive and RF-negative groups. A meta-analysis of the literature on CTLA-4 gene polymorphisms and RA risk revealed that the risk of RA was decreased by CTLA-4 rs231775 gene polymorphisms.
Conclusions: The CTLA-4 rs231775 gene polymorphism decreased the risk of RA, whereas CTLA-4 rs16840252 and CD86 rs17281995 gene polymorphisms were not related to RA risk. A meta-analysis indicated that CTLA-4 rs231775 and rs3087243 gene polymorphisms decreased the risk of RA. To support these analytical results, additional clinical cases should be investigated in further studies.
Keywords: rheumatoid arthritis, polymorphisms, CTLA-4, CD80/86, CD28, meta-analysis
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by persistent inflammation of the synovial membranes of joints and progressive cartilage damage, ultimately leading to severe joint deformity and loss of function (1). According to current research on RA pathogenesis and etiology, 50–60% of its susceptibility is attributed to genetic factors (2). Recently, RA-related genes have been studied intensively, including CTLA-4, CD80/CD86, and CD28 genes.
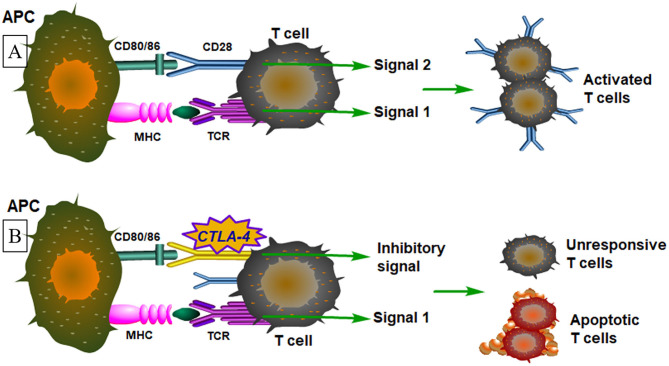
T lymphocytes play an essential role in RA and their activation not only requires recognition of specific antigenic peptides by the T-cell receptor (TCR), but also the co-stimulatory signals provided by accessory surface molecules on T cells (3). CD28, a co-stimulatory receptor, is consecutively expressed on resting T-cells. CTLA-4, a co-inhibitory receptor, is a member of the CD28 family expressed on the surface of T cells soon after its activation (3, 4). CD80 and CD86 (B7 molecules), mainly expressed on B cells, monocytes/macrophages, and dendritic cells, are the ligands of CD28 and CTLA-4, which are upregulated when activated. The binding of the ligands and CD28 promotes the activation of a TCR-stimulated T cell, while the binding of CTLA-4 causes the inhibition of T cell activation (5). CTLA-4 has higher affinity with CD80-CD86 than CD28, although it is homologous to CD28 (6). In a continuous immune response, the expression of CTLA-4 is upregulated to inhibit the proliferation of T-cell and reduce interleukin (IL)-2 production. For B7 molecules, the main function is to augment and sustain T-cell responses by interacting with CD28, while they can also provide inhibitory signals when binding with CTLA-4 (7). The immunological mechanism of T cell activation is illustrated in Figure 1. Lack of CTLA-4 will therefore cause severe lymphoproliferation and harmful destruction of multiorgan tissues, indicating that it plays an vital role in negative regulatory functions (8). Hence, changes in CTLA4 and CD86 genes have the potential to increase the immune response of autoreactive T-cells to self-antigens (9). In mouse experiments, Ewing et al. (10) confirmed that T-cell co-stimulation by connecting with CD28 and its negative regulator CTLA-4 played an important role in accelerating atherosclerosis development. According to recent studies, molecular variants of CTLA-4 were discovered in a number of autoimmune and inflammatory diseases mediated by T-cells, especially in RA (9, 11, 12).
Figure 1.
An illustrative figure shows the immunological mechanism of T cell activation. (A) Signal 1 means T cells are activated by peptide–MHC antigen and the T-cell antigen receptor (TCR) delivers activating signals. Antigen presenting cells (APCs) express costimulatory molecules that bind to receptors on the T-cell simultaneously with antigen recognition. In signal 2, the costimulatory receptors deliver signals, that cooperate with TCR signals to activate the T cell. (B) In a continuous immune response, the expression of CTLA-4 is upregulated which delivers signals that block the activating signals delivered by the T-cell antigen receptor (signal 1 plus coinhibitory signals). The main function will be either functional inactivation of the T cells or T-cell apoptosis.
The association between CTLA-4, CD80/86, and CD28 gene polymorphisms and RA susceptibility may offer new research possibilities to conduct further studies on RA. Although many studies (5, 13–44) have discovered the association between polymorphisms in the CTLA-4 and CD86 genes, the results are contradictory and inconclusive. As different populations have various lifestyles, gene pools and gene-environment interactions, it is impossible to expect that the risk of RA in each population is the same as genotypes. Here, a case-controlled study was conducted to reveal the association between the CTLA-4 rs231775 gene polymorphism, CTLA-4 rs16840252 gene polymorphism, CD86 rs17281995 gene polymorphism, and RA risks in the Chinese Han population. Considering a single case-controlled study may lead to inconclusive results and be less persuasive because of clinical heterogeneity, limited sample sizes, and different ethnic populations, an additional meta-analysis was performed to verify the relevance between known SNPs in the CTLA-4, CD80/86, and CD28 genes and RA to obtain more robust conclusions.
Patients and Methods
Study Population
This study was conducted in accordance with the Helsinki Declaration, and was approved by the Ethics Committee of the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University.
In this study, 574 RA patients (427 women and 147 men), diagnosed using the 2010 ACR/EULAR classification standard for RA (45), were recruited successively from the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University, the Changzhou First Hospital, and the Changzhou Traditional Chinese Medical Hospital (between September 2010 and October 2013). Exclusion criteria included patients from other countries, with other autoimmune diseases, a current or previous history of malignancy, other major systemic diseases, or a family history of autoimmune diseases. The 804 controls (599 women and 205 men) were recruited from the identical institutions during the same period and the majority of them were trauma patients matched for age and sex without RA and other autoimmune diseases. After providing written informed consent, every patient was interviewed separately by using a pre-tested questionnaire to obtain relevant data related to related risk and demographics elements.
Genomic DNA Extraction and Genotyping
All blood samples were preserved by ethylenediaminetetraacetic acid tubes. QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) was performed to extracted genomic DNA from whole blood. The MassARRAY system (Sequenom, San Diego, California, USA) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry were used to genotype SNPs, as previously described (46).
Power Analysis
To evaluate the statistical power of this study, we used the Power and Sample Size Calculations, Version 3.1.6. for power analysis. Our study was performed with the power calculated with a significant value of 0.05 (47).
Statistical Analysis
To assess the correlation between the studied SNPs and RA risk, we calculated the odds ratio (OR) and 95% confidence interval (CI) for five gene models (allele, dominant, recessive, homozygous, and heterozygous). These statistical analyses were all performed using SAS software package (ver. 9.1.3; SAS Institute, Cary, NC, USA) with a significance level of P < 0.05. By applying the goodness-of-fit χ2-test to contrast the observed and expected genotype frequencies among controls, we analyzed the Hardy-Weinberg equilibrium (HWE) of the genotypes.
Meta-Analysis
To thoroughly characterize the association of SNPs in the CTLA-4, CD80/86, and CD28 genes with RA, we searched the databases of Medline, PubMed, Embase, and the Cochrane Library to identify published studies through December 2018. The key words of “polymorphism,” “SNP,” “CTLA-4,” “CD80/86,” “CD28,” “rheumatoid arthritis,” and “RA” were combined for free research. No language or other restrictions were placed on the search. The reference lists of all the related papers were examined to identify any initially omitted studies.
The selection criteria were as follows:
Studies that focused on humans;
Studies that evaluated the association between SNPs of CTLA-4, CD80/86, and CD28 genes and RA;
Studies that included detailed genotype data.
Studies were excluded based on the following exclusion criteria:
Duplication of previous publications;
Review, editorial or other types of studies, which were not focused on detailed genotype research;
Studies that failed to obtain detailed genotype data.
Studies were not conducted on patients who had cancer or other diseases that might have influenced the results. Any studies with questionable inclusion/exclusion criteria were discussed and disagreements were resolved by two reviewers independently evaluating the methodological quality of the included studies. The statistical analyses in our groups were performed using Stata 11.0 software (StataCorp, College Station, TX, USA). Their high methodological quality was evaluated by the Newcastle-Ottawa Scale (NOS) scores. For each meta-analysis, the pooled odds ratio (OR) was calculated for each gene variant, and 95% confidence intervals (CI) were established.
Results
Clinical Details of the Study Population
The clinical characteristics of all patients are summarized in Table 1. In this study, cases and controls were clearly matched for age and sex (P = 0.080 and P = 0.962, respectively). The observed frequency distributions of the rs231775, rs16840252, and rs17281995 genotypes in the control population and the RA patients, which conformed to the HWE. respectively, are presented in Table 2.
Table 1.
Patient demographics and risk factors in rheumatoid arthritis.
| Variable | Cases (n = 574) | Controls (n = 804) | P |
|---|---|---|---|
| Age (years) | 54.5 (± 15.1) | 55.7 (±10.1) | 0.080 |
| Female/male | 427/147 | 599/205 | 0.962 |
| Age at onset, years, mean ± SD |
45.6 (±12.9) | — | — |
| Disease duration, years, mean SD |
8.9 (±9.2) | — | — |
| Treatment duration, years, mean ± SD |
7.6 (±7.8) | — | — |
| RF-positive, no. (%) | 456 (79.4%) | — | — |
| ACPA positive, no. (%) | 300 (52.2%) | — | — |
| CRP-positive, no. (%) | 323 (56.3%) | — | — |
| ESR, mm/h | 33.7 (±25.2) | — | — |
| DAS28 | 4.3 (±1.5) | — | — |
| Functional class, no. (%) | |||
| I | 73 (12.7%) | — | — |
| II | 256 (44.6%) | — | — |
| III | 209 (36.4%) | — | — |
| IV | 36 (6.3%) | — | — |
RF, rheumatoid factor; ACPA, anti-cyclic citrullinated peptide antibodies; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; DAS28, RA disease activity score.
Table 2.
Logistic regression analysis of association between CTLA-4 rs231775, CTLA-4 rs16840252 and CD86 rs17281995 polymorphisms and the risk of rheumatoid arthritis.
| Genotype | Cases (n = 574) | Controls (n = 804) | OR (95% CI) | P | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| CTLA-4 rs231775 | ||||||
| G vs. A | 332/812 | 29.0/71.0 | 478/1,116 | 30.0/70.0 | 0.96 (0.81, 1.13) | 0.585 |
| GG+GA vs. AA | 269/303 | 47.0/53.0 | 410/387 | 51.4/48.6 | 0.84 (0.68, 1.04) | 0.107 |
| GG vs. GA+AA | 63/509 | 11.0/89.0 | 68/729 | 8.5/91.5 | 1.33 (0.93, 1.90) | 0.125 |
| GG vs. AA | 63/303 | 11.0/53.0 | 68/387 | 8.5/48.6 | 1.18 (0.81, 1.72), | 0.378 |
| GA vs. AA | 206/303 | 36.0/53.0 | 342/387 | 42.9/48.6 | 0.77 (0.61, 0.97) | 0.025 |
| CTLA-4 rs16840252 | ||||||
| T vs. C | 158/988 | 13.8/86.2 | 223/1,361 | 14.1/85.9 | 0.98 (0.78, 1.22) | 0.828 |
| TT+TC vs. CC | 145/428 | 25.3/74.7 | 205/587 | 25.9/74.1 | 0.97 (0.76, 1.24) | 0.809 |
| TT vs. TC+CC | 13/560 | 2.3/97.7 | 18/774 | 2.3/97.7 | 0.99 (0.49, 2.05) | 0.996 |
| TT vs. CC | 13/428 | 2.3/74.7 | 18/587 | 2.3/74.1 | 0.99 (0.48, 2.04) | 0.979 |
| TC vs. CC | 132/428 | 23.0/74.7 | 187/587 | 23.6/74.1 | 0.96 (0.75, 1.25) | 0.804 |
| CD86 rs17281995 | ||||||
| C vs. G | 61/1,083 | 5.3/94.7 | 98/1,508 | 6.1/93.9 | 0.87 (0.62, 1.20) | 0.394 |
| CC+GC vs. GG | 60/512 | 10.5/89.5 | 96/707 | 12.0/88.0 | 0.86 (0.61, 1.22) | 0.399 |
| CC vs. GC+GG | 1/571 | 0.2/99.8 | 2/801 | 0.2/99.8 | 0.70 (0.06, 7.75) | 0.772 |
| CC vs. GG | 1/512 | 0.2/89.5 | 2/707 | 0.2/88.0 | 0.69 (0.06, 7.64) | 0.763 |
| GC vs. GG | 59/512 | 10.3/89.5 | 94/707 | 11.8/88.0 | 0.87 (0.61, 1.22) | 0.416 |
Bold values are statistically significant (P < 0.05).
Association Between CTLA-4 Gene Polymorphisms and RA Risk
The genotypic distributions of the CTLA-4 rs231775 gene polymorphisms in the subjects are presented in Table 2. The results showed that the GA genotype was associated with a decreased risk of RA (GA vs. AA, OR = 0.77, P = 0.026) using logistic regression analyses. The effects on RA risk of this SNP were further evaluated according to the characteristics of rheumatoid factor (RF), anti-cyclic citrullinated peptide antibodies (ACPA), C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), RA disease activity score (DAS28), and function class. The American College of Rheumatology's improved grading standards were the following: level 1: life, work, and competitive sports are not restricted; level 2: life, no restrictions on work, and no restrictions on competitive sports; level 3: life is unrestricted, work and competitive sports are restricted; level 4: life, work, and competitive sports are restricted. A significant association was detected among the RF-positive and RF-negative groups (GG, OR = 2.53, P = 0.002; AG+GG, OR = 1.98, P = 0.019, Supplementary Table 1).
The genotypic distributions of the CTLA-4 rs16840252 gene polymorphism in this population are well-delineated in Table 2. Logistic regression analyses suggested that the TT genotype, or T allele carriers of the rs16840252 polymorphism, were not related with the risk of RA (Table 2). The effects on RA risk of this SNP were further tested according to the characteristics of RF, ACPA, CRP, ESR, DAS28, and function class; significant associations were detected among the RF-positive and RF-negative groups, which suggested that the TT genotype of rs16840252 polymorphisms might be associated with the expression of RF (Supplementary Table 1).
Association Between CD86 rs17281995 Gene Polymorphism and RA Risk
The frequencies of the genotypes for the CD86 rs17281995 gene in cases and controls are shown in Table 2. In this study, discrepancies of genotypes for the rs17281995 polymorphisms in the control subjects conformed to the HWE. Then, logistic regression analyses showed that the CD86 rs17281995 gene polymorphisms had no relationship with RA risk. The effects on RA risk of this SNP were further assessed according to the characteristics of RF, ACPA, CRP, ESR, DAS28, and function class; no significant association was found (Supplementary Table 1).
Power Analysis
The power of our study for CTLA-4 rs231775, CTLA-4 rs16840252, and CD86 rs17281995 were 0.668, 0.057, and 0.088, respectively.
Meta-Analysis: General Characteristics of the Included Studies and Quantitative Analysis
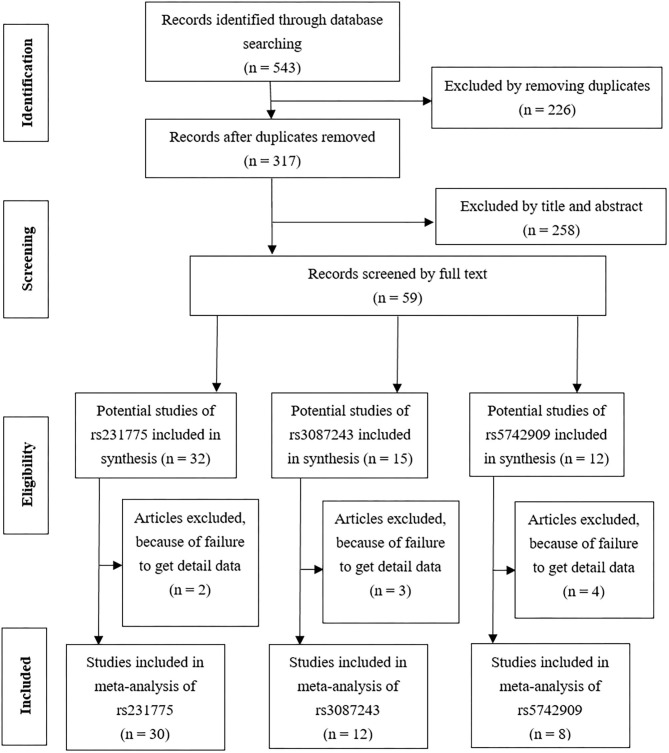
A search of the literature yielded 543 citations. According to the application and refinement of the literature search strategy, 50 related studies were eligible for the data extraction. The flowchart of the reviews, which showed the selection protocol that we used for qualified studies, is presented in Figure 2. In this meta-analysis, these clinical characteristics of the studies exploring the association between the SNPs of CTLA-4, CD80/86, and CD28 genes and RA susceptibility are listed in Supplementary Table 2. A total of 38 citations including 21 SNPs and 75 studies were selected. Finally, three SNPs (CTLA-4 rs231775, CTLA-4 rs3087243, and CTLA-4 rs5742909, Supplementary Table 3) were incorporated into our meta-analysis [32 citations including 43 studies, 24,703 cases and 23,825 controls, (5, 13–43)] because a sufficient number of studies was identified (≥ 5). According to the NOS scores, the included articles ranged from 5 to 7 stars, showing their high methodological quality.
Figure 2.
A flowchart shows the results of the literature search and selection for this study of CTLA-4.
General analysis showed that the rs231775 polymorphism of the CTLA-4 gene decreased RA risk [OR and 95% CI: 0.92 (0.86–0.98) in G vs. A; 0.86 (0.75–0.99) in GG+GA vs. AA; 0.86 (0.78–0.95) in GG vs. GA+AA; and 0.90 (0.83–0.98) in GG vs. AA, Table 3]. Stratification analysis showed that the CTLA-4 gene rs231775 polymorphism decreased the risk of RA among Asians, Caucasians, and Latin-Americans but not among Africans (Table 3).
Table 3.
Meta-analysis of the association between CTAL-4 gene polymorphisms and rheumatoid arthritis risk.
| Comparison | Category | Category | Studies | OR (95% CI) | P-value | P for heterogeneity |
|---|---|---|---|---|---|---|
| CTLA-4 rs231775 | ||||||
| G vs. A | Total | 30 | 0.92 (0.86, 0.98) | 0.015 | <0.001 | |
| Ethnicity | Asian | 12 | 0.88 (0.83, 0.94) | <0.01 | 0.446 | |
| Caucasian | 16 | 0.93 (0.84, 1.04) | 0.201 | <0.001 | ||
| Latin-American | 1 | 0.69 (0.52, 0.91) | 0.010 | – | ||
| African | 1 | 1.25 (0.81, 1.94) | 0.318 | – | ||
| GG+GA vs. AA | Total | 30 | 0.95(0.85, 1.07) | 0.406 | <0.001 | |
| Ethnicity | Asian | 12 | 0.86 (0.75, 0.99) | 0.041 | 0.043 | |
| Caucasian | 16 | 1.02(0.85, 1.23) | 0.821 | 0.001 | ||
| Latin-American | 1 | 0.77 (0.47, 1.27) | 0.308 | – | ||
| African | 1 | 1.33 (0.72, 2.46) | 0.356 | – | ||
| GG vs. GA+AA | Total | 30 | 0.86 (0.78, 0.95) | 0.004 | 0.001 | |
| Ethnicity | Asian | 12 | 0.87 (0.75, 1.01) | 0.065 | 0.270 | |
| Caucasian | 16 | 0.87 (0.76, 0.99) | 0.039 | 0.001 | ||
| Latin-American | 1 | 0.53 (0.35, 0.81) | 0.003 | – | ||
| African | 1 | 1.30 (0.57, 2.97) | 0.534 | – | ||
| GG vs. AA | Total | 30 | 0.90 (0.83, 0.98) | 0.014 | 0.001 | |
| Ethnicity | Asian | 12 | 0.80 (0.69, 0.91) | 0.001 | 0.479 | |
| Caucasian | 16 | 0.99 (0.88, 1.10) | 0.797 | 0.001 | ||
| Latin-American | 1 | 0.54 (0.30, 0.94) | 0.031 | – | ||
| African | 1 | 1.48 (0.60, 3.62) | 0.392 | – | ||
| GA vs. AA | Total | 30 | 1.00 (0.88, 1.14) | 0.992 | <0.001 | |
| Ethnicity | Asian | 12 | 0.89 (0.75, 1.05) | 0.162 | 0.005 | |
| Caucasian | 16 | 1.10 (0.90, 1.33) | 0.350 | 0.001 | ||
| Latin-American | 1 | 1.01 (0.59, 1.72) | 0.980 | – | ||
| African | 1 | 1.29 (0.67, 2.48) | 0.449 | – | ||
| CTLA-4 rs3087243 | ||||||
| A vs. G | Total | 12 | 0.88 (0.85, 0.92) | <0.001 | 0.092 | |
| Ethnicity | Caucasian | 9 | 0.89 (0.85, 0.93) | <0.001 | 0.089 | |
| Latin-American | 1 | 0.79 (0.60, 1.06) | 0.112 | – | ||
| Asian | 2 | 0.82 (0.72, 0.95) | <0.007 | 0.134 | ||
| AA+AG vs. GG | Total | 12 | 0.83 (0.78, 0.88) | <0.001 | 0.040 | |
| Ethnicity | Caucasian | 9 | 0.84 (0.79, 0.89) | <0.001 | 0.083 | |
| Latin-American | 1 | 0.63 (0.42, 0.96) | 0.029 | – | ||
| Asian | 2 | 0.79 (0.66, 0.95) | 0.011 | 0.035 | ||
| AA vs. AG+GG | Total | 12 | 0.88 (0.82, 0.95) | 0.001 | 0.768 | |
| Ethnicity | Caucasian | 9 | 0.89 (0.82, 0.96) | 0.002 | 0.591 | |
| Latin-American | 1 | 0.96 (0.56, 1.65) | 0.891 | – | ||
| Asian | 2 | 0.76 (0.55, 1.06) | 0.104 | 0.985 | ||
| AA vs. GG | Total | 12 | 0.79 (0.73, 0.86) | <0.001 | 0.329 | |
| Ethnicity | Caucasian | 9 | 0.80 (0.74, 0.88) | <0.001 | 0.208 | |
| Latin-American | 1 | 0.72 (0.40, 1.31) | 0.285 | – | ||
| Asian | 2 | 0.67 (0.48, 0.94) | 0.020 | 0.479 | ||
| AG vs. GG | Total | 12 | 0.84 (0.79, 0.90) | <0.001 | 0.076 | |
| Ethnicity | Caucasian | 9 | 0.85 (0.80, 0.91) | <0.001 | 0.193 | |
| Latin-American | 1 | 0.61 (0.39, 0.94) | 0.024 | – | ||
| Asian | 2 | 0.81 (0.67, 0.98) | 0.029 | 0.031 | ||
| CTLA-4 rs5742909 | ||||||
| T vs. C | Total | 8 | 1.19 (0.88, 1.62) | 0.255 | <0.001 | |
| Ethnicity | Caucasian | 4 | 1.22 (0.80, 1.86) | 0.365 | 0.004 | |
| Latin-American | 1 | 1.00 (0.53, 1.89) | 1.000 | – | ||
| Asian | 3 | 1.14 (0.54, 2.42) | 0.733 | 0.003 | ||
| TT+TC vs. CC | Total | 8 | 1.25 (0.86, 1.80) | 0.245 | <0.001 | |
| Ethnicity | Caucasian | 4 | 1.28 (0.79, 2.07) | 0.323 | 0.003 | |
| Latin-American | 1 | 0.89 (0.46, 1.74) | 0.733 | – | ||
| Asian | 3 | 1.26 (0.50, 3.19) | 0.625 | 0.001 | ||
| TT vs. TC+CC | Total | 8 | 1.43 (0.90, 2.28) | 0.128 | 0.519 | |
| Ethnicity | Caucasian | 4 | 1.46 (0.77, 2.78) | 0.245 | 0.394 | |
| Latin-American | 1 | 5.05 (0.24, 105.86) | 0.297 | – | ||
| Asian | 3 | 1.27 (0.63, 2.55) | 0.502 | 0.224 | ||
| TT vs. CC | Total | 8 | 1.71 (0.98, 2.73) | 0.123 | 0.302 | |
| Ethnicity | Caucasian | 4 | 1.62 (0.86, 3.07) | 0.136 | 0.218 | |
| Latin-American | 1 | 4.95 (0.24, 103.73) | 0.303 | – | ||
| Asian | 3 | 1.69 (0.84, 3.43) | 0.143 | 0.131 | ||
| TC vs. CC | Total | 8 | 1.25 (0.86, 1.81) | 0.239 | <0.001 | |
| Ethnicity | Caucasian | 4 | 1.27 (0.81, 1.99) | 0.289 | 0.010 | |
| Latin-American | 1 | 0.79 (0.40, 1.58) | 0.505 | – | ||
| Asian | 3 | 1.33 (0.52, 3.41) | 0.553 | 0.001 | ||
Bold values are statistically significant (P < 0.05).
The meta-analysis also indicated that the CTLA-4 gene rs3087243 polymorphism decreased RA risk [OR and 95% CI: 0.88 (0.85–0.92) in A vs. G; 0.83 (0.78–0.88) in AA+AG vs. GG; 0.88 (0.82–0.95) in AA vs. AG+GG; 0.79 (0.73–0.86) in AA vs. GG; and 0.84 (0.79–0.90) in AG vs. GG, Table 3]. Stratification analysis showed that the CTLA-4 gene rs3087243 polymorphism decreased the risk of RA among Asians, Caucasians, and Latin-Americans (Table 3). No significant association was found between the CTLA-4 gene rs5742909 polymorphism and RA risk.
The results did not change after eliminating studies (32, 35, 42), which did not meet HWE, suggesting that all data arising from the meta-analysis were trustworthy and stable. These results of sensitivity analysis also indicated that the data were stable and credible. Finally, Egger's and Begg's tests did not reveal publication bias.
Discussion
In our study, we conducted a meta-analysis to determine whether SNPs in the CTLA-4, CD80/86, and CD28 genes were associated with RA susceptibility. In addition, our meta-analysis was conducted by combining eight published case-control studies with our study to provide more reliable conclusions. The results indicated that both CTLA-4 rs231775 and rs3087243 reduced the risk of RA. Previously, five meta-analyses (18, 48–51) reported the relationship between CTLA-4 gene polymorphisms and the risk of RA. (1) Daha et al. (51) revealed that CTLA4 gene rs3087243 polymorphisms decreased the risk of RA among East Asians and Caucasians. After including studies from 2009, the present meta-analysis confirmed that rs3087243 polymorphisms decreased the RA susceptibility in Asians, Caucasians, and Latin-Americans populations. (2) Tang et al. (18), Li et al. (49), and Lee et al. (50) all reported that CTLA4 gene rs231775 polymorphisms increased the risk of RA, which was contrary to our results. All studies included in the above three meta-analyses were updated to 2013, whereas the present meta-analysis included five additional case-controlled studies (increased to 1,433 cases and 1,842 controls). Unbiased epidemiological studies and massive samples of the predisposition of gene polymorphisms would offer a more profound understanding of the correlation between diseases and candidate genes. (3) Lee et al. (48) suggested that the CTLA-4 gene rs5742909 was not associated with RA risk, which was not consistent with our findings. In addition to including more citations, we could not extract valid data from one study (32) by Lee et al. Sensitivity analysis indicated that the present data on the three SNPs was trustworthy and robust.
To further characterize the role of CD28-CD80/CD86 and CTLA-4 in the development of RA, our study was initiated. The results indicated that CTLA-4 rs231775 gene polymorphisms decreased the risk of RA, whereas CTLA-4 rs16840252 and CD86 rs17281995 gene polymorphisms were not related to RA risk. In the pathogenesis of RA, genetic and environmental factors are apparently implicated. CTLA-4 impedes IL-2 production and IL-2 receptor expression, and inhibits the function of cytotoxic T-lymphocytes as a vital co-stimulatory molecule. There are two main mRNA transcripts in humans produced by the CTLA-4 gene. CTLA-4 is known as a homolog to CD28. When an immune response continues, CTLA-4 is upregulated and competes with CD28, leading to reduction of IL-2 production and inhibition of T-cell proliferation (52). RA is a complicated disease, and its clinical heterogeneity may account for many additional factors, including the decrease or existence of RF. Our stratification research indicated that the rs231775 TT genotypes and the rs16840252 GG genotypes were more significantly correlated with RF-positive subgroups than with RF-negative subgroups. The data suggested that these genotypes of CTLA-4 gene polymorphisms associated with RA risk might not be associated with a higher risk of yielding rheumatoid factor (RF). Previous genetic association studies yielded conflicting results. Different geographical environments, sample sizes, and study designs may explain these discrepancies.
Previous studies have shown that a soluble, recombinant, and completely humanized fusion protein-Abatacept (CTLA-4Ig) can bind to CD80 and CD86 on antigen presenting cells (APC), thus, blocking interaction with CD28 on T cells (53). In clinical trials, an intravenous (IV) or subcutaneous (SC) Abatacept regimen has beneficial effects on RA signs and symptoms, disease activity, structural damage progression, and body function. Moreover, the current evidence suggested that Abatacept was a useful treatment option for patients with rheumatoid arthritis (54). Therefore, CTLA-4 could be used as a therapeutic target for RA, which proved that the coding gene of CTLA-4 is related to the risk of RA.
There were some limitations in our study. First, due to the lack of corresponding data, subgroups of some confounding factors could not be analyzed. Second, owing to confounding factors, the results of our research were based on unadjusted evaluations. Third, our research cohorts were merely Asians, so it is necessary to study other racial groups. Fourth, due to limited sample sizes, the conclusions obtained by stratification analyses of the rs231775/rs16840252/rs17281995 polymorphisms should be interpreted cautiously. Fifth, it is essential for researchers to investigate clinical cases in further studies to support our results. Finally, all these five genetic models of inheritance were used in the study; hence, type I errors may have appeared as lacking correction for multiple testing.
In conclusion, based on our retrospective analyses, the CTLA-4 rs231775 gene polymorphisms decreased the risk of RA, whereas the CTLA-4 rs16840252 and CD86 rs17281995 gene polymorphisms were not related to RA risk. The present meta-analysis indicated that CTLA-4 rs231775 and rs3087243 gene polymorphisms decreased the risk of RA. Additional investigation of clinical cases should be performed to verify these analytical results.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
RL and YZ designed the study. ZY and YC collected samples. HY and XW conceived and conducted the experiments. RL, WL, and XZ analyzed the data and wrote the manuscript. All authors reviewed the manuscript and gave final approval for the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- CTLA-4
cytotoxic T-lymphocyte-associated protein 4
- CD
costimulatory molecule
- RA
rheumatoid arthritis
- OR
odds ratio
- CI
confidence interval
- HWE
Hardy-Weinberg equilibrium
- SNP
single nucleotide polymorphism.
Footnotes
Funding. This study was supported in part by Changzhou High-Level Medical Talents Training Project (2016CZLJ011) and the Changzhou Sci & Tech Program (CJ20190066).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.598076/full#supplementary-material
References
- 1.Harris ED, Jr. Rheumatoid arthritis. pathophysiology and implications for therapy. N Engl J Med. (1990) 322:1277–89. 10.1056/NEJM199005033221805 [DOI] [PubMed] [Google Scholar]
- 2.van der Helm-van Mil AH, Toes RE, Huizinga TW. Genetic variants in the prediction of rheumatoid arthritis. Ann Rheum Dis. (2010) 69:1694–6. 10.1136/ard.2009.123828 [DOI] [PubMed] [Google Scholar]
- 3.Alegre ML, Frauwirth KA, Thompson CB. T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. (2001) 1:220–8. 10.1038/35105024 [DOI] [PubMed] [Google Scholar]
- 4.Gotsman I, Sharpe AH, Lichtman AH. T-cell costimulation and coinhibition in atherosclerosis. Circ Res. (2008) 103:1220–31. 10.1161/CIRCRESAHA.108.182428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luterek-Puszynska K, Malinowski D, Paradowska-Gorycka A, Safranow K, Pawlik A. CD28, CTLA-4 and CCL5 gene polymorphisms in patients with rheumatoid arthritis. Clin Rheumatol. (2017) 36:1129–35. 10.1007/s10067-016-3496-2 [DOI] [PubMed] [Google Scholar]
- 6.Maszyna F, Hoff H, Kunkel D, Radbruch A, Brunner-Weinzierl MC. Diversity of clonal T cell proliferation is mediated by differential expression of CD152 (CTLA-4) on the cell surface of activated individual T lymphocytes. J Immunol. (2003) 171:3459–66. 10.4049/jimmunol.171.7.3459 [DOI] [PubMed] [Google Scholar]
- 7.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. (2002) 2:116–26. 10.1038/nri727 [DOI] [PubMed] [Google Scholar]
- 8.Kucuk ZY, Charbonnier LM, McMasters RL, Chatila T, Bleesing JJ. CTLA-4 haploinsufficiency in a patient with an autoimmune lymphoproliferative disorder. J Allergy Clin Immunol. (2017) 140:862–4. e4. 10.1016/j.jaci.2017.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Liang WB, Gao LB, Pan XM, Chen TY, Wang YY, et al. CTLA4 and CD86 gene polymorphisms and susceptibility to chronic obstructive pulmonary disease. Hum Immunol. (2010) 71:1141–6. 10.1016/j.humimm.2010.08.007 [DOI] [PubMed] [Google Scholar]
- 10.Ewing MM, Karper JC, Abdul S, de Jong RC, Peters HA, de Vries MR, et al. T-cell co-stimulation by CD28-CD80/86 and its negative regulator CTLA-4 strongly influence accelerated atherosclerosis development. Int J Cardiol. (2013) 168:1965–74. 10.1016/j.ijcard.2012.12.085 [DOI] [PubMed] [Google Scholar]
- 11.Kusztal M, Koscielska-Kasprzak K, Drulis-Fajdasz D, Magott-Procelewska M, Patrzalek D, Janczak D, et al. The influence of CTLA-4 gene polymorphism on long-term kidney allograft function in Caucasian recipients. Transpl Immunol. (2010) 23:121–4. 10.1016/j.trim.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 12.Scalapino KJ, Daikh DI. CTLA-4: a key regulatory point in the control of autoimmune disease. Immunol Rev. (2008) 223:143–55. 10.1111/j.1600-065X.2008.00639.x [DOI] [PubMed] [Google Scholar]
- 13.Fattah SA, Ghattas MH, Saleh SM, Abo-Elmatty DM. Cytotoxic T-lymphocyte-associated protein 4 gene polymorphism is related to rheumatoid arthritis in Egyptian population. Arch Physiol Biochem. (2017) 123:50–3. 10.1080/13813455.2016.1230135 [DOI] [PubMed] [Google Scholar]
- 14.Elshazli R, Settin A, Salama A. Cytotoxic T lymphocyte associated antigen-4 (CTLA-4) +49 A>G gene polymorphism in Egyptian cases with rheumatoid arthritis. Gene. (2015) 558:103–7. 10.1016/j.gene.2014.12.046 [DOI] [PubMed] [Google Scholar]
- 15.Torres-Carrillo N, Ontiveros-Mercado H, Torres-Carrillo NM, Parra-Rojas I, Rangel-Villalobos H, Ramirez-Duenas MG, et al. The−319C/+49G/CT60G haplotype of CTLA-4 gene confers susceptibility to rheumatoid arthritis in Mexican population. Cell Biochem Biophys. (2013) 67:1217–28. 10.1007/s12013-013-9640-6 [DOI] [PubMed] [Google Scholar]
- 16.Liu CP, Jiang JA, Wang T, Liu XM, Gao L, Zhu RR, et al. CTLA-4 and CD86 genetic variants and haplotypes in patients with rheumatoid arthritis in southeastern China. Genet Mol Res. (2013) 12:1373–82. 10.4238/2013.April.25.8 [DOI] [PubMed] [Google Scholar]
- 17.AlFadhli S. Overexpression and secretion of the soluble CTLA-4 splice variant in various autoimmune diseases and in cases with overlapping autoimmunity. Genet Test Mol Biomarkers. (2013) 17:336–41. 10.1089/gtmb.2012.0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang MJ, Zhou ZB. Association of the CTLA-4 +49A/G polymorphism with rheumatoid arthritis in Chinese Han population. Mol Biol Rep. (2013) 40:2627–31. 10.1007/s11033-012-2349-6 [DOI] [PubMed] [Google Scholar]
- 19.El-Gabalawy HS, Robinson DB, Daha NA, Oen KG, Smolik I, Elias B, et al. Non-HLA genes modulate the risk of rheumatoid arthritis associated with HLA-DRB1 in a susceptible North American Native population. Genes Immun. (2011) 12:568–74. 10.1038/gene.2011.30 [DOI] [PubMed] [Google Scholar]
- 20.Benhatchi K, Jochmanova I, Habalova V, Wagnerova H, Lazurova I. CTLA4 exon1 A49G polymorphism in Slovak patients with rheumatoid arthritis and Hashimoto thyroiditis-results and the review of the literature. Clin Rheumatol. (2011) 30:1319–24. 10.1007/s10067-011-1752-z [DOI] [PubMed] [Google Scholar]
- 21.Plant D, Flynn E, Mbarek H, Dieude P, Cornelis F, Arlestig L, et al. Investigation of potential non-HLA rheumatoid arthritis susceptibility loci in a European cohort increases the evidence for nine markers. Ann Rheum Dis. (2010) 69:1548–53. 10.1136/ard.2009.121020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz-Valle JF, Valle Y, Padilla-Gutierrez JR, Parra-Rojas I, Rangel-Villalobos H, Vazquez del Mercado M, et al. The +49A>G CTLA-4 polymorphism is associated with rheumatoid arthritis in Mexican population. Clin Chim Acta. (2010) 411:725–8. 10.1016/j.cca.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 23.Barton A, Eyre S, Ke X, Hinks A, Bowes J, Flynn E, et al. Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum Mol Genet. (2009) 18:2518–22. 10.1093/hmg/ddp177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker EJ, Hirschfield GM, Xu C, Lu Y, Liu X, Lu Y, et al. CTLA4/ICOS gene variants and haplotypes are associated with rheumatoid arthritis and primary biliary cirrhosis in the Canadian population. Arthritis Rheum. (2009) 60:931–7. 10.1002/art.24412 [DOI] [PubMed] [Google Scholar]
- 25.Costenbader KH, Chang SC, De Vivo I, Plenge R, Karlson EW. Genetic polymorphisms in PTPN22, PADI-4, and CTLA-4 and risk for rheumatoid arthritis in two longitudinal cohort studies: evidence of gene-environment interactions with heavy cigarette smoking. Arthritis Res Ther. (2008) 10:R52. 10.1186/ar2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukahara S, Iwamoto T, Ikari K, Inoue E, Tomatsu T, Hara M, et al. CTLA-4 CT60 polymorphism is not an independent genetic risk marker of rheumatoid arthritis in a Japanese population. Ann Rheum Dis. (2008) 67:428–9. 10.1136/ard.2007.079186 [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi F, Kawasugi K, Mori M, Nakaue N, Kobayashi N, Kuwata S, et al. The genetic contribution of CTLA-4 dimorphisms in promoter and exon 1 regions in Japanese patients with rheumatoid arthritis. Scand J Rheumatol. (2006) 35:154–5. 10.1080/03009740500407651 [DOI] [PubMed] [Google Scholar]
- 28.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. (2005) 77:1044–60. 10.1086/498651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suppiah V, O'Doherty C, Heggarty S, Patterson CC, Rooney M, Vandenbroeck K. The CTLA4+49A/G and CT60 polymorphisms and chronic inflammatory arthropathies in Northern Ireland. Exp Mol Pathol. (2006) 80:141–6. 10.1016/j.yexmp.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 30.Lei C, Dongqing Z, Yeqing S, Oaks MK, Lishan C, Jianzhong J, et al. Association of the CTLA-4 gene with rheumatoid arthritis in Chinese Han population. Eur J Hum Genet. (2005) 13:823–8. 10.1038/sj.ejhg.5201423 [DOI] [PubMed] [Google Scholar]
- 31.Orozco G, Torres B, Nunez-Roldan A, Gonzalez-Escribano MF, Martin J. Cytotoxic T-lymphocyte antigen-4-CT60 polymorphism in rheumatoid arthritis. Tissue Antigens. (2004) 64:667–70. 10.1111/j.1399-0039.2004.00318.x [DOI] [PubMed] [Google Scholar]
- 32.Liu MF, Wang CR, Chen PC, Lin TL. CTLA-4 gene polymorphism in promoter and exon-1 regions is not associated with Chinese patients with rheumatoid arthritis. Clin Rheumatol. (2004) 23:180–1. 10.1007/s10067-003-0776-4 [DOI] [PubMed] [Google Scholar]
- 33.Barton A, Jury F, Eyre S, Bowes J, Hinks A, Ward D, et al. Haplotype analysis in simplex families and novel analytic approaches in a case-control cohort reveal no evidence of association of the CTLA-4 gene with rheumatoid arthritis. Arthritis Rheum. (2004) 50:748–52. 10.1002/art.20118 [DOI] [PubMed] [Google Scholar]
- 34.Lee CS, Lee YJ, Liu HF, Su CH, Chang SC, Wang BR, et al. Association of CTLA4 gene A-G polymorphism with rheumatoid arthritis in Chinese. Clin Rheumatol. (2003) 22:221–4. 10.1007/s10067-003-0720-7 [DOI] [PubMed] [Google Scholar]
- 35.Lee YH, Choi SJ, Ji JD, Song GG. No association of polymorphisms of the CTLA-4 exon 1(+49) and promoter(-318) genes with rheumatoid arthritis in the Korean population. Scand J Rheumatol. (2002) 31:266–70. 10.1080/030097402760375142 [DOI] [PubMed] [Google Scholar]
- 36.Vaidya B, Pearce SH, Charlton S, Marshall N, Rowan AD, Griffiths ID, et al. An association between the CTLA4 exon 1 polymorphism and early rheumatoid arthritis with autoimmune endocrinopathies. Rheumatology. (2002) 41:180–3. 10.1093/rheumatology/41.2.180 [DOI] [PubMed] [Google Scholar]
- 37.Hadj Kacem H, Kaddour N, Adyel FZ, Bahloul Z, Ayadi H. HLA-DQB1 CAR1/CAR2, TNFa IR2/IR4 and CTLA-4 polymorphisms in Tunisian patients with rheumatoid arthritis and Sjogren's syndrome. Rheumatology. (2001) 40:1370–4. 10.1093/rheumatology/40.12.1370 [DOI] [PubMed] [Google Scholar]
- 38.Milicic A, Brown MA, Wordsworth BP. Polymorphism in codon 17 of the CTLA-4 gene (+49 A/G) is not associated with susceptibility to rheumatoid arthritis in British Caucasians. Tissue Antigens. (2001) 58:50–4. 10.1034/j.1399-0039.2001.580110.x [DOI] [PubMed] [Google Scholar]
- 39.Yanagawa T, Gomi K, Nakao EI, Inada S. CTLA-4 gene polymorphism in Japanese patients with rheumatoid arthritis. J Rheumatol. (2000) 27:2740–2. 10.1097/00124743-200012000-00009 [DOI] [PubMed] [Google Scholar]
- 40.Matsushita M, Tsuchiya N, Shiota M, Komata T, Matsuta K, Zama K, et al. Lack of a strong association of CTLA-4 exon 1 polymorphism with the susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese: an association study using a novel variation screening method. Tissue Antigens. (1999) 54:578–84. 10.1034/j.1399-0039.1999.540607.x [DOI] [PubMed] [Google Scholar]
- 41.Barton A, Myerscough A, John S, Gonzalez-Gay M, Ollier W, Worthington J. A single nucleotide polymorphism in exon 1 of cytotoxic T-lymphocyte-associated-4 (CTLA-4) is not associated with rheumatoid arthritis. Rheumatology. (2000) 39:63–6. 10.1093/rheumatology/39.1.63 [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Escribano MF, Rodriguez R, Valenzuela A, Garcia A, Garcia-Lozano JR, Nunez-Roldan A. CTLA4 polymorphisms in Spanish patients with rheumatoid arthritis. Tissue Antigens. (1999) 53:296–300. 10.1034/j.1399-0039.1999.530311.x [DOI] [PubMed] [Google Scholar]
- 43.Seidl C, Donner H, Fischer B, Usadel KH, Seifried E, Kaltwasser JP, et al. CTLA4 codon 17 dimorphism in patients with rheumatoid arthritis. Tissue Antigens. (1998) 51:62–6. 10.1111/j.1399-0039.1998.tb02947.x [DOI] [PubMed] [Google Scholar]
- 44.Kim YO, Kim HJ, Kim SK, Chung JH, Hong SJ. Association of the CD28/CTLA4/ICOS polymorphisms with susceptibility to rheumatoid arthritis. Clin Chem Lab Med. (2010) 48:345–53. 10.1515/CCLM.2010.074 [DOI] [PubMed] [Google Scholar]
- 45.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, III, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. (2010) 62:2569–81. 10.1002/art.27584 [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Yin J, Wang L, Wang X, Shi Y, Shao A, et al. Interleukin 1B rs16944 G>A polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Clin Biochem. (2013) 46:1469–73. 10.1016/j.clinbiochem.2013.05.050 [DOI] [PubMed] [Google Scholar]
- 47.Hedges LV, Pigott TD. The power of statistical tests in meta-analysis. Psychol Methods. (2001) 6:203–17. 10.1037/1082-989X.6.3.203 [DOI] [PubMed] [Google Scholar]
- 48.Lee YH, Bae SC, Song GG. Association between the CTLA-4, CD226, FAS polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Hum Immunol. (2015) 76:83–9. 10.1016/j.humimm.2015.01.023 [DOI] [PubMed] [Google Scholar]
- 49.Li X, Zhang C, Zhang J, Zhang Y, Wu Z, Yang L, et al. Polymorphisms in the CTLA-4 gene and rheumatoid arthritis susceptibility: a meta-analysis. J Clin Immunol. (2012) 32:530–9. 10.1007/s10875-012-9650-y [DOI] [PubMed] [Google Scholar]
- 50.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Association between the CTLA-4 +49 A/G polymorphism and susceptibility to rheumatoid arthritis: a meta-analysis. Mol Biol Rep. (2012) 39:5599–605. 10.1007/s11033-011-1364-3 [DOI] [PubMed] [Google Scholar]
- 51.Daha NA, Kurreeman FA, Marques RB, Stoeken-Rijsbergen G, Verduijn W, Huizinga TW, et al. Confirmation of STAT4, IL2/IL21, and CTLA4 polymorphisms in rheumatoid arthritis. Arthritis Rheum. (2009) 60:1255–60. 10.1002/art.24503 [DOI] [PubMed] [Google Scholar]
- 52.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. (1995) 182:459–65. 10.1084/jem.182.2.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonelli M, Scheinecker C. How does abatacept really work in rheumatoid arthritis? Curr Opin Rheumatol. (2018) 30:295–300. 10.1097/BOR.0000000000000491 [DOI] [PubMed] [Google Scholar]
- 54.Blair HA, Deeks ED. Abatacept: a review in rheumatoid arthritis. Drugs. (2017) 77:1221–33. 10.1007/s40265-017-0775-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.