Abstract
A clear definition of developmentally incompetent preimplantation embryo (DIPE) in literature is still missing, while several scientific societies are discussing this challenging topic. From both a clinical and scientific perspective, the identification of embryos unfit for reproductive purpose is crucial. This aim should be pursued in light of all diagnostic technologies for embryo evaluation, encompassing also genetic analyses, of recent implementation in IVF. The Italian context is characterized by an unusual scenario: embryos can be discarded only if not viable and cannot be used for research purposes either. Therefore, thousands of embryos, diagnosed as affected and/or aneuploid as resulting from preimplantation genetic testing (PGT) and clinically not utilizable, are cryopreserved and stored indefinitely, with important psychological, legal, and financial implications. With the aim of updating the definition of DIPE, also on the basis of the embryo genetic status, the Italian Society of Embryology, Reproduction and Research (SIERR) and the Italian Society of Human Genetic (SIGU) reviewed the literature on this topic, found a consensus, and produced a list of relevant criteria.
Keywords: Developmentally incompetent preimplantation embryo, Preimplantation genetic testing, Affected embryo, Arrested embryo, Degenerated embryo
Introduction
How to define a developmentally incompetent preimplantation embryo (hereinafter referred to as DIPE) remains a challenge in the IVF practice. This definition is very important because it may have legal, psychological, ethical, and religious implications. A definition of DIPE is missing in the World Health Organization (WHO) International Committee for Monitoring Assisted Reproductive Technologies (ICMART) glossary of infertility [1, 2]. Some authors reported the use DIPE in their studies, however failing to propose a clear definition. Therefore, the only official document to date which might be used to this end is the Istanbul consensus of the European Society of Human Reproduction and Embryology (ESHRE) and Alpha-Scientists in Reproductive Medicine [3, 4]. In this document, an international panel of experts in clinical embryology stated that “a non-viable embryo is an embryo in which development has been arrested for at least 24 h, or in which all the cells have degenerated or lysed.” In 2018, the American Society for Reproductive Medicine (ASRM) also released its experts’ opinion on “blastocyst culture and transfer” [5], where they stated that “reliable criteria to identify embryos destined to develop to viable blastocysts in vitro remain to be established,” although an “intense investigation to find markers” is ongoing.
Apparently, extended culture to the blastocyst stage, possibly up to day 7 [6, 7], is per se the only practice to identify DIPE by negative selection. In fact, “blastomere fragmentation, degeneration, mitotic arrest, and multinucleation” during early cleavages might associate with a lower competence [8], but they are insufficient to clearly identify non-viable embryos. In 2009, Gavrilov and colleagues defined as non-viable “those embryos that had arrested or failed to divide normally and/or were morphologically abnormal,” thereby supporting that embryo morphological evaluation might suggest the absence of reproductive competence [9]. However, currently, we know that “blastocysts that rate poorly using conventional scoring can result in normal live births, indicating that ART clinics should re-evaluate the threshold criteria used for blastocyst viability” as Morbeck stated in 2017 [10]. The “reduced but modest live birth rates” of poor-quality blastocysts [11] become an even more important issue in light of Hammond’s recent data, which show “significant disagreement for decision to freeze borderline blastocysts among embryologists” [12]. In summary, any embryo that develops to the blastocyst stage, regardless of its morphological quality (including blastocysts graded < BB according to Gardner and Schoolcraft’s classification [13]) and/or the presence of excluded/extruded cells [14–18], might be euploid-diploid and result in a healthy live birth if transferred [11, 19]. Moreover, an excessively sustained embryonic metabolism was found by some authors associated with reproductive incompetence, while embryos with more moderate metabolic rate displayed a higher competence [20, 21]. Apoptosis also may play a physiological role throughout preimplantation development [22, 23]. Conversely, meiotic aneuploidies, although most certainly not compatible with a live birth [24] (except for sex chromosome abnormalities and vital trisomies), only mildly affect embryo development and blastocyst quality [19, 25]. In other terms, the definition of DIPE is still largely unclear, while several new evidences have emerged, especially from genetic testing, in the last decades.
Some countries like Italy have specific laws (for Italy: Law 40/2004) protecting the status of the embryos. Specifically, embryos can only be produced for reproductive purposes and cannot be discarded, donated, or utilized for research. In other terms, only arrested or degenerated embryos can be discarded, while all remaining embryos must be cryopreserved for an indefinite period of time, even if not utilized for treatment. Thus, in Italy, couples are not allowed to make an independent embryo disposition decision (EDD) [26, 27], which is instead imposed by Law. Therefore, regretfully, enforcement of such a law occurs in disregard of crucial information on embryo viability and pregnancy outcome offered by conventional and more novel diagnostic technologies, including those based on genetic testing. In Italy, several thousand embryos diagnosed as affected and/or aneuploid as resulting from preimplantation genetic testing (PGT), and therefore not utilizable clinically, are cryopreserved and must be stored until a new law that will eventually re-define their status. This implies considerable costs for centers and a possible concern for patients. It was reported that the preference of the majority of the couples after a PGT cycle would be, if allowed, to donate affected/aneuploid embryos for research. Couples reported to be strongly motivated by an altruistic will to promote progress in IVF and/or stem cell research, thereby helping other couples to solve their infertility problem [27].
For these reasons, the Italian Society of Embryology, Reproduction and Research (SIERR) and the Italian Society of Human Genetic (SIGU) collaborated to review the definition of DIPE, while also introducing the concept of genetically incompetent embryos.
Material and methods
Study design, size, and duration
Five SIERR embryologists and two SIGU geneticists (a clinician and a biologist), representative of their respective scientific societies, worked as a committee in 6 board meetings through 2018 and 2019, in order to draft the document. The consensus was then approved by the steering committee of the two scientific societies and publicly released through their websites.
Participants/materials, setting, and methods
Over a hundred scientific articles and guidelines were selected through systematic search on the topic of DIPE. The most recent studies on embryo morphology and morphokinetics and the most recent studies reporting data produced with advanced PGT technologies allowed the committee to develop standard criteria to define DIPE.
Glossary
Current definitions and terms of use are derived from the revised glossary on Assisted Reproductive Terminology by the WHO-ICMART [1, 2] and the Istanbul consensus [3, 4].
SIERR and SIGU consensus
SIERR in collaboration with SIGU drafted a consensus document to define DIPE. The two societies agreed on which embryos should be considered not usable for reproduction purpose because they were unable to result in a live birth. These include developmentally arrested and degenerated embryos, zygotes with a number of pronuclei (PN) equal or higher than 3, and embryos affected from lethal genetic and/or chromosomal conditions. The destiny of such embryos is strictly dependent on the specific laws enforced in different countries. Yet, a consensus on their definition is pivotal to build evidence-based directives or future laws and promote research and progress in this important area of medicine and healthcare.
Background
The chance that preimplantation embryos implant in a receptive endometrium and result in a live birth is limited mainly by an impaired developmental/reproductive competence. It is well-known that whole chromosome maternal meiotic aneuploidies produced by advanced female age (> 35 years) [28] and/or lack of factors supporting embryo development [29] affect implantation ability. Unfortunately, though, tools to unquestionably define embryos’ competence are still not available. Embryo development may be affected by metabolic disorders, yet not fully understood or detectable in the context of the daily IVF practice [30]. Still, some abnormal patterns and timings of cell division can represent reliable markers of embryo quality associated with a higher risk for implantation failure. This was already evident from static morphological observations before the advent of time-lapse microscopy (TLM) in the last decade. However, continuous morphological monitoring increased the power of detection of previously described phenomena and the magnitude and precision of morphokinetic analyses [16, 17, 31, 32]. Nevertheless, unequivocal definition of embryo viability based on single morphokinetic features or complex algorithms remains elusive [33]. For instance, not even “direct unequal cleavage” (DUC), a phenomenon consisting of the division of cell directly into 3 daughter cells and that is visible only through TLM, does allow an a priori labelling of an embryo as “non-viable.” In fact, DUC can occur at any stage of early preimplantation development [34]. The resulting cleavage stage embryos are often affected by complex aneuploidies caused by tripolar mitosis [17, 31] and show a lower implantation rate [34]. Yet, although at a significantly lower rate with respect to control embryos, DUC-derived embryos might develop to blastocyst and give rise to normal euploidy and implantation rates when tested and transferred at this stage. The example of DUC highlights how important it is nowadays to complement morphological and morphokinetic evaluations with genetic and/or chromosomal testing to increase our prediction on embryo developmental and reproductive competence. At present, three bioptic specimens are used to test the genetic status of an embryo: (i) polar bodies from oocytes/zygotes, (ii) single blastomeres retrieved at the cleavage stage, and (iii) trophectoderm cells retrieved at the blastocyst stage [35, 36], with the latter representing the current gold standard approach [36]. Trophectoderm biopsy indeed ensures greater accuracy of genetic analysis and less impact on embryo implantation viability after biopsy [37].
The association between daily observations and embryo arrest
Day 1 observation (D1)
Male and female pronuclei appear almost simultaneously 5–12 h post-insemination (hp.i.) and disappear almost 20 hp.i. [38–41]. Whenever no pronuclei or a single pronucleus is visualized (0PN or 1PN) at the fertilization check, especially in the absence of a time-lapse incubator, a further visualization is required 1–2 h later. In any case, the absence of cleavage should be confirmed 24 h after the fertilization check.
Day 2 observation (D2)
The division of the fertilized oocyte into two blastomeres generally occurs starting at 20 hp.i., typically between 23 and 25 hp.i. [38, 41, 42]. Failure to undergo first cleavage between 44 and 46 hp.i. is suggestive of arrested development.
Day 3 observation (D3)
During preimplantation embryo development, the cell cycle becomes progressively slower. Therefore, with reference to D2 observation, in the absence of further mitotic cell division between 44 and 46 and 70–72 hp.i., the embryo can be considered arrested.
Day 5–7 observation (D5–D7)
If the embryo has not developed to the blastocyst stage by days 5–7 post-insemination, it should be considered arrested and non-viable. Clearly, blastocyst culture is the most direct and reliable strategy to assess embryo developmental competence, regardless of the nature of the observations (static or morphodynamic) conducted at the earliest stages.
Definitions
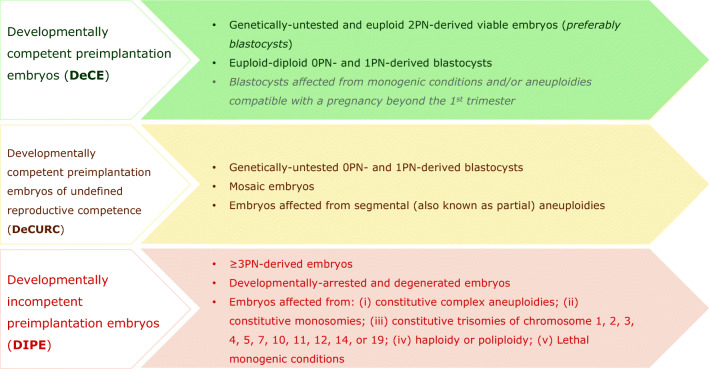
The definitions the authors agreed on and reported in this consensus paper are summarized in Fig. 1. Based on the chances of an embryo to establish a healthy live birth or cause serious adverse events if transferred, we defined three developmental categories: developmentally competent preimplantation embryos (from here onwards defined DeCE), developmentally competent preimplantation embryos of undefined reproductive competence (from here onwards defined DeCURC), and DIPE.
Fig. 1.
Summary of the definitions outlined in this consensus paper by the Italian Society of Embryology, Reproduction and Research (SIERR) and the Italian Society of Human Reproduction (SIGU) to categorize human embryos during IVF cycles as developmentally competent preimplantation embryos (DeCE), developmentally competent preimplantation embryos of undefined reproductive competence (DeCURC), and developmentally incompetent preimplantation embryos (DIPE). Embryos affected from monogenic conditions and/or aneuploidies compatible with a pregnancy beyond the 1st trimester (written among DeCE in gray) might result in a syndromic child therefore cannot be considered neither DeCURC nor DIPE. Only aneuploidies in the uniform range compatible with a meiotic constitutive abnormality were considered to define DIPE. The risk for clinically meaningful false-positive assessments is indeed minimal for them. Any reported chromosomal abnormality other than these outlines a DeCURC. PN pronuclei
Developmentally competent preimplantation embryos
A preimplantation embryo is considered “developmentally competent”:
When derived from a 2PN-zygote, viable at the moment of transfer or cryopreservation and either genetically untested or euploid. These characteristics are more predictive of a live birth when associated with a blastocyst stage embryo.
When derived from a 0PN- or 1PN-zygote and reported euploid-diploid at the blastocyst stage.
Blastocysts affected from monogenic conditions or aneuploidies compatible with a pregnancy beyond the first trimester. Although not compatible with a healthy live birth, these embryos might result in a syndromic child and therefore be reproductively competent.
Developmentally competent embryos of undefined reproductive competence
Some atypical cases deserve further investigation and still represent a gray area in the general question of embryonic competence. Such uncertainties derive from either persistent lack of relevant knowledge or from risk for false-positive results generated by embryological or molecular methodologies of embryo assessment. These embryos were defined as “developmentally competent but of undefined reproductive competence” and correspond to the following cases:
Genetically untested 0PN- and 1PN-derived blastocysts
Mosaic embryos
Embryos affected from segmental (also known as partial) aneuploidies
Developmentally incompetent preimplantation embryos
A preimplantation embryo is considered “developmentally incompetent” if unable to give rise to post-natal life, according to the below criteria:
If the nuclear status is different from diploidy. This evaluation is carried out already at the time of fertilization in day 1 by observing the presence of a number of PN other than 2. However, while ≥ 3PN-derived zygotes are considered DIPE, if 0PN- and 1PN-zygotes develop to the blastocyst stage, they might be categorized as DeCURC when genetically untested, DeCE when tested by PGT and diagnosed euploid-diploid, or DIPE when tested by PGT and showing any chromosomal constitution other than euploid-diploid.
If it does not undergo further cell division up to 48 h after the previous observation and is therefore defined developmentally arrested or degenerated.
If, at the blastocyst stage, it is affected from complex constitutive aneuploidies in the uniform range (trisomy and/or monosomy of 2 or more autosomes).
If, at the blastocyst stage, it is affected from a constitutive monosomy of an autosome in the uniform range.
If, at the blastocyst stage, it is affected from a constitutive trisomy in the uniform range incompatible with a viable pregnancy beyond the first trimester. The chromosomes relevant to this definition are 1, 2, 3, 4, 5, 7, 10, 11, 12, 14, and 19.
If, at the blastocyst stage, it is affected from polyploidy (triploidy/tetraploidy) or haploidy, as ascertained by validated genotyping-based technologies.
If, at the blastocyst stage, it is affected from constitutive in utero lethal monogenic conditions.
Of note, only aneuploidies in the uniform range compatible with a meiotic abnormality were considered to define DIPE. The risk for relevant clinically significant false-positive assessments is indeed minimal. Any chromosomal abnormalities other than those reported above are consistent with a condition of DeCURC.
Discussion
Considerations on embryo observation on day 1 post-insemination to define a DIPE
In the last decades, several groups aimed to define specific criteria to outline embryo viability from the earliest stages of development. Day 1 observation is a relevant step. Male and female PN appear almost simultaneously at 5–12 hp.i. and disappear at ~ 20 hp.i. [38–41]; therefore, zygotes’ morphological evaluation is typically performed at 17 ± 1 hp.i. Different scoring systems were proposed for zygote evaluation [43–45]. These schemes mainly score the number and position of the nucleolar precursor bodies (NPB) in each PN, the size and alignment of the PN, and the quality of the cytoplasm [46]. In fact, several studies reported a positive association between the presence of two equally sized centrally located PN, number and size of the NPBs, and blastocyst formation and/or pregnancy rates [3, 4, 47–52]. By definition, the presence of two polar bodies (PBs) and two PN with two distinct and visible membranes identifies a normally fertilized oocyte. Conversely, the absence of PN (0PN), the presence of a single PN (1PN), or the presence of extra PN (≥ 3PN) are indicative of abnormal fertilization (https://atlas.eshre.eu/es/14546650241811190), and several experts part of the ESHRE agreed that 0PN-, 1PN-, and ≥ 3PN-derived embryos should not be used for clinical purposes [53], but clear international guidelines or good practice recommendations are missing on this topic.
Tables 1 and 2 summarize the results of our systematic search for English manuscripts on the topic of 0PN- and 1PN-derived embryos in terms of reference, study size, workflow, kind of PN assessment (static or morphodynamic), euploidy rate (if assessed), and live birth rate.
Table 1.
Summary of the main published clinical evidence on 0PN-derived embryos. PN pronuclei, D3 day 3, SET single embryo transfer, DET double embryo transfer, LBR live birth rate
| Paper | Size | Workflow | PN assessment | Euploidy rate | LBR |
|---|---|---|---|---|---|
| Destouni 2018 |
268 cycles 301 0PN out of 2337 zygotes (12.9%) 49 D3 embryos out of 301 0PN zygotes (16.3%) |
- ICSI - D3 biopsy - Blastocyst vitrification - Genome-wide haplotyping |
Standard |
Euploid-diploid biparental: 27 out of 49 D3 embryos (55.1%) |
2 out of 13 euploid-diploid blastocyst SET (15%) |
| Liu 2016 |
4424 cycles 4966 0PN out of 43,949 zygotes (11.3%) |
- IVF - D3 culture |
Standard | - | 13 out of 70 untested SET/DET (19%) |
| Hondo 2019 | 11,588 cycles |
- IVF/ICSI - Blastocyst culture |
Standard | - | 22 out of 84 untested SET (26%) |
Table 2.
Summary of the main published clinical evidence on 1PN-derived embryos. PN pronuclei, D3 day 3, STRs short tandem repeats, aCGH array comparative genomic hybridization, qPCR quantitative polymerase chain reaction, SNP single nucleotide polymorphism, NGS next-generation sequencing, TLM mite lapse microscopy, SET single embryo transfer, LBR live birth rate
| Paper | Size | Workflow | PN assessment | Euploidy rate | LBR |
|---|---|---|---|---|---|
| Destouni 2018 |
268 cycles 132 1PN out of 2337 zygotes (6%) 15 D3 embryos out of 132 1PN zygotes (11%) |
- ICSI - D3 biopsy - Blastocyst vitrification - Genome-wide haplotyping |
Standard |
Euploid-diploid biparental: 5 out of 14 D3 embryos (36%) |
1 out of 1 euploid-diploid blastocyst SET (100%) |
| Mateo 2017 |
64 cycles 115 1PN-derived D3 embryos |
- ICSI - D3 biopsy - Blastocyst vitrification - aCGH |
TLM |
Euploid: 15 out of 88 D3 embryos (17%) |
1 out of 2 euploid blastocyst SETs (50%) |
| Capalbo 2017 |
678 cycles 200 1PN out of 3785 zygotes (5%) 13 blastocysts out of 200 1PN zygotes (6.5%) |
- ICSI - TE biopsy - Blastocyst vitrification - qPCR (for aneuploidies) plus SNP-array/NGS (for ploidy) |
Standard/TLM |
Euploid-diploid: 3 out of 13 blastocysts (23%) |
1 out of 2 euploid-diploid blastocyst SETs (50%) |
| Bradley 2017 |
4272 cycles 1192 1PN zygotes 106 blastocysts out of 1192 1PN zygotes (9%) |
- IVF/ICSI - TE biopsy - Blastocyst vitrification - aCGH/NGS (for aneuploidies) plus STRs analysis among female balanced blastocysts (for ploidy) |
Standard |
Euploid-diploid: 55 out of 106 blastocysts (52%) [4 female blastocysts were not tested for ploidy due to absence of parental controls] |
9 out of 26 euploid-diploid blastocyst SETs (35%) |
| Hondo 2019 | 11,588 cycles |
- IVF/ICSI - Blastocyst culture |
Standard | - | 14 out of 73 untested blastocyst SETs (19%) |
0PN-derived embryos
The prevalence of 0PN zygotes is 10–20% [54, 55], as a consequence of an unfertilized oocyte, the precocious disappearance of PN, or the lack of PN formation. In the latter two cases, the fertilization process may have occurred properly, and the related embryos might develop to the blastocyst stage and potentially implant [54–56]. Specifically, Basile et al. outlined that PN might disappear as early as 6 hp.i. or appear as late as 30 hp.i. [57]. Therefore, a zygote might be miscategorized as a 0PN at 17 ± 1 hp.i., even if it is a normally fertilized 2PN embryo. This limitation of static observation might lead to discarding viable embryos. The use of TLM might help overcome this issue [32]. This important limitation of static morphological observation encouraged some groups to transfer 0PN-derived cleavage or blastocyst stage embryos. Although live birth rates variable between 15 and 26% were reported [54–56] (Table 1), the ESHRE recommends not to use them clinically [53]. In this consensus paper, we defined 0PN-derived blastocysts DeCURC if genetically untested and DeCE if tested and diagnosed euploid-diploid.
1PN-derived embryos
The prevalence of 1PN zygotes is 5–9% after conventional IVF or ICSI [55, 58–62]. Of note, although in normally fertilized oocytes, the two PN appear mostly simultaneously, it has been observed that in a small proportion of cases, they are asynchronous [46]. The implementation of TLM incubators helped us discern true 1PN zygotes from embryos that would have been miscategorized as mono-pronuclear via a standard morphological evaluation as a result of an asynchronous appearance of the PN or their fusion [32, 60]. Already in the early 1990s, 1PN-derived zygotes were analyzed and some of them were reported diploid [63, 64]. In particular, some differences were reported depending on the fertilization strategy used with a lower prevalence of diploid 1PN-derived cleavage stage embryos after ICSI (28%) than after conventional IVF (48.7%) [58]. The main explanation for this was the more frequent asymmetrical appearance of PN when IVF is adopted (3 times higher according to Nagy et al. [65]), in turn causing more often the misclassification of 2PN zygotes as 1PN. Nonetheless, in 2017, Bradley and colleagues still reported better outcomes for 1PN-derived zygotes obtained after conventional IVF than after ICSI [61]. In general though, whichever fertilization technique is adopted, ≥ 5% of 1PN zygotes can develop to the blastocyst stage. Therefore, in the last decade, several groups adopted the most recent genetic technologies (genome-wide haplotyping, aCGH, SNP-array, NGS) to rescue these embryos for clinical use, by discriminating the euploid from the aneuploid ones [55, 60, 61, 66]. The rate of clinically usable 1PN-derived embryos ranged 17–52% and their live birth rates were high enough to justify their clinical use. This could suggest a further update of the ESHRE guidelines [53] and led to the recommendation of testing 1PN-derived blastocysts with advanced genetic technologies. However, such option should be explored at least initially in an experimental context, subject to couples’ informed consent, and in the absence of sibling normally fertilized 2PN embryos (Table 2). As for 0PN-derived blastocysts, also 1PN-derived blastocysts were defined DeCURC if genetically untested and DeCE if tested and diagnosed euploid-diploid.
≥ 3PN-derived embryos
Given that the prevalence of triploidy is 1–3% among spontaneous conceptions [67] and 15–18% among miscarriages [68] and that ≥3PN zygotes are supposed to carry a triploid or polyploid chromosomal constitution, these embryos should not be transferred because of their higher probability to result in a miscarriage or molar pregnancy [58]. Indeed, the ESHRE suggested to discard them as not viable [53]. The scenarios leading to 3PN-related abnormal fertilization are digyny (i.e., the presence of two maternal haploid chromosome sets plus one paternal haploid set), which occurs when the fertilized oocyte is diploid due to an error in meiosis, and diandry (i.e., the presence of two paternal haploid chromosome sets plus one maternal haploid set), which occurs when two haploid sperms or one diploid sperm fertilize a haploid oocyte. Although ICSI prevents polyspermy, 3PN zygotes are visualized even when this fertilization technique is adopted.
Due to the aforementioned risks related with the transfer of ≥ 3PN embryos, there is no study that, to the best of our knowledge, analyzed their clinical outcomes and just a few studies provided data to investigate these embryos in general [69, 70]. Some authors reported cases of 3PN-derived blastocysts whose CCT data were compatible with a normal chromosomal constitution, but resulted also in a significantly higher prevalence of uniparental disomy (UPD) [71–73], yet none of these embryos was transferred. In conclusion, the limited evidence in favor of the clinical use of ≥ 3PN-derived embryos and the considerable risks for the patients demanded a careful judgment; hence, these embryos were defined DIPE in this consensus document.
Considerations on genetic and chromosomal abnormalities to define a DIPE
The definition of the genetic state of the embryo is critical in the classification of DIPE. All the alterations listed in this document are linked to pathological conditions that cause embryo arrest, failure to implant in the uterus, or early miscarriage. Some monogenic conditions may be compatible with an ongoing pregnancy, but are certainly incompatible with extrauterine life. The strong conservative criterion “incompatible with extrauterine life” was chosen to define which chromosomal or genetic anomalies define a DIPE. Very serious conditions that are compatible with an extrauterine survival, even if with a poor quality of life and/or an early death (viable syndromes, like the ones involving chromosomes 13, 18, and 21), were defined a DeCE in this consensus document, since they may still result in live births, although syndromic. The chromosomal abnormalities mentioned among DIPE have never been reported in live births, being instead frequently detected in products of conceptions (POCs) or at chorionic villi sampling, and inevitably leading to a non-viable pregnancy [74]. In particular, complex constitutive aneuploidies can cause embryo developmental arrest before implantation, while constitutive trisomy of chromosomes 1, 2, 3, 4, 5, 7, 10, 11, 12, 14, and 19 as well as a monosomy of any autosome is considered incompatible with a viable pregnancy beyond the first trimester. Instead, they are responsible for severe intrauterine growth restriction (chromosomes 2 and 7), blighted ovum (monosomies and trisomies), and early arrest of fetal heartbeat (trisomies). Finally, for trisomies of imprinted chromosomes (e.g., chromosomes 7, 11, 14), there is a minimal risk of trisomy correction (embryo rescue) which can cause UPD with the development of severe fetal pathologies or the birth of a newborn affected from genetic diseases (Prader-Willi/Angelman syndrome, Beckwith-Wiedemann/Silver-Russell syndrome). Nevertheless, in human blastocysts, the prevalence of UPD has been reported exceedingly rare (0.06%) [75].
The possibility to detect embryonic polyploidy adds an important element in the classification of DIPE, as these embryos rarely grow beyond the first gestational weeks (tetraploidy) or the first trimester (triploidy) [76]. Triploidy results from abnormal maternal or paternal genomic contributions to the conceptus. In triploid pregnancies, the parental origin of the extra genome determines the phenotype and outcomes of the fetus and placenta, with differential expression of maternal and paternal genes influencing normal embryonic development [77, 78]. When the third haploid genome is paternally derived, the typical presentation is a partial molar pregnancy (hydatidiform mole, HM), whereas when the third haploid genome is maternally derived, the presentation is a non-molar triploid pregnancy. Up to 5% of women with HM will subsequently require chemotherapy for persistent gestational trophoblastic neoplasia. Another type of molar pregnancy originates from a diploid embryo with a single parent genomic contribution, defined as a complete mole (CM). Unfortunately, with the technologies currently in use in PGT, the analysis of the parental origin of the embryonic genome is not routine. Should this assessment be easier to perform in the future, it would be possible to classify these diploid embryos with only paternally derived genomes as DIPE, avoiding the risk of obtaining a CM pregnancy with a risk of neoplastic degeneration as high as 15%.
Since it is not feasible to provide a definitive and exhaustive list of monogenic diseases that fall in the definition of DIPE after PGT-M, we have defined DIPE as those embryos affected from genetic conditions non-compatible with an extrauterine survival, for instance, incontinentia pigmenti in male embryos, Meckel’s syndrome, complete homozygous defect of the mitochondrial respiratory chain, Friedreich ataxia homozygous for point mutations, spinal muscular atrophy with no copies of the smn2 gene, and homozygous alpha thalassemia with fetal hydrops. All embryos affected from these conditions are at high risk of adverse outcomes during gestation, ranging from failure to implant in the uterus to early miscarriage, intrauterine fetal death, and malignancy.
Considerations on PGT-A and its limitations: implications for the definition of DeCURC
PGT-A at the blastocyst stage is, at present, the most reliable method to assess the cytogenetic constitution of preimplantation embryos. The primary analytical objective of this approach is to identify the presence of meiotically derived chromosomal aneuploidies, which is the main trait associated with the decline in natural fertility in humans [79]. The only technique to define the meiotic origin of chromosomal aneuploidies in a preimplantation embryos is SNP array with the support of parental DNA (i.e., karyomapping) [80]. However, several comprehensive chromosome testing technologies have shown high accuracy in detecting this type of aneuploidies when uniformly present in a single trophectoderm biopsy [81–83]. These include qPCR, aCGH, and SNP array without input from parental DNA and more recently targeted- and whole genome amplification–based NGS protocols with stringent bioinformatic algorithms for aneuploidy calling. Several studies showed that when multiple biopsies are collected from the same blastocyst (multifocal biopsy approach), uniform whole chromosome aneuploidies are consistently detected in all embryonic regions including the inner cell mass [81, 83]. The clinical positive predictive value of uniform aneuploidy classification at the blastocyst stage was also reported in two non-selection studies investigating the developmental potential of aneuploid embryos [82, 84]. In these studies, trophectoderm biopsies were collected during the fresh cycles but analyzed only after the embryo transfers had taken place, thus allowing an unbiased assessment of the reproductive potential of uniformly aneuploid embryos. Collectively, over 98% of uniformly aneuploid embryos that were transferred failed to result in a live birth, highlighting almost perfect concomitance between constitutive aneuploidies and developmental incompetence. The rare normal live births derived from aneuploid embryos are mainly related to inherent diagnostic limitations (mosaicism and technical false positive) that are typical of any form of genetic diagnosis, including those performed at the prenatal stages. Using a different study design, Popovic and colleagues reached similar conclusions. By comparing the chromosomal constitution of clinical trophectoderm biopsies and embryonic outgrowth on day 12 of development, they reported 100% concordance for aneuploidies detected in the uniform range with the cytogenetic analysis of the resulting embryonic outgrowths [85]. Based on this evidence, the detection of uniform constitutive aneuploidies in trophectoderm biopsies is almost certainly associated with embryo developmental incompetence, and the diagnostic result can be considered for embryo de-selection process when involving the specific chromosomal configurations highlighted above. Therefore, we have classified DIPE all those embryos affected from complex aneuploidies, monosomies, and trisomies not compatible with extrauterine life that are detected in the uniform range.
Compared with previous technologies, recent increase in NGS-based PGT-A applications on trophectoderm biopsies has provided greater analytical throughput and cost-effectiveness, extending the employment of PGT-A to far more couples than before. Chromosome copy number variations (CCNV) for whole chromosomes and large deletion/duplication regions (usually above 6 Mb) are identified using custom or commercial algorithms and software. The intrinsic higher sensitivity of NGS for intermediate CCNV, combined with the application of subjective criteria for aneuploidy classification, has led to inconsistency of results across different laboratories, resulting in an extraordinary controversy and debate about the accuracy of PGT-A to correctly detect mosaic aneuploidies in embryos. Unlike uniform aneuploidies, essentially of meiotic origin and present in all cells of the ensuing embryo [28, 86, 87], mosaicism originates from mitotic errors occurring during early cleavage events [74]. The consequence of this type of errors is the simultaneous presence of chromosomally distinct cell lines within the embryo, producing a diploid/aneuploid mosaic (both euploid and aneuploid cells present) or an aneuploid mosaic (cells carrying different aneuploidies) embryo. Following NGS analysis, the presence of intermediate CCNV falling between the disomy and aneuploidy thresholds can be interpreted as chromosomal mosaicism. However, as widely recognized, the lack of standardization in the analytical process employed and technical and biological limitations can severely impact the reliability of mosaic diagnosis derived from a single trophectoderm biopsy. In fact, evidence of inconsistent results was reported both in multifocal biopsy models and in the clinical setting, where the transfer of putative mosaic and fully euploid embryos resulted in similar reproductive and gestational/neonatal outcomes [81, 83, 88]. For these reasons, PGT-A results consistent with diploid/aneuploid mosaicism cannot be used to define DIPE, as they may be in part the result of analytical artifacts [88–90]. Embryos reported “mosaic” according to the current practice should be better called “embryos with a PGT-A result falling in the mosaic range” as suggested by Forman [91] or “embryos with intermediate copy number of individual chromosome” as suggested by Paulson and Treff [92]. Still, assuming that novel protocols might unequivocally identify the presence of both euploid and aneuploid cells in a trophectoderm biopsy, a diagnosis of chromosomal mosaicism is simply not feasible by definition because of the inherent sampling bias (i.e., a genuinely mosaic blastocyst would always result in different diagnoses on multiple trophectoderm biopsies). Based on all these considerations, mosaic embryos were categorized in this consensus as DeCURC.
Lastly, segmental (also known as partial) aneuploidies were the object of a recent comprehensive study carried out in blastocyst stage embryos [81]: (i) these chromosome abnormalities may be detected in about 3–6% of trophectoderm biopsies, affecting any chromosome arm; (ii) their prevalence is independent of maternal age; and (iii) in more than 60% of cases after the analysis of multifocal blastocyst biopsies (including the inner cell mass), they showed a configuration compatible with either a mitotic or artifactual origin. Based on this evidence, the clinical value to report segmental aneuploidies is subject to an ongoing investigation. Regardless, a tentative scheme has been already suggested, based on the size of the segmental aneuploidy and confirmation in two consecutive trophectoderm biopsies. Apparently, if the aneuploidy is larger than 80 Mb and consistent in two clinical samples, it is most probably of meiotic origin and therefore constitutive and present also in the inner cell mass. Future clinical data are required to shed light on this question. For this reason, embryos affected from segmental aneuploidies were classified as DeCURC.
Conclusion
The definition of degenerated and developmentally arrested embryos as non-viable does not have any ethical and/or scientific implication. Instead, the inclusion of both aneuploid embryos and embryos affected from severe monogenic conditions in this definition opens novel perspectives for those countries, like Italy, where these embryos must be cryopreserved indefinitely with all the inherent psychological, legal, and financial implications involving both couples and IVF centers. DIPE might be instead used for research (under couples’ informed consent and approval of an ethic committee) in the field of human development and for the production of embryonic stem cells (ESCs). The Italian situation is unusual also when it comes to ESCs. In fact, such cells may be obtained from other countries and used experimentally, but cannot be produced in Italy for the same purposes. Notably, according to a recent survey promoted by an Italian private IVF clinic, patients considered donation for research more acceptable than indefinite cryo-storage or disposal [27]. From a broader perspective, a recognized definition of DIPE based on embryological and the genetic evidences is essential (i) for clinicians to offer a more complete counseling to the couple with respect to the non-viability of their embryos; (ii) for law-making bodies and institutional ethic committees, to the aim of defining clinically viable and non-viable embryos; and (iii) for couples, to decide with increased awareness the fate of their DIPE based on current knowledge and the consensus of two scientific societies.
While for DIPE, the event of a false-positive classification is remote, DeCURC category includes embryos more subject to false-positive assessments, according to both embryological and molecular criteria. After comprehensive and evidence-based genetic and gynecologic counseling on the realistic short- and long-term prognosis of these embryos (“quoad vitam et quoad valetudinem”), patients shall always be given the possibility to make an independent EDD on the destiny of their embryos. In an experimental setting, DeCURC might even be transferred, while recognizing their negligible chance to result in a healthy live birth and/or the considerable risk of serious adverse events. Finally, among DeCE, we included also blastocysts affected from monogenic conditions and/or aneuploidies compatible with a pregnancy beyond the 1st trimester. As the condition of such embryos might be compatible with a live birth, although syndromic, a couple might even decide for their use after careful genetic and gynecologic counseling [93].
The definitions included in this manuscript are clearly subject to future updates based on the scientific and clinical evidence that might be produced in the coming years in this field.
Acknowledgments
We thank Attilio Anastasi, Emanuele Licata, and Francesca Gioia Klinger for their help in drafting the manuscript and discussing its content. We thank also Dr. Giovanni Coticchio for the constructive comments and general revision of the manuscript.
Authors’ contribution
DC, AC, CS, and LSF contributed equally to this work. All authors participated in drafting the manuscript, discussed the results, and contributed to the final article. LR, RC, MGM, and AN revised the article critically. RC, LR, and LDS coordinated the working group on behalf of SIERR and DZ on behalf of SIGU.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to report related with this manuscript.
Footnotes
Danilo Cimadomo, Antonio Capalbo, Catello Scarica, Laura Sosa Fernandez, Laura Rienzi, Rosanna Ciriminna, Maria Giulia Minasi, and Lucia De Santis on behalf of the Italian Society of Embryology, Reproduction and Research (SIERR)
Antonio Capalbo, Antonio Novelli and Daniela Zuccarello on behalf of the Italian Society of Human Genetics (SIGU)
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Danilo Cimadomo, Antonio Capalbo, Catello Scarica and Laura Sosa Fernandez contributed equally to this work.
References
- 1.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, et al. The international glossary on infertility and fertility care, 2017. Hum Reprod. 2017;32(9):1786–1801. doi: 10.1093/humrep/dex234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. The international glossary on infertility and fertility care, 2017. Fertil Steril. 2017;108(3):393–406. doi: 10.1016/j.fertnstert.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Alpha SIRM, ESHRE SIGE Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Reprod BioMed Online. 2011;22(6):632–646. doi: 10.1016/j.rbmo.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Alpha SiRM, ESHRE SIGoE The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26(6):1270–1283. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 5.ASRM Blastocyst culture and transfer in clinically assisted reproduction: a committee opinion. Fertil Steril. 2018;110(7):1246–1252. doi: 10.1016/j.fertnstert.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Hammond ER, Cree LM, Morbeck DE. Should extended blastocyst culture include day 7? Hum Reprod. 2018;33(6):991–997. doi: 10.1093/humrep/dey091. [DOI] [PubMed] [Google Scholar]
- 7.Tiegs AW, Sun L, Patounakis G, Scott RT. Worth the wait? Day 7 blastocysts have lower euploidy rates but similar sustained implantation rates as day 5 and day 6 blastocysts. Hum Reprod. 2019;34(9):1632–1639. doi: 10.1093/humrep/dez138. [DOI] [PubMed] [Google Scholar]
- 8.Alikani M, Munne S. Nonviable human pre-implantation embryos as a source of stem cells for research and potential therapy. Stem Cell Rev. 2005;1(4):337–343. doi: 10.1385/SCR:1:4:337. [DOI] [PubMed] [Google Scholar]
- 9.Gavrilov S, Prosser RW, Khalid I, MacDonald J, Sauer MV, Landry DW, Papaioannou VE. Non-viable human embryos as a source of viable cells for embryonic stem cell derivation. Reprod BioMed Online. 2009;18(2):301–308. doi: 10.1016/s1472-6483(10)60270-2. [DOI] [PubMed] [Google Scholar]
- 10.Morbeck DE. Blastocyst culture in the era of PGS and FreezeAlls: is a ‘C’ a failing grade? Hum Reprod Open. 2017;2017(3):hox017. doi: 10.1093/hropen/hox017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimadomo D, Soscia D, Vaiarelli A, Maggiulli R, Capalbo A, Ubaldi FM, Rienzi L. Looking past the appearance: a comprehensive description of the clinical contribution of poor-quality blastocysts to increase live birth rates during cycles with aneuploidy testing. Hum Reprod. 2019;34(7):1206–1214. doi: 10.1093/humrep/dez078. [DOI] [PubMed] [Google Scholar]
- 12.Hammond ER, Foong AKM, Rosli N, Morbeck DE. Should we freeze it? Agreement on fate of borderline blastocysts is poor and does not improve with a modified blastocyst grading system. Hum Reprod. 2020;35:1045–1053. doi: 10.1093/humrep/deaa060. [DOI] [PubMed] [Google Scholar]
- 13.Gardner DK, Schoolcraft B. In vitro culture of human blastocysts. In: Jansen R, Mortimer D, editors. Toward reproductive certainty: fertility and genetics beyond. London: Parthenon Publishing; 1999. pp. 378–88. [Google Scholar]
- 14.Lagalla C, Tarozzi N, Sciajno R, Wells D, Di Santo M, Nadalini M, et al. Embryos with morphokinetic abnormalities may develop into euploid blastocysts. Reprod BioMed Online. 2017;34(2):137–146. doi: 10.1016/j.rbmo.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Coticchio G, Lagalla C, Sturmey R, Pennetta F, Borini A. The enigmatic morula: mechanisms of development, cell fate determination, self-correction and implications for ART. Hum Reprod Update. 2019;25(4):422–438. doi: 10.1093/humupd/dmz008. [DOI] [PubMed] [Google Scholar]
- 16.McCollin A, Swann RL, Summers MC, Handyside AH, Ottolini CS. Abnormal cleavage and developmental arrest of human preimplantation embryos in vitro. Eur J Med Genet. 2020;63(2):103651. doi: 10.1016/j.ejmg.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Ottolini CS, Kitchen J, Xanthopoulou L, Gordon T, Summers MC, Handyside AH. Tripolar mitosis and partitioning of the genome arrests human preimplantation development in vitro. Sci Rep. 2017;7(1):9744. doi: 10.1038/s41598-017-09693-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaninovic N, Zhan Q, Norberg C, Ye Z, Clarke R, Rosenwaks Z. Blastomere extrusion and abnormal cleavage behavior in human embryos under time-lapse monitoring: possible way of embryo “self-correction”? Fertil Steril. 2016;106(3, SUPPLEMENT):E353. doi: 10.1016/j.fertnstert.2016.07.1003. [DOI] [Google Scholar]
- 19.Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, Nagy ZP, Ubaldi FM. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014;29(6):1173–1181. doi: 10.1093/humrep/deu033. [DOI] [PubMed] [Google Scholar]
- 20.Baumann CG, Morris DG, Sreenan JM, Leese HJ. The quiet embryo hypothesis: molecular characteristics favoring viability. Mol Reprod Dev. 2007;74(10):1345–1353. doi: 10.1002/mrd.20604. [DOI] [PubMed] [Google Scholar]
- 21.Leese HJ, Sturmey RG, Baumann CG, McEvoy TG. Embryo viability and metabolism: obeying the quiet rules. Hum Reprod. 2007;22(12):3047–3050. doi: 10.1093/humrep/dem253. [DOI] [PubMed] [Google Scholar]
- 22.Betts DH, Madan P. Permanent embryo arrest: molecular and cellular concepts. Mol Hum Reprod. 2008;14(8):445–453. doi: 10.1093/molehr/gan035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy K, Spanos S, Becker D, Iannelli P, Winston RM, Stark J. From cell death to embryo arrest: mathematical models of human preimplantation embryo development. Proc Natl Acad Sci U S A. 2001;98(4):1655–1660. doi: 10.1073/pnas.98.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heffner LJ. Advanced maternal age--how old is too old? N Engl J Med. 2004;351(19):1927–1929. doi: 10.1056/NEJMp048087. [DOI] [PubMed] [Google Scholar]
- 25.Alfarawati S, Fragouli E, Colls P, Stevens J, Gutierrez-Mateo C, Schoolcraft WB, et al. The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil Steril. 2011;95(2):520–524. doi: 10.1016/j.fertnstert.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Bruno C, Dudkiewicz-Sibony C, Berthaut I, Weil E, Brunet L, Fortier C, Pfeffer J, Ravel C, Fauque P, Mathieu E, Antoine JM, Kotti S, Mandelbaum J. Survey of 243 ART patients having made a final disposition decision about their surplus cryopreserved embryos: the crucial role of symbolic embryo representation. Hum Reprod. 2016;31(7):1508–1514. doi: 10.1093/humrep/dew104. [DOI] [PubMed] [Google Scholar]
- 27.Faustini F, Forte M, Capalbo A, Cimadomo D, Ubaldi FM, Rienzi L. The main will of the patients of a private Italian IVF clinic for their aneuploid/affected blastocysts would be donation to research: a currently forbidden choice. J Assist Reprod Genet. 2019;36(8):1555–1560. doi: 10.1007/s10815-019-01465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capalbo A, Hoffmann ER, Cimadomo D, Ubaldi FM, Rienzi L. Human female meiosis revised: new insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum Reprod Update. 2017;23(6):706–722. doi: 10.1093/humupd/dmx026. [DOI] [PubMed] [Google Scholar]
- 29.Bebbere D, Masala L, Albertini DF, Ledda S. The subcortical maternal complex: multiple functions for one biological structure? J Assist Reprod Genet. 2016;33(11):1431–1438. doi: 10.1007/s10815-016-0788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leese HJ, Guerif F, Allgar V, Brison DR, Lundin K, Sturmey RG. Biological optimization, the goldilocks principle, and how much is lagom in the preimplantation embryo. Mol Reprod Dev. 2016;83(9):748–754. doi: 10.1002/mrd.22684. [DOI] [PubMed] [Google Scholar]
- 31.McCoy RC, Newnham LJ, Ottolini CS, Hoffmann ER, Chatzimeletiou K, Cornejo OE, et al. Tripolar chromosome segregation drives the association between maternal genotype at variants spanning PLK4 and aneuploidy in human preimplantation embryos. Hum Mol Genet. 2018;27(14):2573–2585. doi: 10.1093/hmg/ddy147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apter S, Ebner T, Freour T, Guns Y, Kovacic B, Le Clef N, et al. Eshre Working group on Time-lapse technology: Good practice recommendations for the use of time-lapse technology. Hum Reprod Open. 2020;2020(2):hoaa008. doi: 10.1093/hropen/hoaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennetta F, Lagalla C, Borini A. Embryo morphokinetic characteristics and euploidy. Curr Opin Obstet Gynecol. 2018;30(3):185–196. doi: 10.1097/GCO.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 34.Zhan Q, Ye Z, Clarke R, Rosenwaks Z, Zaninovic N. Direct unequal cleavages: embryo developmental competence, genetic constitution and clinical outcome. PLoS One. 2016;11(12):e0166398. doi: 10.1371/journal.pone.0166398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cimadomo D, Rienzi L, Capalbo A, Rubio C, Innocenti F, Garcia-Pascual CM, et al. The dawn of the future: 30 years from the first biopsy of a human embryo. The detailed history of an ongoing revolution. Hum Reprod Update. 2020;26:453–473. doi: 10.1093/humupd/dmaa019. [DOI] [PubMed] [Google Scholar]
- 36.Kokkali G, Coticchio G, Bronet F, Celebi C, Cimadomo D, Goossens V, et al. ESHRE PGT consortium and SIG embryology good practice recommendations for polar body and embryo biopsy for PGT. Hum Reprod Open. 2020;2020(3):hoaa020. doi: 10.1093/hropen/hoaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott RT, Jr, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril. 2013;100(3):624–630. doi: 10.1016/j.fertnstert.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 38.Capmany G, Taylor A, Braude PR, Bolton VN. The timing of pronuclear formation, DNA synthesis and cleavage in the human 1-cell embryo. Mol Hum Reprod. 1996;2(5):299–306. doi: 10.1093/molehr/2.5.299. [DOI] [PubMed] [Google Scholar]
- 39.Nagy ZP, Liu J, Joris H, Devroey P, Van Steirteghem A. Time-course of oocyte activation, pronucleus formation and cleavage in human oocytes fertilized by intracytoplasmic sperm injection. Hum Reprod. 1994;9(9):1743–1748. doi: 10.1093/oxfordjournals.humrep.a138786. [DOI] [PubMed] [Google Scholar]
- 40.Payne D, Flaherty SP, Barry MF, Matthews CD. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod. 1997;12(3):532–541. doi: 10.1093/humrep/12.3.532. [DOI] [PubMed] [Google Scholar]
- 41.Coticchio G, Mignini Renzini M, Novara PV, Lain M, De Ponti E, Turchi D, et al. Focused time-lapse analysis reveals novel aspects of human fertilization and suggests new parameters of embryo viability. Hum Reprod. 2018;33(1):23–31. doi: 10.1093/humrep/dex344. [DOI] [PubMed] [Google Scholar]
- 42.Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26(10):2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 43.Scott LA, Smith S. The successful use of pronuclear embryo transfers the day following oocyte retrieval. Hum Reprod. 1998;13(4):1003–1013. doi: 10.1093/humrep/13.4.1003. [DOI] [PubMed] [Google Scholar]
- 44.Tesarik J, Greco E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear stage morphology. Hum Reprod. 1999;14(5):1318–1323. doi: 10.1093/humrep/14.5.1318. [DOI] [PubMed] [Google Scholar]
- 45.Scott L, Alvero R, Leondires M, Miller B. The morphology of human pronuclear embryos is positively related to blastocyst development and implantation. Hum Reprod. 2000;15(11):2394–2403. doi: 10.1093/humrep/15.11.2394. [DOI] [PubMed] [Google Scholar]
- 46.Machtinger R, Racowsky C. Morphological systems of human embryo assessment and clinical evidence. Reprod BioMed Online. 2013;26(3):210–221. doi: 10.1016/j.rbmo.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 47.Balaban B, Urman B, Isiklar A, Alatas C, Aksoy S, Mercan R, Mumcu A, Nuhoglu A. The effect of pronuclear morphology on embryo quality parameters and blastocyst transfer outcome. Hum Reprod. 2001;16(11):2357–2361. doi: 10.1093/humrep/16.11.2357. [DOI] [PubMed] [Google Scholar]
- 48.Rienzi L, Ubaldi F, Iacobelli M, Ferrero S, Minasi MG, Martinez F, Tesarik J, Greco E. Day 3 embryo transfer with combined evaluation at the pronuclear and cleavage stages compares favourably with day 5 blastocyst transfer. Hum Reprod. 2002;17(7):1852–1855. doi: 10.1093/humrep/17.7.1852. [DOI] [PubMed] [Google Scholar]
- 49.Zollner U, Zollner KP, Hartl G, Dietl J, Steck T. The use of a detailed zygote score after IVF/ICSI to obtain good quality blastocysts: the German experience. Hum Reprod. 2002;17(5):1327–1333. doi: 10.1093/humrep/17.5.1327. [DOI] [PubMed] [Google Scholar]
- 50.Nagy ZP, Dozortsev D, Diamond M, Rienzi L, Ubaldi F, Abdelmassih R, Greco E. Pronuclear morphology evaluation with subsequent evaluation of embryo morphology significantly increases implantation rates. Fertil Steril. 2003;80(1):67–74. doi: 10.1016/s0015-0282(03)00569-7. [DOI] [PubMed] [Google Scholar]
- 51.Wittemer C, Bettahar-Lebugle K, Ohl J, Rongieres C, Nisand I, Gerlinger P. Zygote evaluation: an efficient tool for embryo selection. Hum Reprod. 2000;15(12):2591–2597. doi: 10.1093/humrep/15.12.2591. [DOI] [PubMed] [Google Scholar]
- 52.Montag M, van der Ven H, German pronuclear morphology study G Evaluation of pronuclear morphology as the only selection criterion for further embryo culture and transfer: results of a prospective multicentre study. Hum Reprod. 2001;16(11):2384–2389. doi: 10.1093/humrep/16.11.2384. [DOI] [PubMed] [Google Scholar]
- 53.De los Santos MJ, Apter S, Coticchio G, Debrock S, Lundin K, Plancha CE, et al. Revised guidelines for good practice in IVF laboratories (2015) Hum Reprod. 2016;31(4):685–686. doi: 10.1093/humrep/dew016. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Wang XL, Zhang X, Shen CY, Zhang Z. Live births resulting from 0PN-derived embryos in conventional IVF cycles. J Assist Reprod Genet. 2016;33(3):373–378. doi: 10.1007/s10815-015-0644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Destouni A, Dimitriadou E, Masset H, Debrock S, Melotte C, Van Den Bogaert K, et al. Genome-wide haplotyping embryos developing from 0PN and 1PN zygotes increases transferrable embryos in PGT-M. Hum Reprod. 2018;33(12):2302–2311. doi: 10.1093/humrep/dey325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hondo S, Arichi A, Muramatsu H, Omura N, Ito K, Komine H, et al. Clinical outcomes of transfer of frozen and thawed single blastocysts derived from nonpronuclear and monopronuclear zygotes. Reprod Med Biol. 2019;18(3):278–283. doi: 10.1002/rmb2.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basile N, Morbeck D, Garcia-Velasco J, Bronet F, Meseguer M. Type of culture media does not affect embryo kinetics: a time-lapse analysis of sibling oocytes. Hum Reprod. 2013;28(3):634–641. doi: 10.1093/humrep/des462. [DOI] [PubMed] [Google Scholar]
- 58.Staessen C, Van Steirteghem AC. The chromosomal constitution of embryos developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod. 1997;12(2):321–327. doi: 10.1093/humrep/12.2.321. [DOI] [PubMed] [Google Scholar]
- 59.Kang HJ, Rosenwaks Z. Triploidy--the breakdown of monogamy between sperm and egg. Int J Dev Biol. 2008;52(5–6):449–454. doi: 10.1387/ijdb.082602hk. [DOI] [PubMed] [Google Scholar]
- 60.Mateo S, Vidal F, Parriego M, Rodriguez I, Montalvo V, Veiga A, et al. Could monopronucleated ICSI zygotes be considered for transfer? Analysis through time-lapse monitoring and PGS. J Assist Reprod Genet. 2017;34(7):905–911. doi: 10.1007/s10815-017-0937-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradley CK, Traversa MV, Hobson N, Gee AJ, McArthur SJ. Clinical use of monopronucleated zygotes following blastocyst culture and preimplantation genetic screening, including verification of biparental chromosome inheritance. Reprod BioMed Online. 2017;34(6):567–574. doi: 10.1016/j.rbmo.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 62.Capalbo A, Treff N, Cimadomo D, Tao X, Ferrero S, Vaiarelli A, Colamaria S, Maggiulli R, Orlando G, Scarica C, Scott R, Ubaldi FM, Rienzi L. Abnormally fertilized oocytes can result in healthy live births: improved genetic technologies for preimplantation genetic testing can be used to rescue viable embryos in in vitro fertilization cycles. Fertil Steril. 2017;108:1007–1015.e3. doi: 10.1016/j.fertnstert.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Staessen C, Janssenswillen C, Devroey P, Van Steirteghem AC. Cytogenetic and morphological observations of single pronucleated human oocytes after in-vitro fertilization. Hum Reprod. 1993;8(2):221–223. doi: 10.1093/oxfordjournals.humrep.a138026. [DOI] [PubMed] [Google Scholar]
- 64.Munne S, Tang YX, Grifo J, Cohen J. Origin of single pronucleated human zygotes. J Assist Reprod Genet. 1993;10(4):276–279. doi: 10.1007/BF01204942. [DOI] [PubMed] [Google Scholar]
- 65.Nagy ZP, Janssenswillen C, Janssens R, De Vos A, Staessen C, Van de Velde H, et al. Timing of oocyte activation, pronucleus formation and cleavage in humans after intracytoplasmic sperm injection (ICSI) with testicular spermatozoa and after ICSI or in-vitro fertilization on sibling oocytes with ejaculated spermatozoa. Hum Reprod. 1998;13(6):1606–1612. doi: 10.1093/humrep/13.6.1606. [DOI] [PubMed] [Google Scholar]
- 66.Capalbo A, Ottolini CS, Griffin DK, Ubaldi FM, Handyside AH, Rienzi L. Artificial oocyte activation with calcium ionophore does not cause a widespread increase in chromosome segregation errors in the second meiotic division of the oocyte. Fertil Steril. 2016;105(3):807–14.e2. doi: 10.1016/j.fertnstert.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 67.Jacobs PA, Angell RR, Buchanan IM, Hassold TJ, Matsuyama AM, Manuel B. The origin of human triploids. Ann Hum Genet. 1978;42(1):49–57. doi: 10.1111/j.1469-1809.1978.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 68.McFadden DE, Robinson WP. Phenotype of triploid embryos. J Med Genet. 2006;43(7):609–612. doi: 10.1136/jmg.2005.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joergensen MW, Labouriau R, Hindkjaer J, Stougaard M, Kolevraa S, Bolund L, et al. The parental origin correlates with the karyotype of human embryos developing from tripronuclear zygotes. Clin Exp Reprod Med. 2015;42(1):14–21. doi: 10.5653/cerm.2015.42.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li M, Xue X, Zhao W, Li W, Shi J. Effects of high three pro-nuclei (3PN) proportion incidence on clinical outcomes in the fresh cleavage-stage and blastocyst-stage embryo transfer (ET) cycles. Gynecol Endocrinol. 2017;33(1):53–56. doi: 10.1080/09513590.2016.1190817. [DOI] [PubMed] [Google Scholar]
- 71.Yao G, Xu J, Xin Z, Niu W, Shi S, Jin H, et al. Developmental potential of clinically discarded human embryos and associated chromosomal analysis. Sci Rep. 2016;6:23995. doi: 10.1038/srep23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grau N, Escrich L, Galiana Y, Meseguer M, Garcia-Herrero S, Remohi J, et al. Morphokinetics as a predictor of self-correction to diploidy in tripronucleated intracytoplasmic sperm injection-derived human embryos. Fertil Steril. 2015;104(3):728–735. doi: 10.1016/j.fertnstert.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 73.Xu J, Zhang M, Niu W, Yao G, Sun B, Bao X, et al. Genome-wide uniparental disomy screen in human discarded morphologically abnormal embryos. Sci Rep. 2015;5:12302. doi: 10.1038/srep12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grati FR, Gallazzi G, Branca L, Maggi F, Simoni G, Yaron Y. An evidence-based scoring system for prioritizing mosaic aneuploid embryos following preimplantation genetic screening. Reprod BioMed Online. 2018;36(4):442–449. doi: 10.1016/j.rbmo.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Gueye NA, Devkota B, Taylor D, Pfundt R, Scott RT, Jr, Treff NR. Uniparental disomy in the human blastocyst is exceedingly rare. Fertil Steril. 2014;101(1):232–236. doi: 10.1016/j.fertnstert.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 76.Gardner RJM, Sutherland GR, Schaffer LG. Chromosome abnormalities and genetic counseling. 4. New York: Oxford University Press; 2012. [Google Scholar]
- 77.McFadden DE, Kwong LC, Yam IY, Langlois S. Parental origin of triploidy in human fetuses: evidence for genomic imprinting. Hum Genet. 1993;92(5):465–469. doi: 10.1007/BF00216452. [DOI] [PubMed] [Google Scholar]
- 78.McFadden DE, Langlois S. Parental and meiotic origin of triploidy in the embryonic and fetal periods. Clin Genet. 2000;58(3):192–200. doi: 10.1034/j.1399-0004.2000.580306.x. [DOI] [PubMed] [Google Scholar]
- 79.Gruhn JR, Zielinska AP, Shukla V, Blanshard R, Capalbo A, Cimadomo D, Nikiforov D, Chan ACH, Newnham LJ, Vogel I, Scarica C, Krapchev M, Taylor D, Kristensen SG, Cheng J, Ernst E, Bjørn AMB, Colmorn LB, Blayney M, Elder K, Liss J, Hartshorne G, Grøndahl ML, Rienzi L, Ubaldi F, McCoy R, Lukaszuk K, Andersen CY, Schuh M, Hoffmann ER. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science. 2019;365(6460):1466–1469. doi: 10.1126/science.aav7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thornhill AR, Handyside AH, Ottolini C, Natesan SA, Taylor J, Sage K, et al. Karyomapping-a comprehensive means of simultaneous monogenic and cytogenetic PGD: comparison with standard approaches in real time for Marfan syndrome. J Assist Reprod Genet. 2015;32(3):347–356. doi: 10.1007/s10815-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Girardi L, Serdarogullari M, Patassini C, Poli M, Fabiani M, Caroselli S, Coban O, Findikli N, Boynukalin FK, Bahceci M, Chopra R, Canipari R, Cimadomo D, Rienzi L, Ubaldi F, Hoffmann E, Rubio C, Simon C, Capalbo A. Incidence, origin, and predictive model for the detection and clinical management of segmental aneuploidies in human embryos. Am J Hum Genet. 2020;106:525–534. doi: 10.1016/j.ajhg.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tiegs AW, Tao X, Zhan Y, Whitehead C, Kim J, Hanson B, et al. A multicenter, prospective, blinded, nonselection study evaluating the predictive value of an aneuploid diagnosis using a targeted next-generation sequencing-based preimplantation genetic testing for aneuploidy assay and impact of biopsy. Fertil Steril. 2020. 10.1016/j.fertnstert.2020.07.052. [DOI] [PubMed]
- 83.Popovic M, Dhaenens L, Boel A, Menten B, Heindryckx B. Chromosomal mosaicism in human blastocysts: the ultimate diagnostic dilemma. Hum Reprod Update. 2020;26:313–334. doi: 10.1093/humupd/dmz050. [DOI] [PubMed] [Google Scholar]
- 84.Scott RT, Jr, Ferry K, Su J, Tao X, Scott K, Treff NR. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study. Fertil Steril. 2012;97(4):870–875. doi: 10.1016/j.fertnstert.2012.01.104. [DOI] [PubMed] [Google Scholar]
- 85.Popovic M, Dhaenens L, Taelman J, Dheedene A, Bialecka M, De Sutter P, et al. Extended in vitro culture of human embryos demonstrates the complex nature of diagnosing chromosomal mosaicism from a single trophectoderm biopsy. Hum Reprod. 2019;34(4):758–769. doi: 10.1093/humrep/dez012. [DOI] [PubMed] [Google Scholar]
- 86.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 87.Ottolini CS, Newnham LJ, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK, Sage K, Summers MC, Thornhill AR, Housworth E, Herbert AD, Rienzi L, Ubaldi FM, Handyside AH, Hoffmann ER. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47(7):727–735. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goodrich D, Tao X, Bohrer C, Lonczak A, Xing T, Zimmerman R, et al. A randomized and blinded comparison of qPCR and NGS-based detection of aneuploidy in a cell line mixture model of blastocyst biopsy mosaicism. J Assist Reprod Genet. 2016;33(11):1473–1480. doi: 10.1007/s10815-016-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Capalbo A, Rienzi L. Mosaicism between trophectoderm and inner cell mass. Fertil Steril. 2017;107(5):1098–1106. doi: 10.1016/j.fertnstert.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 90.Capalbo A, Ubaldi FM, Rienzi L, Scott R, Treff N. Detecting mosaicism in trophectoderm biopsies: current challenges and future possibilities. Hum Reprod. 2017;32(3):492–498. doi: 10.1093/humrep/dew250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Forman EJ. Demystifying “mosaic” outcomes. Fertil Steril. 2019;111(2):253. doi: 10.1016/j.fertnstert.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 92.Paulson RJ, Treff N. Isn’t it time to stop calling preimplantation embryos “mosaic”? F&S Reports. 2020. 10.1016/j.xfre.2020.10.009. [DOI] [PMC free article] [PubMed]
- 93.Besser AG, Blakemore JK, Grifo JA, Mounts EL. Transfer of embryos with positive results following preimplantation genetic testing for monogenic disorders (PGT-M): experience of two high-volume fertility clinics. J Assist Reprod Genet. 2019;36(9):1949–1955. doi: 10.1007/s10815-019-01538-2. [DOI] [PMC free article] [PubMed] [Google Scholar]