Abstract
Effects of starter culture composed of Lactobacillus sakei, Pediococcus pentosaceus, Staphylococcus xylosus and Staphylococcus carnosus at the ratios (w/w) of 1:1:1:1 on bacterial community diversity and food safety of Chinese Cantonese sausages were demonstrated by high-throughput sequencing technology. At genus level, spoilage organisms and pathogenic bacteria such as Vibrio spp., Acinetobacter spp., Enterobacter spp., Yersinia spp. accounted for 54.13%, 10.01%, 6.94% and 5.35% of bacterial in the initial fermentation of spontaneous sausage, and the dominant bacteria of Lactobacillus spp. reached 84.61% on day 20. Accordingly, the total proportion of Pediococcus spp., Lactobacillus spp. and Staphylococcus spp. were present higher than 98% during fermentation in fermented sausage by starter culture inoculation, and Pediococcus spp. was dominant genus and increased from 53.53 to 74.09% during whole fermentation process. Moreover, the histamine accumulation was lower 84.17% in sausage fermented by starter culture inoculation than that of spontaneous sausage, suggesting that starter culture could decrease histamine accumulation of sausages significantly (P < 0.01). These results revealed that the starter culture inoculation was conducive to improve the microbial quality and food safety of Chinese Cantonese sausages.
Keywords: Bacterial community diversity, Food safety, High-throughput sequencing, Microbial quality, Starter culture
Introduction
Chinese Cantonese sausages as a Chinese typical fermented meat product is well preferred by most Chinese consumers, especially in Guangdong province in China, for its mellow and delicious flavor and its strong characteristics of beautiful appearance, bright color, thin skin and tender meat (Wang et al. 2015). The traditional Chinese Cantonese sausages is usually made with raw pork meat, salt, sucrose, China liquor, colorless soy sauce, white pepper, fresh ginger juice and nitrite (Cardinali et al. 2018), and then was fermented spontaneously for 25–30 days at 9–14 °C (Wang et al. 2018). The core of producing Cantonese sausages with high quality is the microorganisms which participate in fermentation and regulate the fermentation tendency. It has been reported that suitable starter cultures inoculation could enhance flavor, prolong shelf life and improve quality and safety of produce (Lu et al. 2010). In European countries, lactic acid bacteria(LAB)have been used in fermented sausages, such as Lactobacillus sakei and Pediococcus pentosans in Spanish fermented sausages (Hugas et al. 2002), Lactobacillus plantarum in Italian fermented sausages (di Gioia et al. 2016), which can metabolize carbohydrates to produce organic acids and volatile compounds (Domínguez et al. 2016; de Almeida et al. 2018), and have an ability to produce bacteriocins to inhibit pathogenic bacteria (Woraprayote et al. 2016). Staphylococcus spp. such as Staphylococcus carnosus and Staphylococcus xylosus have been inoculated in Italian-style salami. They exhibit nitrite reducatase activity and protolysis ability, which is conducive to the color improvement, unique flavor formation and delay rancidity of sausages (Blanco-Lizarazo et al. 2018). Due to the spontaneous fermentation which mainly relies on native bacterial population of raw materials, traditional Cantonese sausages is highly susceptible to the growth of undesirable microorganisms, resulting that a uniform product quality and its safety standards is not guaranteed. Therefore, biogenic amines, nitrites and other harmful substances has been detected in traditional Chinese Cantonese sausages recently (Zhang et al. 2017; Liu et al. 2018). Thus, it is essential to perform suitable starter cultures inoculation in Chinese Cantonese sausages to inhibit the growth of undesirable microorganisms by regulating microbial community structure and improve its food safety.
In the study, a starter culture composed of L. sakei, Pediococcus pentosaceus, S. xylosus and S. carnosus was inoculated in Chinese Cantonese sausages. The effects of the starter culture on microbial community structure by high-throughput sequencing technology and its contributions to the microbiological quality, nitrite depletion and histamine decrease, compared to spontaneous fermentation, were evaluated. Additionally, physical and chemical parameters of Chinese Cantonese sausages, namely pH and water activity, were also described.
Materials and methods
Starter culture
The starter culture composed of L. sakei, P. pentosaceus, S. xylosus and S. carnosus, which were cultured to 108–109 cfu/mL and then immediately prepared into freeze-drying powder respectively, at the ratio (w/w) of 1:1:1:1 used to manufacture Chinese Cantonese sausages was supplied by Meat-Processing Application Key Lab of Sichuan Province in China.
Chinese Cantonese sausages preparation and sampling
The Chinese Cantonese sausages were made with lean pork meat (750 g/kg), pork back fat (250 g/kg) and other ingredients including NaCl (20 g/kg), sucrose (50 g/kg), NaNO2 (150 mg/kg), China liquor (40 g/kg), fresh ginger juice (10 mL/kg) and colorless soy sauce (10 mL/kg). The lean pork meat and pork back fat was cut into approximately 2 × 1×0.5 cm pieces, and then was mixed thoroughly with other ingredients. After mixing thoroughly, the mixture was preserved at 4 °C for 24 h. Then the mixture was inoculated with 0.2% (w/w) starter culture labelled GWS and no inoculation labelled GWC as control. After inoculation, the mixture was stuffed into natural casings with a diameter of 10 cm. Raw sausages were fermented at 20 °C for 3 days with 90% relative humidity (RH), and then ripened at 15 °C with 70% RH for 27 days. The sausage sampling (50 g) was preformed in triplicate on 0, 10, 20 and 30 days and immediately stored at − 80 °C for subsequent analysis.
The pH, aw, nitrite concentration and histamine concentration measurement
Histamine concentration in sample was measured in triplicate according to the method reported by Wang et al. (2015) and the determination of the pH, aw and nitrite concentration was performed in triplicate according to the method described by Wang et al. (2018).
DNA extraction
Use the E.Z.N.A™ Mag-Bind Soil DNA Kit (OMEGA, USA) to extract the microbial DNA of samples and monitor DNA quality by agarose gel electrophoresis according to the methods as described by Wang et al. (2018).
Illumina high-throughput sequencing
The V4 region of bacterial 16S rRNA was amplified by PCR using the universal bacterial primers 515F and 806R with Illumina barcoded adapters and extracted DNA was used as the template (Caporaso et al. 2010; Wang et al. 2018). Two step-PCR technology was performed in which the first PCR step ran 25 cycles with untagged primers, and the second PCR step ran 5 cycles using tagged primers and the first step products as template. The PCR amplification was performed according to the methods reported by Wang et al. (2018). After purifying PCR products, sequencing was carried out on the Illumina MiSeq sequencing platform (Illumina, USA) at Sangon Biotech Co., Ltd (Shanghai, China) according to the standard protocol from the manufacturer.
Data analysis
Operational taxonomic unit (OTU) at an identity threshold of 97% using UPARSE software (Edgar 2010) and OTU taxonomy at 97% similarity with the Vegan package (Dixon 2003) were carried out to analyze the sequence data. Set 97% sequence similarity to use RDP calculate estimator values of Shannon index and Chao1 (http://pyro.cme.msu.edu/). Rarefaction curves, Good’s coverage, Simpson and Shannon diversity indices and Chao 1 richness were used to evaluate Alpha diversity (Grice et al. 2009). Microbial communities composition of samples was depicted by Principal component analysis (PCA) (Jiang et al. 2013). Venn diagram was performed to depict the similarity and difference in microbial communities among all samples (Liu et al. 2017).
Results and discussion
Effect of the starter culture on pH and aw in Cantonese sausages
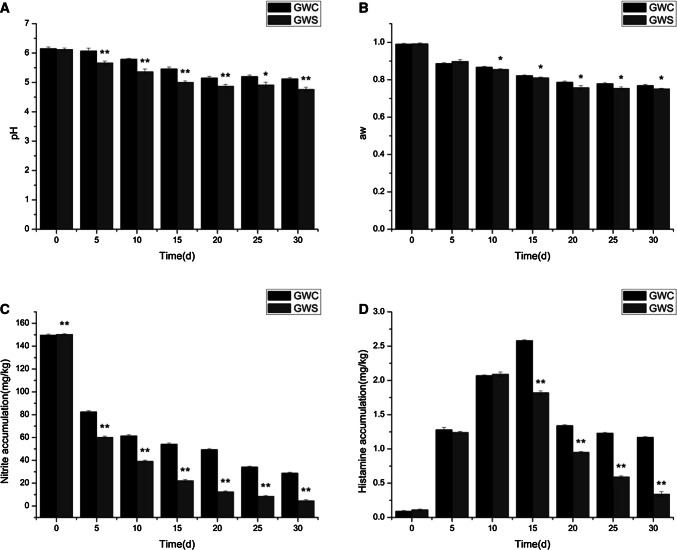
Changes of pH in GWS and GWC (as control) samples are shown in Fig. 1a. The starter culture inoculation had an significant effect (P < 0.01) on the pH in Cantonese sausages and a stronger acidification was found in GWS, compared with the control. During the first 15 days of fermentation, the pH decreased strongly from initial 6.12 to 5.00 in GWS sample and from initial 6.15 to 5.66 in GWC sample, respectively. On the 30th day, the pH in GWS and GWC samples was 4.76 and 5.12, respectively. It has been reported that L. sakei and P. pentosaceus have a strong organic acid production capacity (Oh and Jung 2015; Manini et al. 2016), which may be the reason for a stronger decline in pH in GWS, compared with spontaneous fermentation. The lowering of pH in Cantonese sausages during the fermentation can assist reducing nitrate to nitrite and then to nitric oxide, which contributes to red color of the meat achievement. In addition, the lowering of pH is beneficial to inhibition of food-borne pathogens, which can ensures hygienic stability. Thus, starter culture inoculation will be more beneficial for enhancement of hygienic quality and achievement of red color of Cantonese sausages than that of the spontaneous fermentation.
Fig. 1.
Effect of starter cultures on the pH value (a), water activity (b), nitrite accumulation (c), histamine accumulation (d) in fermented sausages during fermentation and ripening period
Changes in water activity (aw) of GWC and GWS samples during fermentation are given in Fig. 1b. The initial aw were 0.991 and 0.992 in GWC and GWS samples, and then gradually decreased to 0.769 and 0.751 along with the fermentation, respectively. On the 30th day, a slight lower aw with 2.34% was found than that of the control, which could be related to the decrease of water moisture content as a result of rapid growth of the bacterial strains inoculation. Similar results were found in fermented sausages with starter culture inoculation reported by Cavalheiro et al. (2019). It has been observed that some food-borne pathogenic bacteria such as Escherichia coli O157:H7 can survived in presence of organic acids but generally does not survive at a low aw value. Thus, the effective combined factors such as low pH and reduced aw in the preservation of cured meat products has already been employed (Woraprayote et al. 2016).
Effect of the starter culture on nitrite depletion and histamine accumulation in Cantonese sausages
The nitrite concentration in GWS and GWC samples during fermentation are shown in Fig. 1c. The starter culture inoculation had an significant effect (P < 0.05) on the nitrite depletion of Cantonese sausages and a lower level of nitrite was detected in GWS sample than that of the control during the whole fermentation process. The nitrite concentration in GWS sample decreased rapidly from 150.21 to 4.56 mg/kg at the end of ripening. Accordingly, nitrite concentration in GWC sample dropped to 28.81 mg/kg at the end of ripening with a higher level than that of GWS sample. It has been reported that L. sakei has the capacity to directly deplete nitrite accumulation in sausages (Kim et al. 2017). In this study, the starter culture inoculated in Cantonese sausages contains L. sakei that may be the reason for more nitrite depletion than that of the control.
Nitrite is still usually employed as an essential additive for its abilities of fixing color, improving texture and inhibiting the growth of food-borne pathogens in fermented meat industry (Honikel 2008). However, excessive quantities of nitrite consumption leads to methemoglobinemia and increases incidence of cancers, which are harmful to health (Xiao et al. 2018). It is stipulated that the nitrite amount in the final meat products must be controlled below 20 mg/kg (Huang et al. 2011). In this study, the finding indicates that starter culture inoculation contributed to nitrite depletion and safety improvement of Cantonese sausages, which is in agreement with those reported by Cheng (Cheng et al. 2018).
Changes in histamine accumulation in GWS and GWC samples during fermentation are shown in Fig. 1d. A substantial increase in histamine accumulation were observed both in GWS and GWC samples during the initial fermentation process, and the histamine accumulation reached maximum concentration of 2.09 mg/kg in GWS sample on day 10 and 2.58 mg/kg in GWC sample on day 15, respectively. Then the levels of histamine drop rapidly to 0.34 mg/kg in GWS sample and 1.17 mg/kg in GWC sample on day 30, respectively.
It follows that the starter culture inoculation contributed to reduce histamine accumulation. Recent discoveries pointed that biogenic amines can be found in nearly all types of fermented sausages which is formed by the decarboxylation of free histidine by microorganisms in sausages. Appropriate amount of biogenic amines play an irreplaceable role in the growth and development of body, but excessive intake can cause adverse reactions, especially for the presence of nitrite in food. Thus, it is necessary to control the biogenic amines accumulation in sausages. However, several lactic acid bacteria as fermenting microorganisms with decarboxylase activity, such as L. sakei, Lactobacillus brevis and Lactobacillus curvatus have been regarded as potential producers of histamine and other biogenic amines (Moracanin et al. 2015; Bartkiene et al. 2017). In this study, an increasing histamine accumulation was also checked at the fist 10 days in starter culture inoculation sample that might be due to its increasing growth of L. sakei form starter culture. Accordingly, S. xylosus is provided with oxidase activity that contributes to histamine degradation (Martuscelli et al. 2000). In this study, the starter culture was inoculated in Cantonese sausages containing S. xylosus that might be the reason for a lower level of histamine found in starter culture inoculation sample than that of the spontaneous fermentation at the end of ripening of sausages.
Characteristics of the microbial sequencing data comparison
A total of 3061,220 and 284,914 high quality bacterial tags in GWS and GWC samples, respectively, was generated from 8 examined samples sets, namely GWS1 (0 day), GWS2 (10 days), GWS3 (20 days), GWS4 (30 days), GWC1 (0 day), GWC2 (10 days), GWC3 (20 days) and GWC4 (30 days), across the entire sausages fermentation process. The bacterial sequence reads ranged from 119,591 to 60,527 in GWS sample, and in the control sample from 112,318 to 55,569. These sequence reads were clustered into 1215 OTUs with the 97% identity level, in which 753 OTUs belonged to control sample and 720 OTUs was attributed to GWS sample. In total, 252, 290, 207 and 453 OTUs were generated from GWC1, GWC2, GWC3 and GWC4 sample, and 414, 278, 270 and 267 OTUs were generated from GWS1, GWS2, GWS3 and GWS4 sample, respectively. A total of 19 OTUs were shared by the eight samples as shown in Fig. 2, which indicated that fermented sausage by starter culture inoculation and spontaneous fermented sausage have a low level similarity of bacteria diversity during fermentation. The analyses of the alpha indices were present in Table 1, including the Community richness (Chao 1 and ACE index) and Community diversity (Shannon index and Simpson). Good’s coverage was 1.00 for all samples, indicating that captured majority of bacterial phylotypes of sausage samples. Chao 1 richness estimator increase from 0 to 10 days fermentation with a range from 317.83 to 430 in control sample and with a bit down during from 10 to 20 days, then reached its maximum value of 550.87 on day 30. Accordingly, the Chao 1 richness estimator in GWS sample reached its maximum value of 690.23 on day 0, and then decreased to 397.5 on day 30. In addition, the bacterial Shannon index was the lowest value of 1.63 on day 20 in control sample, however the lowest value reached 1.00 at the end of fermentation in GWS sample, which indicated that the Shannon index of spontaneous fermentation was higher than that of starter culture inoculation. These results showed that other bacteria from raw meat and environment will be inhibited by dominant bacteria from starter culture due to interspecies competition.
Fig. 2.
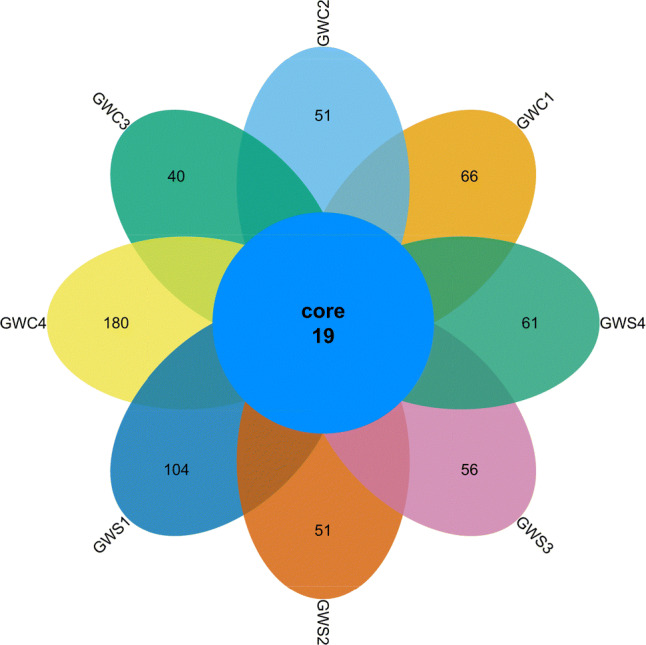
GWC1: control sample fermented for 0 day, GWC2: control sample fermented for 10 days, GWC3: control sample fermented for 20 days, GWC4: control sample fermented for 30 days, GWS1: sample was inoculated starter culture fermented for 0 day, GWS2: sample was inoculated starter culture fermented for 10 days, GWS3: sample was inoculated starter culture fermented for 20 days, GWS4: sample was inoculated starter culture fermented for 30 days
Table 1.
The operational taxonomic units (OTUs) for GWS and GWC (as control) sample
| Fermentation time (days) | Reads | Observed OTUs | ACE index | Chao1 index | Good’s coverage | Shannon index | Simpson | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GWC | GWS | GWC | GWS | GWC | GWS | GWC | GWS | GWC | GWS | GWC | GWS | GWC | GWS | |
| 0 | 112,318 | 119,591 | 252 | 414 | 350.45 | 883.94 | 317.83 | 690.23 | 1.00 | 1.00 | 1.91 | 1.32 | 0.31 | 0.38 |
| 10 | 55,569 | 60,527 | 290 | 278 | 500.11 | 750.49 | 430 | 655.5 | 1.00 | 1.00 | 2.04 | 1.43 | 0.28 | 0.37 |
| 20 | 55,891 | 65,441 | 207 | 270 | 435.12 | 514.61 | 333.04 | 408.96 | 1.00 | 1.00 | 1.63 | 1.38 | 0.36 | 0.37 |
| 30 | 61,136 | 60,561 | 453 | 267 | 601.89 | 553.04 | 550.87 | 397.5 | 1.00 | 1.00 | 2.88 | 1.00 | 0.11 | 0.57 |
Richness estimators (Chao and Ace) and Diversity indices (Shannon and Simpson) of the bacterial 16S rRNA for each sample were determined using OTU based analysis
GWC, sample was no-inoculated starter culture as control sample; GWS, sample was inoculated starter culture
*P < 0.05, **P < 0.01
Effect of the starter culture on bacterial community composition
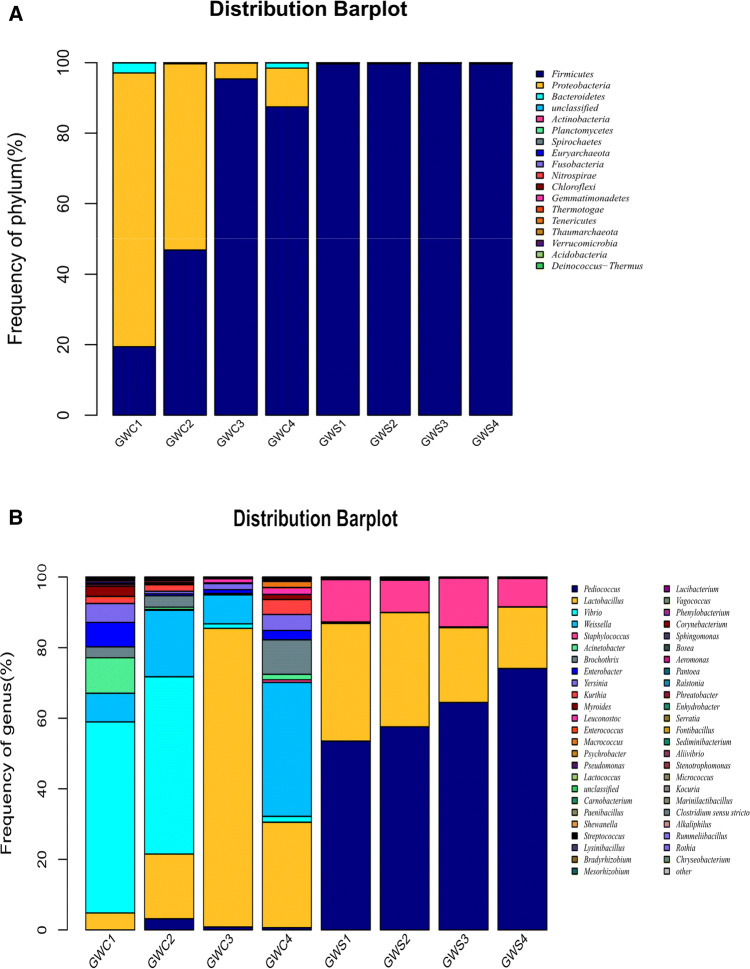
In starter culture inoculation sausage (GWS sample), the shared bacterial OTUs mainly belong to Firmicutes at phylum level, and Pediococcus spp., Lactobacillus spp. and Staphylococcus spp. at the genus level. Accordingly, in spontaneous fermentation sausage (GWC sample), the shared bacterial OTUs mainly belong to Firmicutes and Proteobacteria at phylum level, and Vibrio spp., Lactobacillus spp. and Weissella spp. at the genus level. The bacterial community of GWS and GWC samples were analyzed at the phylum (Fig. 3a) and genus level (Fig. 3b), respectively. A total of 4 identified phyla, namely Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria, and 52 identified genus were found in GWC samples during fermentation, in which Firmicutes and Proteobacteria were the dominant phyla with a total proportion of above 90% at all fermentation stages.
Fig. 3.
Relative abundance of bacteria community proportions at phylum (a) and genus (b) level. Phyla and genera occurred at < 1% abundance in all the samples are defined as “Others”. Taxonomic classification of 97% sequence identity clusters demonstrated at genera classification levels. GWC1: control sample fermented for 0 day, GWC2: control sample fermented for 10 days, GWC3: control sample fermented for 20 days, GWC4: control sample fermented for 30 days, GWS1: sample was inoculated starter culture fermented for 0 day, GWS2: sample was inoculated starter culture fermented for 10 days, GWS3: sample was inoculated starter culture fermented for 20 days, GWS4: sample was inoculated starter culture fermented for 30 days
At genus levels, 35, 40, 32 and 37 genera were determined in GWC1, GWC2, GWC3 and GWC4 sample, respectively. In the initial fermentation (sample GWC1), Vibrio spp. was the most abundant bacterial genus, comprising about 54.13% of bacterial, followed by Acinetobacter spp., accounting for 10.01% of bacterial, and Lactobacillus spp. and Weissella spp. were with a low ratio, accounting for 4.79% and 8.15% of bacterial, respectively. Vibrio spp. and Acinetobacter spp. had a drastic drop in bacterial relative abundance from day 0 to day 20 in GWC sample during fermentation, which the proportion of bacterial declined from 54.13 to 1.25% and from 10.01 to 0.19%, respectively. Accordingly, Lactobacillus spp. underwent a sharp increase from 4.79 to 84.61% and become the most dominant genus, and Weissella spp. increased from 8.15 to 18.82% during the first 10 days fermentation in GWC sample. Meanwhile, Pediococcus spp. also rapidly increased and reached its maximum proportion of 3.19% on day 10 in GWC sample.
Moreover, other undesirable genera with a low ration were found in starter culture inoculation sausage (GWS sample) including Acinetobacter spp., Weissella spp., Enterococcus spp. and Vibrio spp., Enterobacter spp. Accordingly, Enterobacter spp.(6.94%), Yersinia spp. (5.35%), Brochothrix spp. (3.09%) and Myroides spp. (2.88%) with a higher ration were present in spontaneous fermentation sausage (GWC sample) at initial stage of fermentation. Thus, the starter culture inoculation could effectively inhibit the growth of undesirable bacteria, which is beneficial to the fermentation of sausage.
Effect of the starter culture on bacterial succession
The similarities in bacterial communities evaluated by PCA analysis of starter culture inoculation sausage (GWS sample) and spontaneous fermentation sausage (GWC sample) are shown in Fig. 4, which showed an obvious separation of the bacterial communities among the eight different samples. The first and second axes showed values of cumulative percentage variance of species equal to 59% and 23%, respectively. In total, 82% variances of species were explained by the two axes. According to PCA analysis, bacterial communities of spontaneous sausage fluctuated wildly during fermentation, but starter culture inoculation sausage was relatively stable.
Fig. 4.
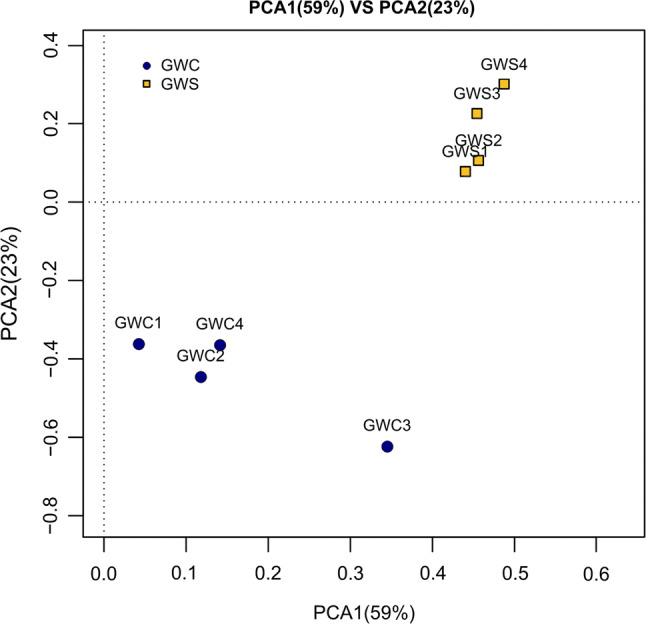
Multiple samples of PCA analysis according to the bacterial diversity. GWC1: control sample fermented for 0 day, GWC2: control sample fermented for 10 days, GWC3: control sample fermented for 20 days, GWC4: control sample fermented for 30 days, GWS1: sample was inoculated starter culture fermented for 0 day, GWS2: sample was inoculated starter culture fermented for 10 days, GWS3: sample was inoculated starter culture fermented for 20 days, GWS4: sample was inoculated starter culture fermented for 30 days
The differences and similarity of the bacterial community structures in starter culture inoculation sausage (GWS sample) and spontaneous fermentation sausage (GWC sample) were compared by heat map analysis as shown in Fig. 5. The bacterial distribution of GWS sample was not abundant, showing mainly bacterial microflora were Pediococcus spp., Lactobacillus spp. and Staphylococcus spp., which primarily participate to enhance food safety and sensory of sausage. Contrarily, the bacterial distribution in spontaneous fermented sausage was more abundant than fermented sausage, and apart from the functional bacteria, namely Weissella spp., Lactobacillus spp. and Pediococcus spp., the undesirable bacteria, such as Enterobacter spp., Yersinia spp. and Myroides spp., from raw meat with a higher abundance, were determined. Furthermore, a competitive relationship between Weissella spp. and Lactobacillus spp. was found in the spontaneous fermented sausage. The proportion of Weissella spp. and Lactobacillus spp. was 8.15% and 4.79% respectively in initial fermentation. Then Weissella spp. decreased from 18.82 to 8.2% from day 10 to day 20, while Lactobacillus spp. increased from 18.33 to 84.61% from day 10 to day 20. At the end of the fermentation, Weissella spp. was the dominant genus due to the decrease of Lactobacillus spp. In addition, the results showed that the starter culture inoculation was conducive to improve microbial quality by inhibiting the growth of native microflora originated from raw meat, such as Vibrio spp., Brochothrix spp., Yersinia spp. and Enterobacter spp., which are pathogenic bacteria and can cause acute gastroenteritis diarrhea, headache, vomiting and so on (Elbashir et al. 2018; Wu et al. 2018), threatening the health of consumers.
Fig. 5.

Bacterial community heatmap analysis of GWS and GWC (as control) sample during the fermentation. GWC1: control sample fermented for 0 day, GWC2: control sample fermented for 10 days, GWC3: control sample fermented for 20 days, GWC4: control sample fermented for 30 days, GWS1: sample was inoculated starter culture fermented for 0 day, GWS2: sample was inoculated starter culture fermented for 10 days, GWS3: sample was inoculated starter culture fermented for 20 days, GWS4: sample was inoculated starter culture fermented for 30 days
Acknowledgements
The research was supported by National Natural Science Foundation of China (31772093) and Sichuan Science and Technology Research Project (2020YFS0504).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinhui Wang, Yalin Zhang and Tian Tian have contributed equally to this work.
References
- Bartkiene E, Bartkevics V, Mozuriene E, Krungleviciute V, Novoslavskij A, Santini A, Rozentale I, Juodeikiene G, Cizeikiene D. The impact of lactic acid bacteria with antimicrobial properties on biodegradation of polycyclic aromatic hydrocarbons and biogenic amines in cold smoked pork sausages. Food Control. 2017;71:285–292. doi: 10.1016/j.foodcont.2016.07.010. [DOI] [Google Scholar]
- Blanco-Lizarazo CM, Sotelo-Díaz I, Arjona-Roman JL, Llorente-Bousquets A, Miranda-Ruvalcaba R. Effect of starter culture and low concentrations of sodium nitrite on fatty acids, color, and Escherichia coli behavior during salami processing. Int J Food Sci. 2018;10:1–10. doi: 10.1155/2018/5934305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali F, Milanović V, Osimani A, Aquilanti L, Taccari M, Garofalo C, Polverigiani S, Clementi F, Franciosi E, Tuohy K, Mercuri ML, Altissimi MS, Haouet MN. Microbial dynamics of model Fabriano-like fermented sausages as affected by starter cultures, nitrates and nitrites. Int J Food Microbiol. 2018;278:61–72. doi: 10.1016/j.ijfoodmicro.2018.04.032. [DOI] [PubMed] [Google Scholar]
- Cavalheiro CP, Ruiz-Capillas C, Herrero AM, Jiménez-Colmenero F, Pintado T, de Menezes CR, Leadir LMF. Effect of different strategies of Lactobacillus plantarum incorporation in Chorizo sausages. J Sci Food Agric. 2019;99(15):6706–6712. doi: 10.1002/jsfa.9952. [DOI] [PubMed] [Google Scholar]
- Cheng JR, Liu XM, Zhang YS. Characterization of Cantonese sausage fermented by a mixed starter culture. J Food Process Preserv. 2018;42(6):e13623. doi: 10.1111/jfpp.13623. [DOI] [Google Scholar]
- de Almeida MA, Saldaña E, da Silva Pinto JS, Palacios J, Contreras-Castillo CJ, Sentandreu MA, Fadda SG. A peptidomic approach of meat protein degradation in a low-sodium fermented sausage model using autochthonous starter cultures. Food Res Int. 2018;109:368–379. doi: 10.1016/j.foodres.2018.04.042. [DOI] [PubMed] [Google Scholar]
- Di Gioia D, Mazzola G, Nikodinoska I, Aloisio I, Langerholc T, Rossi M, Raimondi S, Melero B, Rovira J. Lactic acid bacteria as protective cultures in fermented pork meat to prevent Clostridium spp. growth. Int J Food Microbiol. 2016;235:53–59. doi: 10.1016/j.ijfoodmicro.2016.06.019. [DOI] [PubMed] [Google Scholar]
- Dixon P. Vegan, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- Domínguez R, Agregán R, Lorenzo JM. Role of commercial starter cultures on microbiological, physicochemical characteristics, volatile compounds and sensory properties of dry-cured foal sausage. Asian Pac J Trop Dis. 2016;6(5):396–403. doi: 10.1016/S2222-1808(15)61055-6. [DOI] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Elbashir S, Parveen S, Schwarz J, Rippen T, Jahncke M, DePaola A. Seafood pathogens and information on antimicrobial resistance: a review. Food Microbiol. 2018;70:85–93. doi: 10.1016/j.fm.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honikel KO. The use and control of nitrate and nitrite for the processing of meat products. Meat Sci. 2008;78(1–2):68–76. doi: 10.1016/j.meatsci.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Huang A, Sirisansaneeyakul S, Ge C, Chen Z, Huang Q, Qin W, Chisti Y. Physicochemical changes during processing of Chinese Xuanwei ham. Kasetsart J (Nat Sci) 2011;45:539–550. [Google Scholar]
- Hugas M, Garriga M, Pascual M, Aymerich MT, Monfort JM. Enhancement of sakacin K activity against Listeria monocytogenes in fermented sausages with pepper or manganese as ingredients. Food Microbiol. 2002;19(5):519–528. doi: 10.1006/fmic.2002.0497. [DOI] [Google Scholar]
- Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, Tam NFY. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol. 2013;66(1):96–104. doi: 10.1007/s00248-013-0238-8. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kang KH, Kim SH, Lee S, Lee SH, Ha ES, Sung NJ, Kim JG, Chung MJ. Lactic acid bacteria directly degrade N-nitrosodimethylamine and increase the nitrite-scavenging ability in kimchi. Food Control. 2017;71:101–109. doi: 10.1016/j.foodcont.2016.06.039. [DOI] [Google Scholar]
- Liu W, Yang D, Chen W, Gu X. High-throughput sequencing-based microbial characterization of size fractionated biomass in an anoxic anammox reactor for low-strength wastewater at low temperatures. Biores Technol. 2017;231:45–52. doi: 10.1016/j.biortech.2017.01.050. [DOI] [PubMed] [Google Scholar]
- Liu SJ, Xu JJ, Ma CL, Guo CF. A comparative analysis of derivatization strategies for the determination of biogenic amines in sausage and cheese by HPLC. Food Chem. 2018;266:275–283. doi: 10.1016/j.foodchem.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Lu S, Xu X, Zhou G, Zhu Z, Meng Y, Sun Y. Effect of starter cultures on microbial ecosystem and biogenic amines in fermented sausage. Food Control. 2010;21(4):444–449. doi: 10.1016/j.foodcont.2009.07.008. [DOI] [Google Scholar]
- Manini F, Casiraghi MC, Poutanen K, Brasca M, Erba D, Plumed-Ferrer C. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT-Food Sci Technol. 2016;66:275–283. doi: 10.1016/j.lwt.2015.10.045. [DOI] [Google Scholar]
- Martuscelli M, Crudele MA, Gardini F, Suzzi G. Biogenic amine formation and oxidation by Staphylococcus xylosus strains from artisanal fermented sausages. Lett Appl Microbiol. 2000;31(3):228–232. doi: 10.1046/j.1365-2672.2000.00796.x. [DOI] [PubMed] [Google Scholar]
- Moracanin SV, Stefanovic S, Radicevic T, Borovic B, Djukic D. Production of biogenic amines by lactic acid bacteria isolated from Uzicka sausages. Procedia Food Sci. 2015;5:308–311. doi: 10.1016/j.profoo.2015.09.068. [DOI] [Google Scholar]
- Oh YJ, Jung DS. Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT-Food Sci Technol. 2015;63(1):437–444. doi: 10.1016/j.lwt.2015.03.005. [DOI] [Google Scholar]
- Wang X, Ren H, Wang W, Zhang Y, Bai T, Li J, Zhu W. Effects of inoculation of commercial starter cultures on the quality and histamine accumulation in fermented sausages. J Food Sci. 2015;80(2):377–384. doi: 10.1111/1750-3841.12765. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang Y, Ren H, Zhan Y. Comparison of bacterial diversity profiles and microbial safety assessment of salami, Chinese dry-cured sausage and Chinese smoked-cured sausage by high-throughput sequencing. LWT. 2018;90:108–115. doi: 10.1016/j.lwt.2017.12.011. [DOI] [Google Scholar]
- Woraprayote W, Malila Y, Sorapukdee S, Swetwiwathana A, Benjakul S, Visessanguan W. Bacteriocins from lactic acid bacteria and their applications in meat and meat products. Meat Sci. 2016;120:118–132. doi: 10.1016/j.meatsci.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Wu W, Jing Z, Yu X, Yang Q, Sun J, Liu C, Zhang W, Zeng L, He H. Recent advances in screening aquatic products for Vibrio spp. TrAC Trends Anal Chem. 2018;111:239–251. doi: 10.1016/j.trac.2018.11.043. [DOI] [Google Scholar]
- Xiao Y, Li P, Zhou Y, Ma F, Chen C. Effect of inoculating Lactobacillus pentosus R3 on N-nitrosamines and bacterial communities in dry fermented sausages. Food Control. 2018;87:126–134. doi: 10.1016/j.foodcont.2017.12.025. [DOI] [Google Scholar]
- Zhang QQ, Jiang M, Rui X, Li W, Chen XH, Dong MS. Effect of rose polyphenols on oxidation, biogenic amines and microbial diversity in naturally dry fermented sausages. Food Control. 2017;78:324–330. doi: 10.1016/j.foodcont.2017.02.054. [DOI] [Google Scholar]