NLRX1 is an underappreciated member of the NOD‐like receptor family. Functions in pathogen response are responsible for the regulation of multiple immune pathways. Emergence in disease highlights the potential in exploiting NLRX1 as a therapeutic target.
Keywords: autoimmunity, cancer, inflammation, mitochondria, NLRX1, NOD‐like receptors, SARS‐CoV‐2
Summary
NLRX1 is a member of the NOD‐like receptor family, a set of pattern recognition receptors associated with innate immunity. Interestingly, NLRX1 exists in somewhat of an exile from its NLR counterparts with unique features that mediate atypical functions compared with traditional NOD‐like receptors (NLRs). Aside from a mitochondrial targeting sequence, the N‐terminal region is yet to be characterized. Mitochondrially located, NLRX1 sits within a subgroup of regulatory NLRs responsible for negatively regulating cellular inflammatory signalling. As well as modulating pathogen response, emerging evidence is implicating NLRX1 as a central homeostatic gatekeeper between mitochondrial biology and immunological response. More recently, NLRX1 has been implicated in a wide range of disease, both pathogen‐driven and otherwise. Emerging links of NLRX1 in cancer biology, autoimmunity and other inflammatory conditions are raising the potential of targeting NLRX1 therapeutically, with recent studies in inflammatory bowel disease showing great promise. Within this review, we address the unique features of NLRX1, its roles in innate immune signalling and its involvement in a range of inflammatory, metabolic and oncology disease indications with a focus on areas that could benefit from therapeutic targeting of NLRX1.
Abbreviations
- CGAS
cyclic GMP‐AMP synthase
- IAV
influenza A virus
- IFN
interferon
- IR
ischaemia–reperfusion
- IRF
interferon regulatory factor
- MAVS
mitochondrial antiviral signalling
- MTS
mitochondrial targeting sequence
- NBD
nucleotide‐binding domain
- NLR
NOD‐like receptor
- PRR
pattern recognition receptor
- RIG‐I
retinoic acid‐inducible gene I
- ROS
reactive oxygen species
- STING
stimulator of interferon genes
- TLR
Toll‐like receptor
- TNF
tumour necrosis factor
INTRODUCTION
Throughout evolution, higher‐order organisms have developed two, somewhat intertwined, branches of immunity: the adaptive and innate systems. Whilst the primarily lymphocyte‐driven adaptive system relies on clonal expansion from epitope‐driven recognition, the innate immune system is much less specific. The innate system uses germ line‐encoded receptors termed pattern recognition receptors (PRRs), to drive a generic inflammatory response consisting of cytokines and type I interferons (IFN). 1 The PRRs are categorized into the membrane‐bound, consisting of the Toll‐like receptors (TLRs), and the cytosolic receptors, including retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs), absent in melanoma 2 (AIM2), cyclic GMP‐AMP synthase (cGAS) stimulation of interferon gene (STING) system and the NOD‐like receptors (NLRs). These PRRs are responsible for sensing and directing signalling pathways in response to damage‐associated, pathogen‐associated or, newly termed, homeostatic‐altering molecular patterns (DAMPs, PAMPs and HAMPs, respectively). 2 Of the PRRs, the NLR family is the largest, comprising over 20 members and responsible for directing important signalling pathways in pathogen response, host development and disease. NLRs typically consist of a tripartite domain structure comprising an N‐terminal effector domain (predominantly a caspase activation and recruitment domain (CARD) or pyrin), a central NACHT domain and a C‐terminal leucine‐rich repeat (LRR) domain (Figure 1). 3 Whilst the majority of NLRs possess this typical domain architecture, NLRX1 is unique with several atypical features that impart distinct functions and contribute to its unique roles in health and disease. NLRX1 is mitochondrially located, cannot form inflammasomes and acts to dampen inflammatory responses, unlike the majority of its NLR relatives. That said, NLRX1 is still implicated in disease. Much like other PRRs and specifically NLRs, NLRX1 is critical in the regulation of pathogen response and plays an important role in multiple inflammatory diseases and cancer. Here, we review NLRX1 from the perspectives of cellular location(s) and structural intricacies before discussing pathogen sensing/regulation and links to disease.
Figure 1.
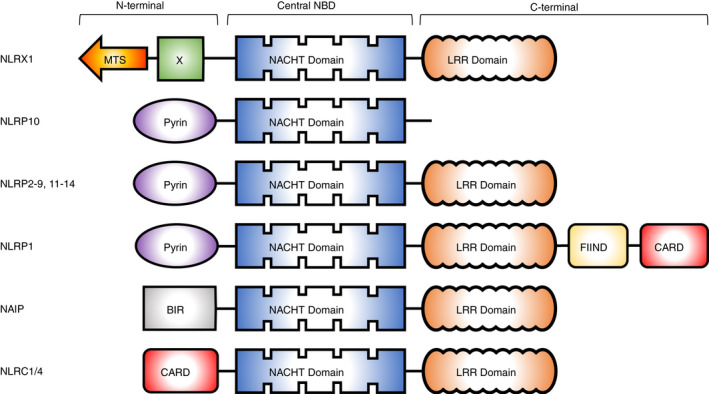
Domain architecture of NLR proteins. BIR, baculovirus inhibitor repeat; CARD, caspase activation recruitment domain; FIIND, function to find domain; LRR, leucine‐rich repeat; MTS, mitochondrial targeting sequence; NBD, nucleotide‐binding domain
Mitochondrial localization
NLRX1 (NOD5, NOD9 and CLR11.3) is ubiquitously expressed in mammalian tissues, with the strongest expression reported in the heart, muscle and mammary gland. 4 , 5 NLRX1 comprises two of the three typical NLR domains, a central NACHT domain and a carboxy‐terminus LRR domain but lacks a recognizable N‐terminal effector domain found in other NLRs (Figure 1). Instead, NLRX1 contains an unconventional N‐terminal domain, that has no current assigned function, giving rise to the ‘X’ in the nomenclature. 6 However, unique to the NLR family, NLRX1 possesses a mitochondrial targeting sequence (MTS) encoded in the first 39 amino acids of the protein. 7 This sequence means NLRX1 is the only mitochondrially targeted NLR family member and allows NLRX1 to possess some unique functions amongst NLRs. 4 , 5 , 7 The precise mitochondrial localization of NLRX1 is highly controversial, with suggestions including both the mitochondrial inner and outer membranes and the mitochondrial matrix. 5 , 7 It is plausible that NLRX1 may be distributed amongst different subcellular and sub‐mitochondrial components, in a context‐ and cell‐type‐dependent manner. Furthermore, the distribution of NLRX1 may be dynamic. Indeed, other NLR family members have been reported to shuttle between different cellular compartments, 8 and it has been proposed that NLRX1 may behave similarly, and shuttle from the cytosol to mitochondria. 9 , 10 , 11 NLRX1 has been shown to interact with proteins within the mitochondrial matrix (UQCRC2 7 and FASTKD5 12 ), mitochondrial outer membrane (MAVS 13 ), endoplasmic reticulum (STING 14 ) and cytoplasm (TRAF6 and IKK complex 13 , 15 ), which adds further complexity to the location of the protein. Apart from the identification of the MTS in NLRX1, much of the biology around the processing, regulation and trafficking of NLRX1 to the mitochondria is unknown. Further context‐dependent investigation into interacting partners will provide insights into the cellular location and functions in different scenarios.
Structure and function
Phylogenetic analysis of the LRR domains of human NLRs shows that NLRX1 shares the most homology with a subgroup of NLRs composed of NOD1 (NLRC1), NOD2 (NLRC2), NOD3 (NLRC3), NOD27 (NLRC5) and CIITA. 4 The crystal structure of the carboxy‐terminus of NLRX1 revealed that it comprises of seven LRR modules flanked by an N‐terminal helical domain and an uncharacterized C‐terminal three‐helix bundle. 16 Furthermore, studies showed that the LRR and its α‐helical domains assemble into a compact hexameric platform, composed of a trimer of three dimers, stabilized by extensive intermolecular and intramolecular interactions. 16 ssRNA and dsRNA, but not DNA, have been shown to interact with a large continuous positive patch thought to form part of the ligand‐binding region of the LRR. 16 , 17 , 18 Mutagenesis of the highly conserved Arg699 residue in this region disrupted RNA binding. 16 Interestingly, several fatty acids were both modelled and experimentally shown to bind to the C‐terminal region with dependence on Asp677, Phe680, Phe681 and Glu684. 18 Zhang et al. reported that the LRR domain of NLRX1 was important to maintain NLRX1 in an inactive confirmation, much like NLRC4, 19 with cells expressing an LRR deletion construct exhibiting constitutively formed oligomers of NLRX1. 20
NLRX1 has been implicated in the modulation of immune responses, including the inhibition of NF‐κB and IRF signalling pathways downstream of TLR activation and viral infection, 13 , 14 , 15 , 21 the modulation of metabolism and reactive oxygen species (ROS) production, 4 , 22 , 23 as well as the induction of autophagy, 24 and the regulation of cell death. 25 , 26 , 27 , 28 Interestingly, NLRX1 is emerging as a key player in viral immune evasion and several viruses including human immunodeficiency virus (HIV), influenza A (IAV) and human papillomavirus virus (HPV) 16/18 (discussed below) have been shown to interact with and exploit NLRX1 to benefit viral invasion. Topically, the recent interactome of SARS‐CoV‐2 highlighted an interaction of the viral protein Orf9c with NLRX1, thrusting this NLR into the limelight once more. 29 Considering the multiple functions of NLRX1, and other reviews covering these topics in detail, 6 , 30 here we will not focus on these roles specifically. Instead, we will discuss these functions within the wider contexts of pathogen sensing and pathophysiology.
PATHOGEN SENSING
The innate response relies on the detection of PAMP/DAMPs by cytosolic or endosomal receptors, such as the NLRs. These sensors survey cellular compartments for a diverse array of exogenous ligands (PAMPs), or host ligands which are mis‐localized following infection (DAMPs). Following recognition, they activate downstream signalling cascades leading to the activation of transcription factor families, which subsequently induce the production of cytokines and IFN. IFNs are produced by both immune and non‐immune cells following infection, but do not have any direct effector functions. Rather, they are secreted and signal through IFN receptors in an autocrine and paracrine manner, activating signal transduction pathways that induce the expression of interferon‐stimulated genes, which encode an array of effector proteins that restrict infection. 31 Below, we will discuss the ways in which NLRX1 participates in responding to, or interacting with, viral, bacterial and fungal infection (Figure 2).
Figure 2.
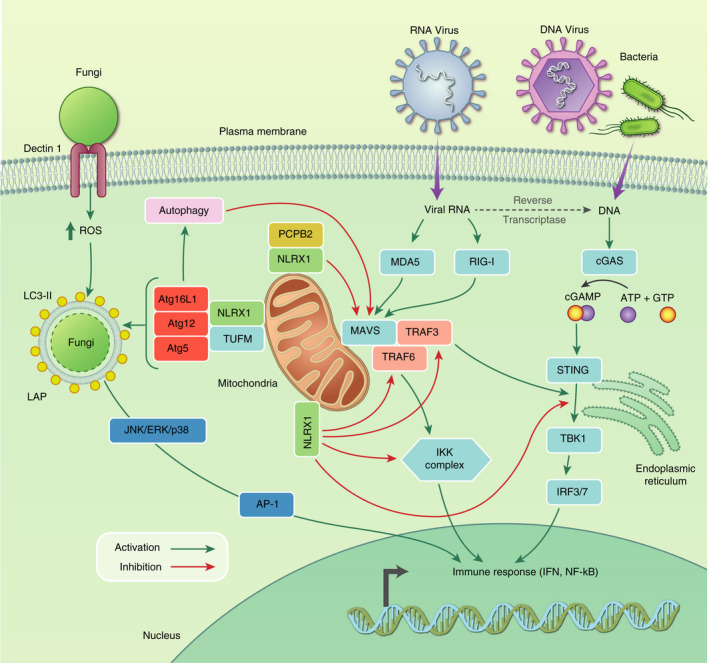
NLRX1 interactions with pathogen sensing pathways. NLRX1 can interact with multiple pathways of pathogen sensing. RNA viruses can be detected by the RIG‐1/MDA5‐MAVS pathway, which can be inhibited by NLRX1 at multiple steps including PCPB2‐mediated degradation of MAVS itself, 21 NLRX1‐TUFM‐driven autophagy, 24 NLRX1‐TRAF6 sequestering and NLRX1‐mediated ubiquitination of the IKK complex downstream of MAVS signalling. 13 , 15 Additionally, NLRX1 can inhibit the DNA sensing pathway of cGAS‐STING. NLRX1 can sequester the interaction between STING and TBK1, 14 as well as TRAF3‐mediated signalling downstream of MAVS. 13 NLRX1 can also interact with autophagy proteins to control fungal infection via LC3‐associated phagocytosis. 72 All of these interactions are in place as negative regulators of inflammation and can be exploited by pathogens to aid immune avoidance
Viruses
RIG‐I/MAVS
RIG‐I and melanoma differentiation‐associated protein 5 (MDA5) are cytosolic PRRs that recognize RNA viruses. 32 , 33 Following activation, these sensors interact with mitochondrial antiviral signalling protein (MAVS; also known as Cardif, IPS‐1 and VISA). 34 , 35 Following engagement, activated MAVS drives activation of transcription factors of the interferon regulatory factor (IRF) and NF‐κB families, which subsequently induce expression of IFNs (Figure 2; reviewed in Ren et al. 36 ). Viruses have evolved strategies to subvert the RLR‐MAVS antiviral immunity. For instance, hepatitis C virus (HCV) encodes a NS3/4A serine protease, which cleaves MAVS to attenuate antiviral immune responses. 35 , 37 Multiple studies have reported that NLRX1 is a negative regulator of the innate immune response to viral infections that are sensed by RIG‐I/MAVS, including IAV, Sendai virus (SeV) and HCV. 5 , 13 , 21 Interestingly, a genetic association between patients harbouring the NLRX1 p.Arg707Cys variant and chronic hepatitis B virus infection has been reported, though the functional significance of this mutation remains to be determined/is unknown. 38 Crucially, Arg707 resides close to critical positive patches within NLRX1 with responsibility for ligand binding and structural stabilization. 16 , 18 Early reports showed that NLRX1‐deficient fibroblasts stimulated with a synthetic dsRNA analogue or RNA viruses had significantly enhanced type I IFN and IL‐6 production. 13 By using a panel of viruses that differentially activate RIG‐I and MDA5, NLRX1 was demonstrated to negatively regulate RIG‐I‐MAVS signalling but not MDA5‐dependent signalling, and that NLRX1 sequestered MAVS, preventing its interaction with downstream signalling molecules (Figure 2). Later studies further showed that the NACHT domain of NLRX1 has been shown to be critical for the attenuation of RLR‐MAVS signalling, due to its interaction with poly(rC) binding protein 2 (PCBP2), which drives polyubiquitination and subsequent degradation of MAVS, attenuating downstream signalling and IFN production. 21 Conversely, other studies have reported that NLRX1 did not attenuate MAVS‐dependent antiviral signalling. 39 , 40 Interestingly, NLRX1 activity appears to be cell‐type‐dependent. In response to SeV, NLRX1‐deficient fibroblasts exhibited enhanced IFN production. However, this enhanced response was not replicated in macrophages nor plasmacytoid dendritic cells (pDCs), highlighting the cell‐type‐specific antiviral functions of NLRX1. 13 Interestingly, RIG‐I and MAVS deficiency abolishes the viral induction of NF‐κB and IRF signalling in multiple cell types except for pDCs, 41 which predominantly detect viruses through the TLR system as opposed to the RIG‐I/MAVS pathway. 41 , 42 pDCs have been reported to upregulate RIG‐I expression following TLR engagement and subsequently respond to RIG‐I ligands, 43 suggesting that the temporal expression of signalling components may influence how NLRX1 differentially regulates signalling during the course of an infection. NLRX1 has also been shown to have distinct and opposing effects in early IRF1 and IRF3 antiviral responses. 44 Whilst NLRX1 attenuated MAVS‐dependent IRF3 activation, it was also required for increased IRF1 abundance and thus IRF1‐mediated antiviral responses. Interestingly, viral infection leads to the activation of IRF3 and IRF7 in most cell types, whereas other IRF family members are selectively activated in response to different stimuli in a cell‐type‐dependent manner. 45 , 46 , 47 , 48 Together, these studies demonstrate how NLRX1 can have seemingly opposing effects on viral immune responses, and how a single regulatory molecule can exert different functional consequences as a result of cell type, stimulus and temporally dependent mechanisms.
Influenza, PB1‐F1 and NLRX1
Influenza A virus is an enveloped single‐stranded RNA virus, which causes significant annual global mortality and significant socioeconomic burden. The virulence of IAV is partly determined by its ability to subvert and attenuate host antiviral immune responses. 49 IAV encodes several virulence factors that inhibit type I IFN signalling and/or the induction of early apoptotic cell death. One such factor, protein 1‐frame 2 (PB1‐F2), has been shown to translocate into the mitochondrial inner membrane space via Tom40, 50 disrupting mitochondrial membrane potential, leading to mitochondrial fragmentation and ultimately the induction of apoptosis. 50 , 51 NLRX1 has been reported to negatively regulate IFN production downstream of IAV infection by attenuating MAVS signalling. 13 In contrast, another study reported that NLRX1 binds to PB1‐F2, preventing IAV‐mediated early apoptosis, leading to increased type I IFN production in macrophages. 26 Consequently, NLRX1‐deficient mice infected with IAV had diminished type I IFN production, increased pulmonary viral replication and dysfunction, and morbidity. 26 Interestingly, PB1‐F1 can induce apoptosis in monocytic cells, whilst epithelial cells remain resistant, and PB1‐F1‐deficient IAV could still drive IFN‐β production in macrophages and lymphocytes, and not epithelial lines, 52 , 53 suggesting cell‐type‐specific differences in the regulation and consequence of IAV infection.
cGAS‐STING
NLRX1 has been shown to regulate immune responses to DNA detected by cGAS, which couples to the ER‐resident STING protein to drive type I IFN production (Figure 2). 54 , 55 NLRX1 was identified in an unbiased siRNA screen of host factors required for HIV infection, 56 and further studies elucidated that NLRX1 interacts with and sequesters STING through the NACHT domain, preventing its interaction with TANK‐binding kinase 1 (TBK1), which attenuates the production of type I IFNs, ISGs and proinflammatory cytokines downstream of HIV‐1 infection (Figure 2). 14 Accordingly, NLRX1‐deficient cells have enhanced the levels of IFNs and ISGs, which restrict HIV‐1 infection. Importantly, treatment with anti‐IFNR1 promoted HIV‐1 infection of NLRX1‐deficient cells. 14 However, the role of NLRX1 in CD4 T cells, the predominant cell type in which HIV‐1 replicates in vivo, remains to be determined. NLRX1 was also shown to modulate the cGAS‐STING pathway in response to DNA viruses including herpes simplex virus 1 and vaccinia virus, which has important clinical considerations. 14 NLRX1 expression was also shown to be upregulated in rhesus monkeys following infection with simian immunodeficiency virus, which directly correlated with the levels of viral RNA. 57 Furthermore, the expression of NLRX1 expression was inversely correlated with the expression of antiviral restriction factors. 57
Bacteria
Listeria monocytogenes is a facultative intracellular Gram‐positive bacterium responsible for the life‐threatening foodborne disease listeriosis. L. monocytogenes encodes numerous virulence factors that promote its survival. One of these factors, listeriolysin O (LLO), a secreted pore‐forming toxin, has been shown to promote the escape of L. monocytogenes from phagosomes, 58 as well as modulating mitochondrial morphology and function. 59 , 60 Recently, Zhang et al. reported that L. monocytogenes infection induced mitophagy, 20 a selective form of autophagy that is targeted by various pathogens to subvert immune responses and promote pathogen survival. 61 The induction of mitophagy by L. monocytogenes was shown to be dependent on LLO, which was required for the oligomerization of NLRX1. The NACHT domain of NLRX1 was found to contain an LC3‐interacting region (LIR), which mediates the interaction of mitochondria with microtubule‐associated protein light chain 3 (LC3) to promote mitophagy. Consequently, NLRX1 deficiency or deletion of the LIR motif attenuated L. monocytogenes‐induced mitophagy, leading to an accumulation of damaged mitochondria, an increase in mitochondrial ROS and the suppression of bacterial growth and survival. 20 Whilst the molecular mechanisms underlying the oligomerization of NLRX1 by LLO remain to be determined, LLO has been shown to trigger Ca2+ influx into the host cytoplasm, 60 , 62 and from there into the mitochondria by the mitochondrial calcium uniporter (MCU) complex. Interestingly, MCU protein 1, a component of the MCU complex, was shown to be enriched following L. monocytogenes infection. 63 Although further study is warranted, it is possible that LLO‐mediated oligomerization of NLRX1 indirectly or directly drives mitochondrial matrix Ca2+ overload, which can lead to mitochondrial dysfunction.
The intracellular bacterium Helicobacter pylori is one of the most common causes of gastritis, a significant risk factor for the development of gastric cancer. 64 An intronic polymorphism in NLRX1 (rs10790286) was reported to be significantly associated with increased risk of H. pylori infection. 65 Furthermore, the expression of NLRX1 was significantly reduced in a monocytic cell line following challenge with a virulent H. pylori clinical isolate, but not a reference strain of H. pylori. Similarly, murine bone marrow‐derived macrophages (BMDMs) challenged with a mouse‐adapted strain of H. pylori exhibited decreased NLRX1 expression . 23 NLRX1‐deficient BMDMs infected with H. pylori had lower bacterial burdens and increased levels of IFN‐γ and ROS. Additionally, NLRX1‐deficient mice infected with H. pylori had significantly reduced gastric bacterial loads, supporting the observations in BMDMs, although the lack of conditional NLRX1 knockouts did not permit the role of specific cell types to be addressed in vivo. 23 Further work evaluating a larger number of H. pylori strains encoding different virulence factors may help to identify the mechanisms through which H. pylori infection leads to a decrease in NLRX1 expression, and how this may influence bacterial burden and pathogenesis.
Fungi
Aspergillus fumigatus is a ubiquitous airborne mould that can cause opportunistic fungal infections in immunocompromised patients, as well as those with pre‐existing airway disease who are otherwise immunocompetent, with exceptionally high mortality rates. 66 Recently, Kastelberg et al. showed that Nlrx1−/− mice inoculated with A. fumigatus clinical isolates had significantly enhanced fungal burden, pulmonary inflammation and immune cell recruitment compared with wild‐type mice. 67 The survival of immunocompetent wild type and Nlrx1−/− mice inoculated with A. fumigatus was comparable to mock‐infected controls. Conversely, in specific immunosuppressive models of invasive pulmonary aspergillosis (IPA) or when challenged with high inoculums of A. fumigatus, Nlrx1−/− had significantly reduced survival compared with wild‐type mice. The decreased survival of Nlrx1−/− mice in immunosuppressive models of IPA was attributed to increased recruitment of CD103+ DCs, which had elevated IL‐4 expression facilitated by enhanced activation of c‐Jun N‐terminal kinase (JNK)/JunB pathways. 67 Consequently, adoptive transfer of Nlrx1−/− CD103+ DCs into neutrophil‐depleted NRG mice enhanced mortality in IPA models. Interestingly, Nlrx1−/− non‐hematopoietic stem cell populations had enhanced IL‐6 and IL‐8 production mediated by enhanced activation of p38, culminating in elevated neutrophil and monocyte recruitment. Accordingly, in models of IPA, adoptive transfer of wild‐type bone marrow into irradiated Nlrx1−/− mice diminished mortality, whereas transfer of Nlrx1−/− bone marrow into wild‐type mice significantly decreased survival. 67
Histoplasmosis is a disease caused by the fungus Histoplasma capsulatum, which causes significant morbidity and mortality worldwide. 68 LC3‐associated phagocytosis (LAP) is a non‐canonical autophagy pathway that has emerged as an important innate defence mechanism. 69 , 70 LAP is initiated following the recognition of pathogens by PRRs, leading to the recruitment of LC3 to the single‐membraned phagosome resulting in the maturation of the phagosome and degradation of internalized cargo. 71 NLRX1 has been reported to have a critical role in LAP induction in macrophages in response to H. capsulatum through its association with Tu translation elongation factor mitochondrial (TUFM). 72 Furthermore, LAP induction was shown to induce inflammatory cytokine production through enhanced mitogen‑activated protein kinases (MAPKs) and activator protein‑1 (AP‐1) signalling cascades, which was attenuated in macrophages from Nlrx1−/− mice. 72
NLRX1 IN DISEASE
NOD‐like receptors are critical for traditional host defence, as discussed above, but alternative implications in multiple pathologies have placed them in a novel light for therapeutic targeting. 73 , 74 , 75 NLRP3 has emerged to be a central player in a plethora of diseases with several therapeutic strategies in development. 76 , 77 , 78 NLRX1 has been linked to disease of the central nervous system (CNS), ischaemia–reperfusion (IR) injury, autoimmunity and cancer to name but a few (Figure 3). Below, we will discuss the work associated with linking this enigmatic NLR to diseases where new therapeutic strategies are in desperate need.
Figure 3.
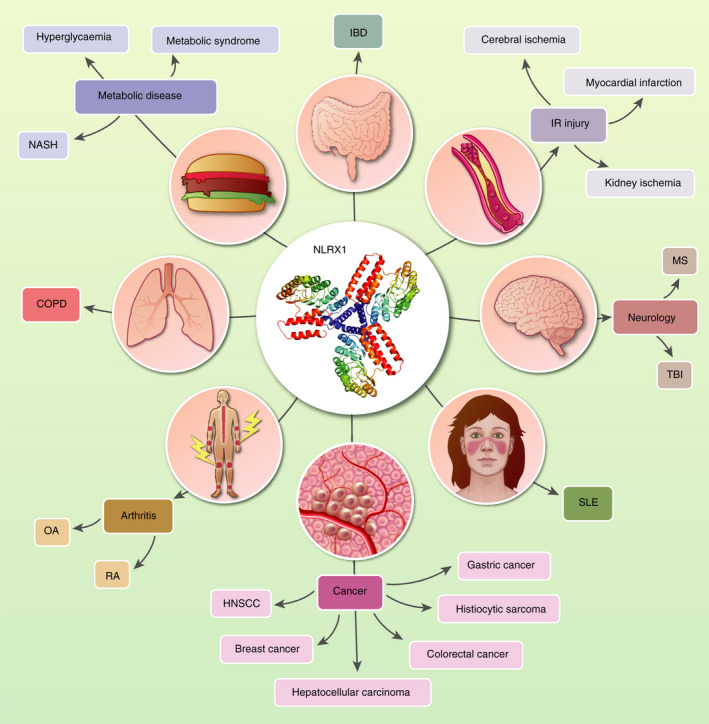
Disease associated with NLRX1. MS: multiple sclerosis, TBI: traumatic brain injury, COPD: chronic obstructive pulmonary disease, 125 HNSCC, head and neck small cell carcinoma; IBD, inflammatory bowel disease; IR, ischaemia–reperfusion; NASH, non‐alcoholic steatohepatitis; OA, osteoarthritis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus. 126 Diseases not referred to in the text are referenced here. All other diseases are referenced within the relevant section of text
Neuroinflammatory disorders
NLRX1 has been implicated in CNS inflammation and disease, with a focus on multiple sclerosis (MS), a disease with mitochondrial dysfunction central to its pathophysiology. 79 The link between sterile inflammation and diseases of the CNS led the Ting laboratory to hypothesize that NLRX1 may be involved in MS‐associated microglial/macrophage activation, especially as other NLRs exacerbate MS phenotypes. 80 In this study, the authors exploit the experimental autoimmune encephalomyelitis (EAE) mouse model to mimic MS and show NLRX1 suppresses inflammation and macrophage/microglial activation, protecting against excessive CNS inflammation that drives limb paralysis, tissue damage and immune cell activation. Nlrx1−/− mice exhibited worse clinical outcomes, higher T‐cell infiltration and increased microglial expression of NOS2 and MHC class II. Additionally, these knockout mice were also more susceptible to myelin‐reactive T cells upon adoptive transfer. Interestingly, as well as furthering the evidence for NLRX1 playing a critical role in the subclinical onset of EAE, six genetic mutations in MS‐susceptible families were also discovered including ten patients harbouring a Glu192Ter truncation. 81 A recent study exploited the blood–brain‐permeable properties of the peptide dNP2 to deliver both the NBD and LRR regions of NLRX1 separately to the CNS of EAE mice at multiple stages of disease. 82 Curiously, only the LRR‐dNP2 strategy had beneficial effects on T‐cell infiltration and tissue inflammation at all disease phases.
Glutamate regulation is critical for neurological cell homeostasis with glutamate dysregulation seen in several neurological disorders, including Alzheimer's disease, MS and Parkinson's disease. 83 In primary astrocyte cultures, Mahmoud et al. 84 observed NLRX1 having a vital role in maintaining glutamate transport capabilities and mitochondrial function. Interestingly, NLRX1 is also implicated in brain injury with decreased NLRX1 expression in human and mouse brain injury situations. 85 In a controlled cortical injury mouse model, NLRX1 was found to regulate response to brain injury, which includes ischaemia, inflammation, excitotoxicity and ROS bursts. Mice lacking NLRX1 fared considerably worse by measured outcomes including cerebral brain lesions and motor ability. Additionally, human patients that had suffered brain aneurysms exhibited significant reductions in NLRX1 expression, which correlated with inflammation driven by dysregulated NF‐κB signalling. 85 Collectively, these studies of NLRX1 in MS, glutamate homeostasis and brain injury provide an excellent insight into the importance of NLRX1 in neurological homeostasis. Further work to elucidate the underpinning biology and functions of NLRX1 in neurological conditions may yield much needed novel therapeutic opportunities in areas with a large and growing unmet medical need. Targeting other NLRs, NLRP3 for example, has already shown great promise in the neurological space. 86 , 87 , 88 , 89
Inflammatory bowel disease
Given the mitochondrial location and proposed role in metabolic regulation, the function of NLRX1 in diseases linked to immunometabolism is unsurprising. Accordingly, a set of recent publications linked NLRX1 to inflammatory bowel disease (IBD) through immunometabolic regulation of CD4+ T cells. 90 , 91 , 92 Early work indicated that dextran sodium sulphate (DSS)‐treated Nlrx1−/− mice exhibited increases in disease severity, upregulated Th1 and Th17 response and heightened inflammatory markers (IL‐17, TNF‐α and IFN‐γ) with T cells primed to differentiate into inflammatory phenotypes with higher proliferative capabilities and decreased response to checkpoint pathways. 90 Adoptive transfer of T‐cell subtypes into Rag2−/− mice confirmed the phenotype to be T‐cell‐specific, with only NLRX1‐deficient T effector or naïve cells causing increased disease severity, and pathogenic T cells preferentially leaning towards an aerobic glycolytic tendency. To this effect, NLRX1 deletion exacerbated the progression of IBD and so the authors postulated NLRX1 activation may be of benefit. To test this, a NLRX1 agonist termed NX13 was developed and confirmed to have favourable safety and pharmacokinetics in rodents. 91 NX13 was utilized in DSS, mdr1a−/− and CD45RBhi adoptive transfer models of IBD in mice, and in primary PBMCs from human ulcerative colitis patients. 92 NX13 decreased disease severity, limited colonic immune cell infiltration and reduced cytokines, which was determined to be driven by major immunometabolic changes in CD4+ T cells specifically. Importantly, NX13 treatment limited the ability of in vitro T‐cell differentiation into Th1 and Th17 subsets by increasing oxidative phosphorylation, whilst decreasing ROS production and NF‐κB signalling. Collectively, this work elucidates a critical role for NLRX1 in restricting the adoption of a disease phenotype by modulating metabolism in a specific subtype of pathogenic T cells. These fundamental studies and any future work will only aid in deciphering the underlying relationship between NLRX1 and mitochondrial metabolic state, throwing light onto the potential in targeting the metabolic regulatory role of NLRX1 in other diseases.
Autoimmunity
There has been a recent emergence of the role of NLRX1 in multiple autoimmune conditions. Aside from IBD as discussed above, NLRX1 has been implicated in systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and, most recently, atopic dermatitis and allergic sensitization. 9 , 93 , 94 Given the innate immune function of NLRX1, this is hardly surprising, but the suggested mechanisms of NLRX1 in autoimmunity are intriguing. A study in SLE patients observed MAVS aggregating in a prion‐like fashion and despite NLRX1 levels not changing between aggregate‐positive and aggregate‐negative patients, there was evidence of cytoplasmic NLRX1. 9 Although the authors did not directly address NLRX1 in the context of SLE, the potential cytoplasmic function of a typically mitochondrial protein in this specific disease context is curious. Additionally, the expression of NLRX1, along with other NLRs, has been reported to be altered in RA. 93 The downregulation of regulatory NLRs, such as NLRX1, coupled with the increase in inflammation‐associated NLRs could drive pathogenic phenotypes in this scenario. Further studies manipulating the in vitro expression of these are essential to understand the relative contribution of individual NLRs, including NLRX1, in a relevant disease setting. More recently, a study has uncovered a genetic association (rs4245191) between NLRX1 and both allergic sensitization and atopic dermatitis. 94 Intriguingly, when considering the interaction with mtDNAs such as mt‐ND6, the odds ratio associated with these NLRX1 variants increased, with the authors of this study suggesting a ROS‐based mechanism at play. Together, these autoimmune conditions collectively highlight the lack of understanding of NLRX1 in autoimmunity with regard to cellular location, interaction with other NLRs and mitochondrial ROS homeostasis and justify efforts to understand NLRX1 further with these disease areas in mind.
Metabolic disease
In mouse models of non‐alcoholic steatohepatitis, NLRX1 mRNA levels were decreased in mice fed on high‐fat diets and lower still on high‐fat fed mice treated with lipopolysaccharide. 95 Another study observed that genetic ablation of NLRX1 resulted in protection against the onset of diet‐induced non‐alcoholic fatty liver disease and metabolic syndrome, with the proposed mechanism of metabolic flux away from fatty acid oxidation and towards glycolysis. 96 Similarly, a partial protection from hyperglycaemia in a high‐fat fed Nlrx1−/− mouse model was observed, with a specific decrease in pancreatic lipid accumulation, and although the mechanism was not clear, this may also be due to a hepatocyte‐specific rewiring of metabolic flux dependant on NLRX1. 97 Additionally, a missense variant in NLRX1 (rs4245191) has been implicated in vascular complications associated with type 2 diabetes mellitus (T2DM) in a select population, but a streptozotocin model of T2DM found NLRX1 not to have a role. 98 , 99 Despite these findings in T2DM, NLRX1 may have a function in metabolic rewiring in other metabolic diseases. These early observations certainly justify further efforts to elucidate whether NLRX1 is of importance in metabolic disease and whether it could be a bona fide therapeutic target in this area.
Ischaemia–reperfusion injury
Ischaemia–reperfusion injury, a leading cause of disabling disease and death worldwide, 100 , 101 is intrinsically linked to mitochondrial biology, with succinate build‐up and reverse‐electron transport‐mediated ROS production responsible for cellular damage. 102 , 103 Considering this mitochondrial link, it is therefore no surprise NLRX1 has been implicated. 104 , 105 , 106
Ischaemia–reperfusion injury is most associated with myocardial infarction, stroke and kidney scenarios such as transplantation and hypovolaemic shock. 107 In acute ischaemic myocardial human tissue and hypoxic rat cardiac cells, NLRX1 expression is decreased and NLRX1 depletion significantly increases the damage associated with hypoxia‐induced ischaemic injury via a MAVS/NLRP3‐dependent mechanism. 108 NLRX1 is also a critical regulator in both human and mouse kidney IR models. 27 Like the heart, 108 the expression of NLRX1 was decreased in human kidney during both acute ischaemic injury and cellular rejection phases, and depletion of NLRX1 in IR injury models resulted in increased oxygen consumption, ROS production and apoptosis. 27 In cerebral ischaemia, a type of stroke that accounts for ~85% of stroke‐like disease and disables 80% of survivors, the associated neuroinflammation is driven by multiple mechanisms. 109 One study had highlighted the protein DJ‐1 (PARK7 and GATD2) in regulating CNS inflammation by NF‐κB signalling. 110 , 111 Recently, Peng et al. 112 discovered this DJ‐1‐mediated regulation of inflammation is dependent on NLRX1 and an interactor protein SHP1. The authors use an oxygen–glucose deprivation IR model in primary astrocytes, as well as an in vivo cerebral occlusion model to assess the relationship of DJ‐1, SHP1 and NLRX1. In these models, DJ‐1 can activate SHP1, a cytoplasmic protein tyrosine phosphatase associated with innate immune regulation, 113 , 114 which can sequester TRAF6 from NLRX1, allowing NLRX1 to negatively regulate the production of TNF‐α, IL‐6 and IL‐1β. Activation of DJ‐1, or/and modulation of SHP1‐TRAF6‐NLRX1 interactions, may be an interesting therapeutic angle considering the tissue‐specific expression of DJ‐1. 115 Manipulation of the relationship between NLRX1 and interacting partners may provide novel strategies to control neuroinflammation across multiple neurological diseases, including MS, as we discuss below. Collectively, these studies in heart, kidney and the CNS highlight the role of NLRX1 in the regulation of mitochondrial biology and therefore IR injury.
Cancer
Regulators of inflammatory homeostasis are critical for limiting contribution towards cancer progression, and the NLRs are no exception. Their control and contribution in the oncology field were nicely summarized in a recent review. 74 Recent interest in (and acquisitions of) NLRP3 modulators highlights the awareness of and value in exploiting the natural innate system as a therapeutic option for cancer. 116 NLRX1 has been implicated in several oncology settings and typically acts as a tumour suppressor, much like its immunological functions as discussed above. Conversely, NLRX1 has also been suggested to promote tumour growth in some models and contexts. These contradictory roles for NLRX1 in cancer suggest that NLRX1 has divergent functions, dependent on the tissue‐ and context‐specific environment.
As a tumour suppressor
One such example of cancer model‐specific differences in NLRX1 function is seen in colitis‐associated cancer (CAC) models of colorectal cancer (CRC). The effect of azoxymethane (AOM) to drive tumour progression was enhanced by NLRX1, whereas tumorigenesis in a DSS/AOM model was suppressed by NLRX1. 25 The differences in these models were suggested to be driven by the response to DSS‐induced inflammation, which can alter the balance of extrinsic and intrinsic apoptotic signals. The authors suggest that the response to these signals can be dictated by NLRX1, explaining the conflicting phenotypes observed in the two models. 25 Two further studies, including one by the original authors of the above publication, address NLRX1 in the context of CRC, albeit in subtly different mouse models of NLRX1 deletion. 117 , 118 One study utilized an intestinal epithelial cell (IEC)‐specific NLRX1 knockout model, which heightened DSS/AOM‐mediated CAC progression. 118 In the other study, the authors observed that only in a non‐haematopoietic compartment knockout of NLRX1 was DSS/AOM‐driven CAC progression exacerbated. 117 Away from inflammation‐mediated tumour progression, both studies also confirmed NLRX1 deletion, in either system, accelerated disease in a mouse model of CRC based on the multiple intestinal neoplasia mutant allele in the adenomatous polyposis coli locus (APCmin model). However, the precise mechanisms as to how NLRX1 was regulating oncogenic signalling pathways were not aligned across the studies. The group using the IEC‐specific model focussed on the alterations in wound‐healing pathways, whereas the non‐haematopoietic compartment NLRX1 knockout mouse had abhorrent STAT3/IL‐6 signalling, with beneficial use of anti‐IL‐6R therapy. Despite these discrepancies, the roles of NLRX1 on TNF/IL‐6 signalling cascades are curious, and differences likely lie in the use of very different NLRX1 knockout models, especially considering the contribution of myeloid cells regarding IL‐6 production. Regardless, in multiple models of CRC, NLRX1 deletion, in either tissue‐specific or non‐haematopoietic compartments, drove a progression of CRC due to the tumour‐suppressive function of NLRX1.
NLRX1 has also been implicated in the liver, breast and gastric cancer, as well as the rare histiocytic sarcoma (HS). Interestingly, in hepatocellular carcinoma (HCC), RA and H. pylori‐infection‐related gastric cancer, NLRX1 expression is downregulated, whilst NOD2 is upregulated. 65 , 93 , 119 , 120 A study of NLR expression in primary liver tumours highlighted that NOD2high/NLRX1low tumours corresponded to a significant reduction in survival, as well as an increase in disease recurrence. 119 , 120 Further mechanistic work implicated NLRX1 in the regulation of the epithelial‐to‐mesenchymal transition (EMT) associated with increased cancer progression. 119 In this study, NLRX1 depletion in multiple HCC cell lines promoted tumour growth in vivo, driven by decreased Akt phosphorylation, reduced Snail1 expression and altered E‐cadherin levels – all indicative of EMT. Interestingly, the authors show that only the C‐terminal LRR region (residues 556–974) was essential for this tumour‐suppressive function, drawing parallels with the LRR‐based therapy used in MS. 82 Additionally, given the observations surrounding reduced Akt phosphorylation, it would be interesting to assess interacting pathways such as mTOR in this system. Furthering our understanding of signalling alterations downstream of NLRX1 will provide further insights into the relevance of NLRX1 in oncology.
In a urethane‐treated HS model, Nlrx1−/− mice exhibited enhanced tumorigenesis and poorer survival compared with wild‐type controls. 121 Upon assessing the transcriptome associated with enhanced tumorigenesis in Nlrx1−/− mice, the authors discovered an upregulation in autophagy‐related proteins including Atg5, Atg16L1 and Atg12, which are known to interact with NLRX1 via TUFM and regulate NF‐κB signalling by promotion of autophagy (Figure 2). 24 Additionally, an upregulation in genes associated with TNF‐α‐mediated apoptosis was observed, which aligns with findings in other cancer cell lines and a genome‐wide CRISPR screen in human endothelial cells where NLRX1 was deemed essential for TNF‐α‐mediated apoptosis. 24 , 122 Like HCC above, NLRX1 was also found to negatively regulate Akt signalling in HS with downstream nodes upregulated in the absence of NLRX1. Interestingly, IL‐6 was also seen to be upregulated upon Nlrx1−/−. 117 Assessing the signatures associated with tumour suppression by NLRX1 across CRC, HCC and HS, some common themes emerge. The central axis of Akt and IL‐6 appears repeatedly, suggesting a pathway of signal regulation by NLRX1 in these settings. Further studies are certainly warranted to uncover the regulatory links between NLRX1 as a tumour suppressor in these cancers and the key cancer‐driving signalling pathways observed.
In tumour progression
Although many studies are aligned in agreeing upon the tumour suppressor role of NLRX1, this observation may be tissue‐specific and even cancer‐type‐specific. One recent study addresses the role of NLRX1 in breast cancer cell lines, elucidating the key role NLRX1 plays in regulation of mitochondria–lysosomal communication in TNF‐α‐treated cell lines. 123 Interestingly, the loss of NLRX1 from triple‐negative breast cancer cells resulted in a decrease in mitophagy due to aberrant lysosomal function and dysregulation of mitochondrial electron transport chain components. Together, the depletion of NLRX1 drove a reduction in cancer cell migration and proliferation. Although the same study highlights a tumour‐suppressive role of NLRX1 in benign and ER/PR‐positive tumours, in aggressive cancers NLRX1 acts to maintain cancer‐promoting energy phenotypes, acting to drive tumour progression and proliferation. The dependency of different tumours on divergent metabolic pathways may dictate the regulatory balance that NLRX1 can provide.
In an interesting parallel to viral‐mediated immune evasion discussed above, a recent publication has implicated NLRX1 in potentiating autophagy‐mediated inhibition of STING in HPV16+ head and neck squamous cell carcinoma (HSNCC). 124 Much like HPV18, which uses E7 to directly target STING, HPV16 uses E7 to instead hijack NLRX1 to induce post‐transcriptional inhibition of STING, which limits immune detection of E7‐expressing tumours in vivo. Critically, depletion of NLRX1 in this model reverses many of the phenotypes of HPV16‐positive HSNCC, including limiting tumour infiltrating lymphocytes, increased rejection of spontaneous tumours and reduction in exhausted T cells. Although not strictly an example of NLRX1 independently driving cancer progression, this highlights the involvement of NLRX1 in the immune‐suppressive function of autophagy‐mediated turnover of key immune surveillance pathways and how they can be exploited to drive disease progression, something to be considered in other scenarios.
NLRX1 – A PROMISING FUTURE?
More than a decade since the discovery of NLRX1 and its implications in innate immunology, NLRX1 is emerging to be a critical regulator of multiple cellular processes, and its dysfunction or dysregulation has been implicated in several human diseases. However, despite steady progress in deciphering the fundamental functions of this unique NLR, several conflicting and controversial topics remain. Aside from the MTS, mystery still surrounds the identity and function of the N‐terminal domain. Equally, functional knowledge about the C‐terminal LRR domain is limited, particularly around the drivers and downstream effects of oligomerization. Aside from questions around the domain characteristics, one major issue that remains somewhat debated is NLRX1’s cellular and subcellular location. Numerous studies, as discussed, have claimed to prove the location of NLRX1; however, few have appreciated what is likely to be dynamic interplay across multiple subcellular compartments, which may be entirely context‐ and cell‐type‐dependent. Therapeutically, NLRX1 has good prospects in multiple disease scenarios, as discussed above. However, discovering the missing links in signal transduction beyond NLRX1 activation and deciphering biological complexities that surround NLRX1 remain an obstacle in developing targeted and successful strategies. That said, early work using NX13 has shown great promise in altering immunometabolic balance to positively benefit gut autoimmunity, and similar strategies should be employed in other NLRX1‐related diseases. The development of novel technologies, tool compounds and assessing NLRX1 with a view not often considered with NLRs will accelerate both the understanding of NLRX1 and the development of targeted therapies. The ability of NLRX1 to function as both an antiviral factor and proviral factor, as an example of its polarized behaviours, highlights the need for context and cell‐type appreciation when considering its biology. Fundamentally, our understanding of the processes NLRX1 is implicated in is still poorly defined. Coupled with complex and overlapping signalling pathways, furthering our knowledge of this somewhat exiled NLR will require novel and thorough approaches.
CONFLICT OF INTEREST
The authors declare no competing financial interests. L.M.B is currently an employee of GlaxoSmithKline.
L.M.B and R.J.P were employees of GlaxoSmithKline at the timing of writing. R.J.P is currently an employee of the University of Cambridge.
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
Enigmatic inflammasomes. Immunology 2021, 162: 249‐251.
MHC class I transactivator NLRC5 in host immunity, cancer and beyond. Immunology 2021, 162: 252‐261.
NLRP9 in innate immunity and inflammation. Immunology 2021, 162: 262‐267.
The NLRP6 inflammasome. Immunology 2021, 162: 281‐289.
DATA AVAILABILITY STATEMENT
No original data are present in this review article.
REFERENCES
- 1. Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13:551–65. [DOI] [PubMed] [Google Scholar]
- 2. Liston A, Masters SL. Homeostasis‐altering molecular processes as mechanisms of inflammasome activation. Nat Rev Immunol. 2017;17:208–14. [DOI] [PubMed] [Google Scholar]
- 3. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell 2014;157:1013–22. [DOI] [PubMed] [Google Scholar]
- 4. Tattoli I, Carneiro LA, Jéhanno M, Magalhaes JG, Shu Y, Philpott DJ, et al. NLRX1 is a mitochondrial NOD‐like receptor that amplifies NF‐κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 2008;451:573–7. [DOI] [PubMed] [Google Scholar]
- 6. Nagai‐Singer MA, Morrison HA, Allen IC. NLRX1 is a multifaceted and enigmatic regulator of immune system function. Front Immunol. 2019;10:2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnoult D, Soaras F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. An N‐terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122:3161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–91. [DOI] [PubMed] [Google Scholar]
- 9. Shao WH, Shu DH, Zhen Y, Hilliard B, Priest SO, Cesaroni M, et al. Prion‐like aggregation of mitochondrial antiviral signaling protein in lupus patients is associated with increased levels of type I interferon. Arthritis Rheumatol. 2016;68:2697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coutermarsh‐Ott S, Eden K, Allen IC. Beyond the inflammasome: regulatory NOD‐like receptor modulation of the host immune response following virus exposure. J Gen Virol. 2016;97:825–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orlandi C, Forlani G, Tosi G, Accolla RS. Molecular and cellular correlates of the CIITA‐mediated inhibition of HTLV‐2 Tax‐2 transactivator function resulting in loss of viral replication. J Transl Med. 2011;9:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh K, Sripada L, Lipatova A, Roy M, Prajapati P, Gohel D, et al. NLRX1 resides in mitochondrial RNA granules and regulates mitochondrial RNA processing and bioenergetic adaptation. Biochim Biophys Acta Mol Cell Res. 2018;1865:1260–76. [DOI] [PubMed] [Google Scholar]
- 13. Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG‐I‐MAVS and TRAF6‐NF‐κB signaling pathways. Immunity 2011;34:854–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo H, König R, Deng M, Riess M, Mo J, Zhang L, et al. NLRX1 sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV‐1 and DNA viruses. Cell Host Microbe. 2016;19:515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, et al. NLRX1 negatively regulates TLR‐induced NF‐κB signaling by targeting TRAF6 and IKK. Immunity 2011;34:843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong M, Yoon S‐I, Wilson IA. Structure and functional characterization of the RNA‐binding element of the NLRX1 innate immune modulator. Immunity 2012;36:337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Deng M, Petrucelli AS, Zhu C, Mo J, Zhang L, et al. Viral DNA binding to NLRC3, an inhibitory nucleic acid sensor, unleashes STING, a cyclic dinucleotide receptor that activates type I interferon. Immunity 2019;50:591–9.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu P, Hontecillas R, Abedi V, Kale S, Leber A, Heltzel C, et al. Modeling‐enabled characterization of novel NLRX1 ligands. PLoS One 2016;10:e0145420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 2013;341:172–5. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Yao Y, Qiu X, Wang G, Hu Z, Chen S, et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat Immunol. 2019;20:433–46. [DOI] [PubMed] [Google Scholar]
- 21. Qin Y, Xue B, Liu C, Wang X, Tian R, Xie Q, et al. NLRX1 mediates MAVS degradation to attenuate the hepatitis C virus‐induced innate immune response through PCBP2. J Virol. 2017;91:e01264–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdul‐Sater AA, Saïd‐Sadier N, Lam VM, Singh B, Pettengill MA, Soares F, et al. Enhancement of reactive oxygen species production and chlamydial infection by the mitochondrial nod‐like family member NLRX1. J Biol Chem. 2010; 285:41637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Philipson CW, Bassaganya‐Riera J, Viladomiu M, Kronsteiner B, Abedi V, Hoops S, et al. Modeling the regulatory mechanisms by which NLRX1 modulates innate immune responses to Helicobacter pylori infection. PLoS One 2015;10:e0137839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 2012;36:933–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soares F, Tattoli I, Rahman MA, Robertson SJ, Belcheva A, Liu D, et al. The mitochondrial protein NLRX1 controls the balance between extrinsic and intrinsic apoptosis. J Biol Chem. 2014;289:19317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jaworska J, Coulombe F, Downey J, Tzelepis F, Shalaby K, Tattoli I, et al. NLRX1 prevents mitochondrial induced apoptosis and enhances macrophage antiviral immunity by interacting with influenza virus PB1‐F2 protein. Proc Natl Acad Sci USA. 2014;111:E2110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stokman G, Kors L, Bakker PJ, Rampanelli E, Claessen N, Teske GJD, et al. NLRX1 dampens oxidative stress and apoptosis in tissue injury via control of mitochondrial activity. J Exp Med. 2017;214:2405–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Killackey SA, Rahman MA, Soares F, Zhang AB, Abdel‐Nour M, Philpott DJ, et al. The mitochondrial Nod‐like receptor NLRX1 modifies apoptosis through SARM1. Mol Cell Biochem. 2019;453:187–96. [DOI] [PubMed] [Google Scholar]
- 29. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parvatiyar K, Cheng G. NOD so fast: NLRX1 puts the brake on inflammation. Immunity 2011;34:821–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider WM, Chevillotte MD, Rice CM. Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature 2006;441:101–5. [DOI] [PubMed] [Google Scholar]
- 33. Brisse M, Ly H. Comparative structure and function analysis of the RIG‐I‐like receptors: RIG‐I and MDA5. Front Immunol. 2019;10:1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seth RB, Sun L, Ea C‐K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF‐κB and IRF3. Cell 2005;122:669–82. [DOI] [PubMed] [Google Scholar]
- 35. Horner SM, Liu HM, Park HS, Briley J, Gale M. Mitochondrial‐associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:14590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ren Z, Ding T, Zuo Z, Xu Z, Deng J, Wei Z. Regulation of MAVS expression and signaling function in the antiviral innate immune response. Front Immunol. 2020;11:1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X‐D, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Q, Peng L, Huang W, Li Q, Pei Y, Yuan P, et al. Rare inborn errors associated with chronic hepatitis B virus infection*. Hepatology 2012;56:1661–70. [DOI] [PubMed] [Google Scholar]
- 39. Soares F, Tattoli I, Wortzman ME, Arnoult D, Philpott DJ, Girardin SE. NLRX1 does not inhibit MAVS‐dependent antiviral signalling. Innate Immun. 2012;19:438–48. [DOI] [PubMed] [Google Scholar]
- 40. Rebsamen M, Vazquez J, Tardivel A, Guarda G, Curran J, Tschopp J. NLRX1/NOD5 deficiency does not affect MAVS signalling. Cell Death Differ. 2011;18:1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Q, Sun L, Liu H‐H, Chen X, Seth RB, Forman J, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 2006; 24:633–42. [DOI] [PubMed] [Google Scholar]
- 42. Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, et al. Cell type‐specific involvement of RIG‐I in antiviral response. Immunity 2005;23:19–28. [DOI] [PubMed] [Google Scholar]
- 43. Szabo A, Magyarics Z, Pazmandi K, Gopcsa L, Rajnavolgyi E, Bacsi A. TLR ligands upregulate RIG‐I expression in human plasmacytoid dendritic cells in a type I IFN‐independent manner. Immunol Cell Biol. 2014;92:671–8. [DOI] [PubMed] [Google Scholar]
- 44. Feng H, Lenarcic EM, Yamane D, Wauthier E, Mo J, Guo H, et al. NLRX1 promotes immediate IRF1‐directed antiviral responses by limiting dsRNA‐activated translational inhibition mediated by PKR. Nat Immunol. 2017;18:1299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weiss G, Maaetoft‐Udsen K, Stifter SA, Hertzog P, Goriely S, Thomsen AR, et al. MyD88 drives the IFN‐β response to Lactobacillus acidophilus in dendritic cells through a mechanism involving IRF1, IRF3, and IRF7. J Immunol. 2012;189:2860–8. [DOI] [PubMed] [Google Scholar]
- 46. Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity 2007;27:228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li P, Wong JJ‐Y, Sum C, Sin W‐X, Ng K‐Q, Koh MBC, et al. IRF8 and IRF3 cooperatively regulate rapid interferon‐β induction in human blood monocytes. Blood 2011;117:2847–54. [DOI] [PubMed] [Google Scholar]
- 48. Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, et al. IRF‐3, IRF‐5, and IRF‐7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLOS Pathog. 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schrauwen EJA, de Graaf M, Herfst S, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. Determinants of virulence of influenza A virus. Eur J Clin Microbiol Infect Dis. 2014;33:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, et al. Influenza A virus protein PB1‐F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun. 2014;5:4713. [DOI] [PubMed] [Google Scholar]
- 51. Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. The influenza A virus PB1‐F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol. 2003;77:7214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–12. [DOI] [PubMed] [Google Scholar]
- 53. Le Goffic R, Bouguyon E, Chevalier C, Vidic J, Da Costa B, Leymarie O, et al. Influenza A Virus protein PB1‐F2 exacerbates IFN‐β expression of human respiratory epithelial cells. J Immunol. 2010;185:4812–23. [DOI] [PubMed] [Google Scholar]
- 54. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008;455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gao D, Wu J, Wu Y‐T, Du F, Aroh C, Yan N, et al. Cyclic GMP‐AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013;341:903–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. König R, Zhou Y, Elleder D, Diamond TL, Bonamy GMC, Irelan JT, et al. Global analysis of host‐pathogen interactions that regulate early‐stage HIV‐1 replication. Cell 2008;135:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barouch DH, Ghneim K, Bosche WJ, Li Y, Berkemeier B, Hull M, et al. Rapid inflammasome activation following mucosal SIV infection of rhesus monkeys. Cell 2016;165:656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. 2018; 16:32–46. [DOI] [PubMed] [Google Scholar]
- 59. Osborne SE, Brumell JH. Listeriolysin O: from bazooka to Swiss army knife. Philos Trans R Soc B Biol Sci. 2017;372:20160222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stavru F, Bouillaud F, Sartori A, Ricquier D, Cossart P. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proc Natl Acad Sci USA. 2011;108:3612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pickles S, Vigié P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Repp H, Pamukçi Z, Koschinski A, Domann E, Darji A, Birringer J, et al. Listeriolysin of Listeria monocytogenes forms Ca2+‐permeable pores leading to intracellular Ca2+ oscillations. Cell Microbiol. 2002;4:483–91. [DOI] [PubMed] [Google Scholar]
- 63. Carvalho F, Spier A, Chaze T, Matondo M, Cossart P, Stavru F. Listeria monocytogenes exploits mitochondrial contact site and cristae organizing system complex subunit mic10 to promote mitochondrial fragmentation and cellular infection. MBio 2020;11:e03171–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Castaño‐Rodríguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM. The NOD‐like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer: a case‐control study and gene expression analyses. PLoS One 2014;9:e98899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. [DOI] [PubMed] [Google Scholar]
- 67. Kastelberg B, Tubau‐Juni N, Ayubi T, Leung A, Leber A, Hontecillas R, et al. NLRX1 is a key regulator of immune signaling during invasive pulmonary aspergillosis. PLOS Pathog. 2020;16:e1008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Horwath MC, Fecher RA, Deepe GS Jr. Histoplasma capsulatum, lung infection and immunity. Future Microbiol. 2015;10:967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Heckmann BL, Boada‐Romero E, Cunha LD, Magne J, Green DR. LC3‐associated phagocytosis and inflammation. J Mol Biol. 2017;429:3561–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Herb M, Gluschko A, Schramm M. LC3‐associated phagocytosis ‐ The highway to hell for phagocytosed microbes. Semin Cell Dev Biol. 2020;101:68–76. [DOI] [PubMed] [Google Scholar]
- 71. Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, et al. Toll‐like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007;450:1253–7. [DOI] [PubMed] [Google Scholar]
- 72. Huang J‐H, Liu C‐Y, Wu S‐Y, Chen W‐Y, Chang T‐H, Kan H‐W, et al. NLRX1 facilitates Histoplasma capsulatum‐induced LC3‐associated phagocytosis for cytokine production in macrophages. Front Immunol. 2018;9:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kanneganti TD, Lamkanfi M, Núñez G. Intracellular NOD‐like receptors in host defense and disease. Immunity 2007;27:549–59. [DOI] [PubMed] [Google Scholar]
- 74. Velloso FJ, Trombetta‐Lima M, Anschau V, Sogayar MC, Correa RG. NOD‐like receptors: major players (and targets) in the interface between innate immunity and cancer. Biosci Rep. 2019;39:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhong Y, Kinio A, Saleh M. Functions of NOD‐like receptors in human diseases. Front Immunol. 2013;4:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fusco R, Siracusa R, Genovese T, Cuzzocrea S, Di Paola R. Focus on the role of NLRP3 inflammasome in diseases. Int J Mol Sci. 2020;21:4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pirzada RH, Javaid N, Choi S. The roles of the NLRP3 inflammasome in neurodegenerative and metabolic diseases and in relevant advanced therapeutic interventions. Genes. 2020;11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shao BZ, Xu ZQ, Han BZ, Su DF, Liu C. NLRP3 inflammasome and its inhibitors: a review. Front Pharmacol. 2015;6:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Patergnani S, Fossati V, Bonora M, Giorgi C, Marchi S, Missiroli S, et al. Mitochondria in multiple sclerosis: molecular mechanisms of pathogenesis. Int Rev Cell Mol Biol. 2017;328:49–103. [DOI] [PubMed] [Google Scholar]
- 80. Eitas TK, Chou WC, Wen H, Gris D, Robbins GR, Brickey J, et al. The nucleotide‐binding leucine‐rich repeat (NLR) family member NLRX1 mediates protection against experimental autoimmune encephalomyelitis and represses macrophage/ microglia‐induced inflammation. J Biol Chem. 2014;289:4173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gharagozloo M, Mahmoud S, Simard C, Yamamoto K, Bobbala D, Ilangumaran S, et al. NLRX1 inhibits the early stages of CNS inflammation and prevents the onset of spontaneous autoimmunity. PLOS Biol. 2019;17:e3000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Koo JH, Kim DH, Cha D, Kang MJ, Choi JM. LRR domain of NLRX1 protein delivery by dNP2 inhibits T cell functions and alleviates autoimmune encephalomyelitis. Theranostics. 2020;10:3138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miladinovic T, Nashed MG, Singh G. Overview of glutamatergic dysregulation in central pathologies. Biomolecules. 2015;5:3112–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mahmoud S, Gharagozloo M, Simard C, Amrani A, Gris D. NLRX1 enhances glutamate uptake and inhibits glutamate release by astrocytes. Cells 2019;8:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Theus MH, Brickler T, Meza AL, Coutermarsh‐Ott S, Hazy A, Gris D, et al. Loss of NLRX1 exacerbates neural tissue damage and NF‐κB signaling following brain injury. J Immunol. 2017;199:3547–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dempsey C, Rubio Araiz A, Bryson KJ, Finucane O, Larkin C, Mills EL, et al. Inhibiting the NLRP3 inflammasome with MCC950 promotes non‐phlogistic clearance of amyloid‐β and cognitive function in APP/PS1 mice. Brain Behav Immun. 2017;61:306–16. [DOI] [PubMed] [Google Scholar]
- 87. Khan N, Kuo A, Brockman DA, Cooper MA, Smith MT. Pharmacological inhibition of the NLRP3 inflammasome as a potential target for multiple sclerosis induced central neuropathic pain. Inflammopharmacology 2018;26:77–86. [DOI] [PubMed] [Google Scholar]
- 88. Gordon R, Albornoz EA, Christie DC, Langley MR, Kumar V, Mantovani S, et al. Inflammasome inhibition prevents ‐synuclein pathology and dopaminergic neurodegeneration in mice. Sci Transl Med. 2018;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kuwar R, Rolfe A, Di L, Xu H, He L, Jiang Y, et al. A novel small molecular NLRP3 inflammasome inhibitor alleviates neuroinflammatory response following traumatic brain injury. J Neuroinflammation. 2019;16:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Leber A, Hontecillas R, Tubau‐Juni N, Zoccoli‐Rodriguez V, Hulver M, McMillan R, et al. NLRX1 regulates effector and metabolic functions of CD4 + T cells. J Immunol. 2017;198:2260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leber A, Hontecillas R, Zoccoli‐Rodriguez V, Ehrich M, Chauhan J, Bassaganya‐Riera J. Exploratory studies with NX‐13: oral toxicity and pharmacokinetics in rodents of an orally active, gut‐restricted first‐in‐class therapeutic for IBD that targets NLRX1. Drug Chem Toxicol. 2019;3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Leber A, Hontecillas R, Zoccoli‐Rodriguez V, Bienert C, Chauhan J, Bassaganya‐Riera J. Activation of NLRX1 by NX‐13 alleviates inflammatory bowel disease through immunometabolic mechanisms in CD4 + T cells. J Immunol. 2019;203:3407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kim HW, Kwon YJ, Park BW, Song JJ, Park YB, Park MC. Differential expressions of NOD‐like receptors and their associations with inflammatory responses in rheumatoid arthritis. Clin Exp Rheumatol. 2017;35:630–7. [PubMed] [Google Scholar]
- 94. Jang H, Kim M, Hong JY, Cho H‐J, Kim C‐H, Kim YH, et al. Mitochondrial and nuclear mitochondrial variants in allergic diseases. Allergy Asthma Immunol Res. 2020;12:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang YG, Fang WL, Wei J, Wang T, Wang N, Ma JL, et al. The involvement of NLRX1 and NLRP3 in the development of nonalcoholic steatohepatitis in mice. J Chinese Med Assoc. 2013;76:686–92. [DOI] [PubMed] [Google Scholar]
- 96. Kors L, Rampanelli E, Stokman G, Butter LM, Held NM, Claessen N, et al. Deletion of NLRX1 increases fatty acid metabolism and prevents diet‐induced hepatic steatosis and metabolic syndrome. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1883–95. [DOI] [PubMed] [Google Scholar]
- 97. Costford SR, Tattoli I, Duan FT, Volchuk A, Klip A, Philpott DJ, et al. Male mice lacking nlrx1 are partially protected from high‐fat diet‐induced hyperglycemia. J Endocr Soc. 2018;2:336–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Scantlebery AML, Uil M, Butter LM, Poelman R, Claessen N, Girardin SE, et al. NLRX1 does not play a role in diabetes nor the development of diabetic nephropathy induced by multiple low doses of streptozotocin. PLoS One 2019;14:e0214437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zeng C, Zhou Z, Han Y, Wen Z, Guo C, Huang S, et al. Interactions of TRAF6 and NLRX1 gene polymorphisms with environmental factors on the susceptibility of type 2 diabetes mellitus vascular complications in a southern Han Chinese population. J Diabetes Complications. 2017;31:1652–7. [DOI] [PubMed] [Google Scholar]
- 100. Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. New Engl J Med. 2007;357:1121–35. [DOI] [PubMed] [Google Scholar]
- 102. Chouchani ET, Pell VR, Gaude E, Aksentijević D, Sundier SY, Robb EL, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014;515:431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Prag HA, Pala L, Kula‐Alwar D, Mulvey JF, Luping D, Beach TE, et al. Ester prodrugs of malonate with enhanced intracellular delivery protect against cardiac ischemia‐reperfusion injury in vivo. Cardiovasc Drugs Ther. 2020;1–13. https://link.springer.com/article/10.1007%2Fs10557‐020‐07033‐6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Toldo S, Abbate A. The NLRP3 inflammasome in acute myocardial infarction. Nat Rev Cardiol. 2018;15:203–14. [DOI] [PubMed] [Google Scholar]
- 105. Arslan F, De Kleijn DP, Pasterkamp G. Innate immune signaling in cardiac ischemia. Nat Rev Cardiol. 2011;8:292–300. [DOI] [PubMed] [Google Scholar]
- 106. Zuurbier CJ, Abbate A, Cabrera‐Fuentes HA, Cohen MV, Collino M, De Kleijn DP, et al. Innate immunity as a target for acute cardioprotection. Cardiovasc Res. 2018;115:1131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Beach TE, Prag HA, Pala L, Logan A, Huang MM, Gruszczyk AV, et al. Targeting succinate dehydrogenase with malonate ester prodrugs decreases renal ischemia reperfusion injury. Redox Biol. 2020;36:101640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li H, Zhang S, Li F, Qin L. NLRX1 attenuates apoptosis and inflammatory responses in myocardial ischemia by inhibiting MAVS‐dependent NLRP3 inflammasome activation. Mol Immunol. 2016;76:90–7. [DOI] [PubMed] [Google Scholar]
- 109. Jayaraj RL, Azimullah S, Beiram R, Jalal FY, Rosenberg GA. Neuroinflammation: friend and foe for ischemic stroke. J Neuroinflammation. 2019;16:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kim HS, Nam ST, Mun SH, Lee SK, Kim HW, Park YH, et al. DJ‐1 controls bone homeostasis through the regulation of osteoclast differentiation. Nat Commun. 2017;8:1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kim J‐H, Choi D‐J, Jeong H‐K, Kim J, Kim DW, Choi SY, et al. DJ‐1 facilitates the interaction between STAT1 and its phosphatase, SHP‐1, in brain microglia and astrocytes: a novel anti‐inflammatory function of DJ‐1. Neurobiol Dis. 2013;60:1–10. [DOI] [PubMed] [Google Scholar]
- 112. Peng L, Zhou Y, Jiang N, Wang T, Zhu J, Chen Y, et al. DJ‐1 exerts anti‐inflammatory effects and regulates NLRX1‐TRAF6 via SHP‐1 in stroke. J Neuroinflammation. 2020;17:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kanwal Z, Zakrzewska A, den Hertog J, Spaink HP, Schaaf MJM, Meijer AH. Deficiency in hematopoietic phosphatase Ptpn6/Shp1 hyperactivates the innate immune system and impairs control of bacterial infections in zebrafish embryos. J Immunol. 2013;190:1631–45. [DOI] [PubMed] [Google Scholar]
- 114. Marengere LEM, Pawson T. Structure and function of SH2 domains. J Cell Sci. 1994;107:97–104. [DOI] [PubMed] [Google Scholar]
- 115. Bandopadhyay R, Kingsbury AE, Cookson MR, Reid AR, Evans IM, Hope AD, et al. The expression of DJ‐1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain 2004;127:420–30. [DOI] [PubMed] [Google Scholar]
- 116. Mullard A. NLRP3 inhibitors stoke anti‐inflammatory ambitions. Nat Rev Drug Discov. 2019;18:405–7. [DOI] [PubMed] [Google Scholar]
- 117. Koblansky AA, Truax AD, Liu R, Montgomery SA, Ding S, Wilson JE, et al. The innate immune receptor NLRX1 functions as a tumor suppressor by reducing colon tumorigenesis and key tumor‐promoting signals. Cell Rep. 2016;14:2562–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tattoli I, Killackey SA, Foerster EG, Molinaro R, Maisonneuve C, Rahman MA, et al. NLRX1 acts as an epithelial‐intrinsic tumor suppressor through the modulation of TNF‐mediated proliferation. Cell Rep. 2016;14:2576–86. [DOI] [PubMed] [Google Scholar]
- 119. Hu B, Ding GY, Fu PY, Zhu XD, Ji Y, Shi GM, et al. NOD‐like receptor X1 functions as a tumor suppressor by inhibiting epithelial‐mesenchymal transition and inducing aging in hepatocellular carcinoma cells. J Hematol Oncol. 2018;11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang X, Yang C, Liao X, Han C, Yu T, Huang K, et al. NLRC and NLRX gene family mRNA expression and prognostic value in hepatocellular carcinoma. Cancer Med. 2017;6:2660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Coutermarsh‐Ott S, Simmons A, Capria V, LeRoith T, Wilson JE, Heid B, et al. NLRX1 suppresses tumorigenesis and attenuates histiocytic sarcoma through the negative regulation of NF‐ΛB signaling. Oncotarget. 2016;7:33096–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cai M, Li S, Shuai Y, Li J, Tan J, Zeng Q. Genome‐wide CRISPR‐Cas9 viability screen reveals genes involved in TNF‐α‐induced apoptosis of human umbilical vein endothelial cells. J Cell Physiol. 2019;234:9184–93. [DOI] [PubMed] [Google Scholar]
- 123. Singh K, Roy M, Prajapati P, Lipatova A, Sripada L, Gohel D, et al. NLRX1 regulates TNF‐α‐induced mitochondria‐lysosomal crosstalk to maintain the invasive and metastatic potential of breast cancer cells. Biochim Biophys Acta Mol Basis Dis. 2019;1865:1460–76. [DOI] [PubMed] [Google Scholar]
- 124. Luo X, Donnelly CR, Gong W, Heath BR, Hao Y, Donnelly LA, et al. HPV16 drives cancer immune escape via NLRX1‐mediated degradation of STING. J Clin Invest. 2020;130:1635–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kang MJ, Yoon CM, Kim BH, Lee CM, Zhou Y, Sauler M, et al. Suppression of NLRX1 in chronic obstructive pulmonary disease. J Clin Invest. 2015;125:2458–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ma D, Zhao Y, She J, Zhu Y, Zhao Y, Liu L, et al. NLRX1 alleviates lipopolysaccharide‐induced apoptosis and inflammation in chondrocytes by suppressing the activation of NF‐κB signaling. Int Immunopharmacol. 2019;71:7–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No original data are present in this review article.


