S. japonicum infection leads to a dramatic decrease in hepatocyte CD1d level, while overexpression of CD1d in hepatocytes significantly attenuates liver damage in infected mice. Our findings highlight the potential of hepatocyte CD1d‐targeted therapies for liver immunopathology control in schistosomiasis.
Keywords: CD1d, hepatocyte, schistosomiasis, immunopathology
Summary
Schistosomiasis is a neglected tropical disease with over 250 million people infected worldwide. The main clinically important species Schistosoma mansoni (S. mansoni) and Schistosoma japonicum (S. japonicum) cause inflammatory responses against tissue‐trapped eggs, resulting in formation of granulomas mainly in host liver. Persistent granulomatous response results in severe fibrosis in the liver, leading to irreversible impairment of the liver and even death of the host. CD1d, a highly conserved MHC class I‐like molecule, is expressed by both haematopoietic and non‐haematopoietic cells. CD1d on antigen‐presenting cells (APCs) of haematopoietic origin presents pathogen‐derived lipid antigens to natural killer T (NKT) cells, which enables them to rapidly produce large amounts of various cytokines and facilitate CD4+ T helper (Th) cell differentiation upon invading pathogens. Noteworthy, hepatocytes of non‐haematopoietic origin have recently been shown to be involved in maintaining liver NKT cell homeostasis through a CD1d‐dependent manner. However, whether hepatocyte CD1d‐dependent regulation of NKT cell homeostasis also modulates CD4+ Th cell responses and liver immunopathology in murine schistosomiasis remains to be addressed. Here, we show in mice that CD1d expression on hepatocytes was decreased dramatically upon S. japonicum infection, accompanied by increased NKT cells, as well as upregulated Th1 and Th2 responses. Overexpression of CD1d in hepatocytes significantly decreased local NKT numbers and cytokines (IFN‐γ, IL‐4, IL‐13), concomitantly with downregulation of both Th1 and Th2 responses and alleviation in pathological damage in livers of S. japonicum‐infected mice. These findings highlight the potential of hepatocyte CD1d‐targeted therapies for liver immunopathology control in schistosomiasis.
Abbreviations
- S. mansoni
Schistosoma mansoni
- S. japonicum
Schistosoma japonicum
- APCs
antigen‐presenting cells
- NKT
natural killer T cells
- Th
CD4+ T helper cells
- IFN‐γ
interferon gamma
- IL‐4
interleukin‐4
- IL‐13
interleukin‐13
- AAV8
adeno‐associated virus serotype 8
- FCM
flow cytometry
- MNCs
mononuclear cells
INTRODUCTION
Schistosomiasis is a neglected tropical disease and affects over 250 million people worldwide. The schistosome species considered to be of major medical importance are Schistosoma mansoni (S. mansoni), Schistosoma japonicum (S. japonicum) and Schistosoma haematobium. During S. mansoni and S. japonicum infections, the host liver is the principal site that is affected. The adult parasites reside in mesenteric veins and lay eggs. Many of the eggs are carried by blood flow into liver and trapped in the microvasculature of the liver. Consequently, immunopathological responses against trapped eggs in liver lead to formation of granulomas, which are progressively replaced by fibrotic deposits eventually resulting in cirrhosis, portal hypertension and even death of the host. 1 , 2 , 3 , 4
CD1d is a highly conserved MHC class I‐like molecule and expressed by both professional antigen‐presenting cells (APCs) of haematopoietic origin (dendritic cells, macrophages and B cells) and non‐haematopoietic cells (hepatocytes, epithelial cells, adipocytes, etc.). 5 , 6 , 7 In particular, CD1d mostly presents lipid antigens to natural killer T (NKT) cells. 8 Studies show that CD1d on APCs presents pathogen‐derived lipid antigens to NKT cells, which enables them to rapidly produce large amounts of various cytokines and facilitate CD4+ T helper (Th) cell differentiation upon invading pathogens. 9 , 10 During murine schistosomiasis, NKT cell subset is one of the important mediators of hepatic inflammation during schistosome infection. Schistosome‐derived lipid antigens are presented to NKT cells by CD1d on APCs, leading to amplification of Th2 responses and egg‐induced pathology in liver. 11 , 12
Noteworthy, CD1d is highly expressed on hepatocytes and a recent study has revealed an unanticipated role of parenchymal cells that hepatocyte CD1d can control local NKT cell numbers and maintain immunological homeostasis in the liver through directly inducing NKT cell apoptosis. 13 , 14 However, whether hepatocyte CD1d‐dependent regulation of NKT cell homeostasis has modulating effects on CD4+ Th cell responses and immunopathology during murine schistosomiasis remains to be addressed.
Therefore, we aimed to determine the function and mechanism of hepatocyte CD1d in hepatic schistosomiasis. Here, we found that S. japonicum infection leads to a dramatic decrease in hepatocyte CD1d expression. Overexpression CD1d in hepatocytes significantly decreased local NKT numbers and Th1/Th2 cytokines (IFN‐γ, IL‐4, IL‐13) in liver, downregulated both Th1 and Th2 responses and attenuated liver immunopathological damage in S. japonicum‐infected mice. Thus, hepatocyte CD1d is suggested as a promising regulator of liver immunopathology in hepatic schistosomiasis.
MATERIALS AND METHODS
Ethics statement and mice
All the animals were handled in strict accordance with Regulations for the Administration of Affairs Concerning Experimental Animals. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (Approval Number: IACUC‐1905064).
Specific pathogen‐free (SPF) 6‐ to 8‐week‐old male mice were purchased from the Animal Center of Nanjing Medical University. All mice were maintained in an SPF breeding facility with standard environment conditions (ambient temperature, 22°C; humidity, 40%; 12‐h light–dark cycle). All efforts were made to minimize suffering.
Infection of mice with S. japonicum
Mice were infected percutaneously with 12 cercariae of the Chinese mainland strain of S. japonicum from infected snails (Oncomelania hupensis), which were purchased from the Jiangsu Institute of Parasitic Diseases.
Production of CD1d‐expressing AAV8 vector and in vivo delivery methods
A recombinant adeno‐associated viral serotype 8 vector (AAV8) containing mouse CD1d1 gene sequence and an empty AAV8 vector control were purchased from Hanbio Biotechnology. The recombinant vector was named as AAV8‐CMV‐m‐CD1d1‐3xflag‐ZsGreen (AAV‐CD1d) or AAV8‐ZsGreen (AAV‐Ctrl).
A week before S. japonicum cercariae infection, each mouse was injected via tail vein containing 1 × 1011 particles of either AAV‐CD1d or AAV‐Ctrl vectors. At 8 weeks post‐infection, mice were killed, and spleens and liver samples were collected for analysis.
Western blot
Western blot analysis was performed according to previous standard procedures. Liver tissues or isolated primary hepatocytes were lysed with RIPA buffer (Invitrogen). Total protein lysates were heat‐denatured, separated by 12% SDS–PAGE, transferred onto nitrocellulose membranes (Whatman, Maidstone) and then detected with the following primary antibodies: anti‐CD1d (Cat. No. AF4884, R&D Systems), anti‐IL‐33 (clone Nessy‐1, Abcam), anti‐β‐actin (clone D6A8, CST) and anti‐GAPDH (clone D16H11, CST). Secondary antibodies used in this study are as follows: HRP‐linked goat anti‐rabbit IgG (CST) and HRP‐linked rabbit anti‐sheep IgG (BioVision). Protein bands were detected using Immun‐Star HRP Substrate (Bio‐Rad), digitally captured with the Bio‐Rad Gel Doc XR System (Bio‐Rad), and quantified with the ImageJ software (NIH; http://imagej.nih.gov/ij).
Quantitative real‐time PCR (qPCR)
Tissue or cell samples were homogenized or lysed in TRIzol™ Reagent (Invitrogen), and total RNA was isolated with a phenol–chloroform extraction. Isolated RNA (1 µg) from each sample was reverse‐transcribed using the SuperScript III First‐Strand cDNA Synthesis System (Invitrogen). RT‐PCRs were performed on the ABI PRISM 7300 Real‐Time PCR System (Applied Biosystem) with FastStart SYBR Green Master Mix (Roche Applied Science). Gene expression levels were calculated using the following formula: Fold change = 2−ΔΔCt, ΔΔCt = ΔCtsample – ΔCtcontrol, and ΔCt = Cttarget – Ctgapdh. Primers used were as follows:
Cd1d1, forward, 5’‐GCAGCCAGTACGCTCTTTTC‐3’, and reverse, 5’‐ACAGCTTGTTTCTGGCAGGT‐3’;
PU.1, forward, 5’‐AGAAGCTGATGGCTTGGAGC‐3’, and reverse, 5’‐GCGAATCTTTTTCTTGCTGCC‐3’;
Lef‐1, forward, 5’‐TCCTGAAATCCCCACCTTCT‐3’, and reverse, 5’‐TGGGATAAACAGGCTGACCT‐3’;
Sp1, forward, 5’‐CTCCAGACCATTAACCTCAGTGC‐3’, and reverse, 5’‐CAAGTAACGGCATGGAGGAC‐3’;
GAPDH, forward, 5’‐TGGTGAAGGTCGGTGTGAAC‐3’, and reverse, 5’‐CCATGTAGTTGAGGTCAATGAAGG‐3’;
IFN‐γ, forward, 5’‐TGCTGATGGGAGGAGATGTCT‐3’, and reverse, 5’‐TGCTGTCTGGCCTGCTGTTA‐3’;
IL‐13, forward, 5’‐CCTGGCTCTTGCTTGCCTT‐3’, and reverse, 5’‐GGTCTTGTGTGATGTTGCTCA‐3’;
IL‐4, forward, 5’‐ACAGGAGAAGGGACGCCAT‐3’, and reverse, 5’‐GAAGCCCTACAGACGAGCTCA‐3’;
IL‐10, forward, 5’‐ACTTTAAGGGTTACTTGGGTTGC‐3’, and reverse, 5’‐ATTTTCACAGGGGAGAAATCG‐3’;
TNF‐α, forward, 5’‐AACCACCAAGTGGAGGAGCAGCT‐3’, and reverse, 5’‐GGAAGACTCCTCCCAGGTATATGG G‐3’; and
TGF‐β1, forward, 5’‐ATGCTAAAGAGGTCACCCGC‐3’, and reverse, 5’‐CCAAGGTAACGCCAGGAATT‐3’.
Serum ALT/AST analysis
The levels of serum ALT and AST were tested by Olympus AU2700 Chemical Analyzer (Olympus) according to the manufacturer's guide.
Flow cytometry (FCM) analysis
Mouse hepatic mononuclear cells (MNCs) were isolated using Percoll density gradient centrifugation as described previously. 15 For NKT cell analysis, MNCs were surface‐stained with CD3e‐Percp‐Cy5.5 (clone 145‐2C11, BD Bioscience, San Diego, CA) and NK1.1‐FITC (clone PK136, eBioscience). To analyse CD1d expression on macrophages, MNCs were surface‐stained with CD11b‐FITC (clone M1/70, BD Bioscience), F4/80‐APC (clone BM8, eBioscience) and CD1d‐PE (clone 1B1, Invitrogen). For CD1d analysis on dendritic cells (DCs), MNCs were surface‐stained with F4/80‐Percp‐Cy5.5 (clone BM8, Invitrogen), MHCⅡ‐PE‐Cy7 (clone M5/114.15.2, eBioscience), CD11c‐APC (clone N418, eBioscience) and CD1d‐PE (clone 1B1, Invitrogen).
Mouse hepatic lymphocytes were purified using lympholyte M (Cedarlane) according to the manufacturer's instructions. To analyse CD1d expression on T cells, cells were surface‐stained with CD3e‐Percp‐Cy5.5 (clone 145‐2C11, BD Bioscience) and CD1d‐PE (Invitrogen). To analyse CD1d expression on B cells, cells were surface‐stained with CD19‐FITC (clone 6D5, Invitrogen) and CD1d‐PE (Invitrogen). To determine intracellular cytokine expression, cells were stimulated with 25 ng/ml of phorbol myristate acetate (PMA, Sigma‐Aldrich) and 1 µg/ml of ionomycin (Sigma‐Aldrich) in the presence of 1 μl/ml of GolgiStop (BD Bioscience). After 6 h, the cells were collected and surface‐stained with CD3e‐APC (clone 145‐2C11, eBioscience) and CD4‐FITC (clone GK1.5, BioLegend), and washed, fixed and permeabilized with Cytofix/Cytoperm buffer (BD Bioscience). Next, the cells were intracellularly stained with PE‐conjugated antibodies against IFN‐γ‐PE (clone XMG1.2, BioLegend) and IL‐4‐PE (clone 11B11, eBioscience). To analyse regulatory T cells, hepatic lymphocytes were surface‐stained with CD3e‐Percp‐Cy5.5 (clone 145‐2C11, BD Bioscience), CD4‐FITC (clone GK1.5, BioLegend) and CD25‐APC (clone PC 61.5, eBioscience). The cells were then permeabilized with cold Fix/Perm Buffer (eBioscience™ Foxp3/Transcription Factor Staining Buffer Set, Cat. No. 00‐5523‐00), and the Fc receptors were blocked with anti‐mouse CD16/32 (Fc Block, Invitrogen). Finally, cells were stained with Foxp3‐PE (clone FJK‐16 s, eBioscience).
To analyse CD1d expression on primary hepatocytes from normal and infection mice, hepatocytes were prepared as below and cells were washed and then stained with PE‐conjugated antibodies against CD1d (clone 1B1, Invitrogen) or the isotype control antibody (clone eB149/10H5, eBioscience).
Following staining, cells were examined using a FACSVerse instrument (BD Bioscience) and analysed using the FlowJo software (version 10.0.7; Tree Star).
Extraction of primary hepatocytes
Primary hepatocytes were separated as previously described. 16 Briefly, mice were anaesthetized and their livers were perfused with HBSS containing 0.5 m EGTA via portal vein, followed by 0.05% collagenase IV digestive fluid. The digested liver suspension was filtered through a 200‐gauge steel mesh, and the hepatocytes in the flow through were isolated by 40% Percoll.
Statistical analysis
Statistical analysis was performed using the SPSS program (version 26 for Windows, IBM Corp.). The comparisons of two group were analysed by Student's t‐test, while multiple group analysis was carried out by one‐way analysis of variance (ANOVA) followed by an LSD post hoc test. For all statistical analyses, P < 0.05 was considered significant.
RESULTS
CD1d expression dramatically decreases in liver of S. japonicum‐infected mice
To examine CD1d expression in livers of S. japonicum‐infected mice, RT‐PCR and Western blot were performed, respectively. We found that infected mice livers showed a significant decline in CD1d mRNA levels as early as 1 week after S. japonicum infection and a more than 15‐fold decline at 3 weeks post‐infection, and thereafter remained at a very low level (Figure 1A). A corresponding significant, progressive decrease in CD1d expression was also shown by Western blotting starting from 1 week after infection (Figure 1B). These data demonstrated a dramatic decrease in hepatic CD1d expression in mice with S. japonicum infection.
Figure 1.
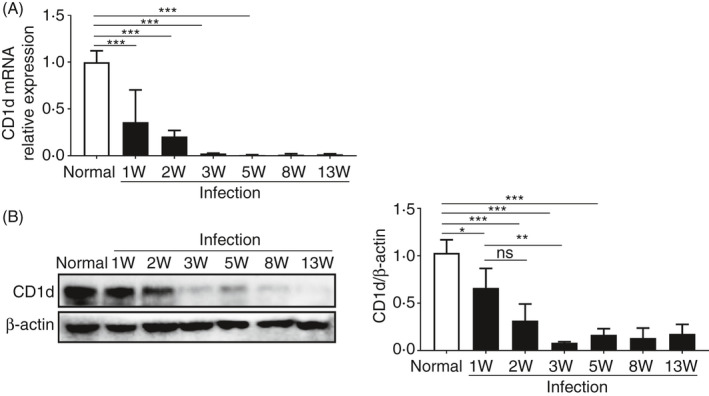
CD1d expression dramatically decreases in liver of S. japonicum‐infected mice. (A and B) CD1d mRNA and protein expression levels in liver samples of normal or S. japonicum‐infected mice (1, 2, 3, 5, 8 and 13 weeks post‐infection) were analysed by RT‐PCR (A) and Western blot (B), respectively. The graphs (B) show the quantification of the Western blot. Data are means ±SD of 4 mice and representative of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001; ns, not significant.
Hepatocytes are the major contributor to the dramatic reduction in hepatic CD1d expression in S. japonicum‐infected mice
Hepatocytes account for the vast majority of cells in liver and express a high level of CD1d among liver parenchymal cells. 13 , 17 We investigated the expression of CD1d on hepatocytes from S. japonicum‐infected mice. Results showed a sharp decline in CD1d expression on hepatocytes after S. japonicum infection (Figure 2A,B). To rule out non‐specific experimental factors that may repress gene expression in hepatocytes, IL‐33, which is an alarmin and increases after S. japonicum infection, 18 was used as a control. Result showed that IL‐33 expression increased (Figure S1). In addition, FCM analysis showed that surface expression of CD1d on hepatocytes dramatically decreased in infected mice (Figure 2C). We also detected CD1d expression by FCM on some other cells of haematopoietic origin in the liver of S. japonicum‐infected mice. Results showed no significant changes in CD1d expression on F4/80−CD11c+MHCII+ DCs, CD11b+F480+ macrophages and CD3+ T cells after S. japonicum infection (Figure 2D–F). However, CD19+ B cells presented a mild decline in CD1d expression after infection (Figure 2G). Collectively, these results suggest that hepatocytes are the major contributor to the dramatic reduction in hepatic CD1d expression in S. japonicum‐infected mice.
Figure 2.
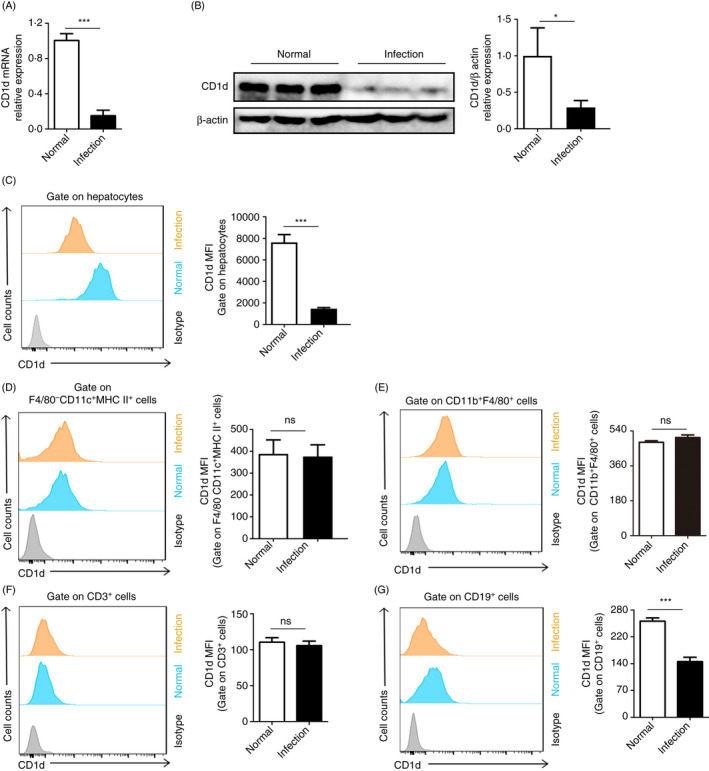
Hepatocytes are the major contributor to the dramatic reduction in hepatic CD1d expression in S. japonicum‐infected mice. (A and B) CD1d mRNA and protein expression levels in hepatocytes from livers of normal or S. japonicum‐infected (8 weeks post‐infection) mice were determined by RT‐PCR (A) and Western blot (B), respectively. Representative images of Western blots are shown, and protein level of CD1d normalized against β‐actin was quantified by ImageJ software. (C–G) CD1d expression on hepatocytes (C), F4/80−CD11c+MHCII+ DCs (D), CD11b+F480+ macrophages (E), CD3+ T cells (F) and CD19+ B cells (G) from livers of normal or S. japonicum‐infected (8 weeks post‐infection) mice were analysed by FCM, and representative histograms are shown. The bar graphs show the average MFI of CD1d expression in each cell population. Data are means ±SD from 4 mice per group, and similar results were obtained in two independent experiments. *P < 0.05 and ***P < 0.001; ns, not significant.
Furthermore, hepatocytes from S. japonicum‐infected mice showed a dramatic decrease in SP1 mRNA level and significant increase in both PU.1 and LEF‐1 mRNA levels (Figure S2A–C), which are transcriptional activator (SP1) or repressors (PU.1 and LEF‐1) of CD1d gene. 19 , 20 , 21
AAV8‐mediated overexpression of CD1d leads to protection against pathological damage in S. japonicum‐infected mouse livers
Given the dramatic decrease in CD1d expression observed in hepatocytes from S. japonicum‐infected mice, we used an AAV8 vector, which is proven to have high affinity for hepatocytes and achieves ≈95% hepatocyte transduction rate, 22 , 23 , 24 to overexpress CD1d in hepatocytes to investigate its role in pathological damage during schistosomiasis. The detailed experimental design is shown in Figure 3A. Nine weeks after tail vein injection of AAV8 vector encoding CD1d gene, efficient CD1d overexpression was mainly observed in the liver but not detected in the spleen (Figure 3B,C). S. japonicum‐infected mice with CD1d overexpression showed decreased size of isolated hepatic granulomas (Figure 3D,E). In addition, both ALT and AST levels were significantly reduced in infected mice with AAV8‐CD1d injection (Figure 3F). These results suggest that hepatocyte CD1d may be involved in protection against pathological damage in S. japonicum‐infected mice.
Figure 3.
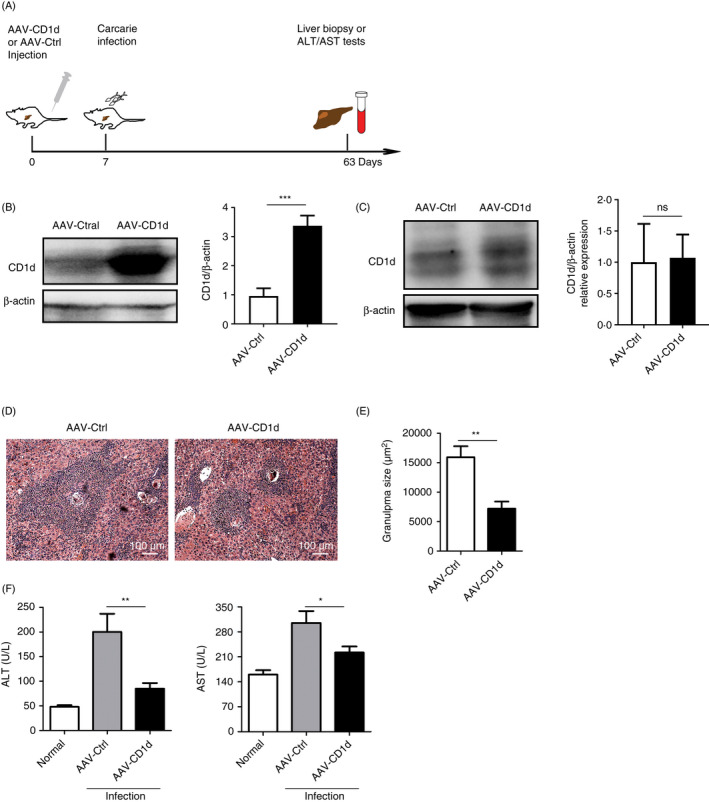
AAV8‐mediated overexpression of CD1d leads to protection against pathological damage in S. japonicum‐infected mice. (A) Experimental design for AAV8 vector (AAV‐CD1d or AAV‐Ctrl) injection, cercariae infection and further analysis. (B–C) Effect of AAV vectors on CD1d protein expression in livers (B) and spleens (C) of mice injected with AAV‐CD1d or AAV‐Ctrl was analysed by Western blot. The graphs show the quantification of the Western blot. (D) Representative liver sections of S. japonicum‐infected mice injected with AAV‐CD1d or AAV‐Ctrl were H&E‐stained to show granulomas (original magnifications, ×100). (E) Granuloma areas with a single egg were measured. (F) Levels of alanine transaminase (ALT) and aspartate transaminase (AST) in the serum of mice with injection of AAV‐CD1d or AAV‐Ctrl were detected by an automatic biochemical analyser. Data are means ±SD from 5 mice per group and representative of two independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001; ns, not significant.
Decreased NKT cells accompanying downregulation of both Th1 and Th2 cytokines in CD1d‐overexpressing liver of S. japonicum‐infected mice
Following S. japonicum infection, despite decreased frequency of CD3+NK1.1+ NKT cells (Figure 4A), the absolute numbers of mononuclear cells (MNCs) and NKT cells were increased in the livers of infected mice (Figure 4B). Hepatocyte CD1d can maintain immunological homeostasis in the liver by controlling local NKT cell numbers. 14 We next investigated whether overexpression of CD1d affects the homeostasis of hepatic NKT cells in S. japonicum‐infected mice. FCM analysis revealed significantly decreased frequencies and absolute numbers of hepatic NKT cells in S. japonicum‐infected mice with AAV‐CD1d injection compared with those in S. japonicum‐infected mice with AAV vector injection, but without changes in the total numbers of liver MNCs between groups (Figure 4C,D). As expected, the decrease in NKT cells was mainly in the liver but not in the spleen of AAV‐CD1d‐injected mice (Figure S3).
Figure 4.
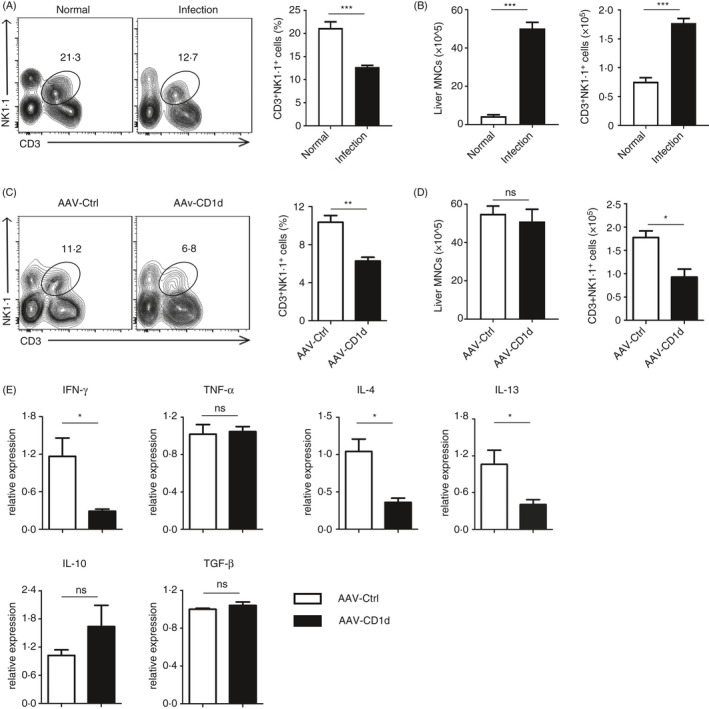
NKT cells are decreased in CD1d‐overexpressing liver of S. japonicum‐infected mice. (A) Representative FCM dot plots and average percentages of CD3+NK1.1+ NKT cells from the liver of normal or S. japonicum‐infected mice. (B) The bar graphs show the absolute numbers of liver MNCs (left) and CD3+NK1.1+ NKT cells (right)in the liver from mice in (A). (C) Representative FCM dot plots and average percentages of CD3+NK1.1+ NKT cells from the liver of S. japonicum‐infected mice injected with AAV‐CD1d or AAV‐Ctrl. (D) The bar graphs show the absolute numbers of liver MNCs (left) and CD3+NK1.1+ NKT cells (right) in the liver from mice in (C). (E) Relative mRNA expression levels of IFN‐γ, TNF‐α, IL‐4, IL‐13, IL‐10 and TGF‐β in livers from S. japonicum‐infected mice injected with AAV‐CD1d or AAV‐Ctrl. Data are means ±SD from 5 mice per group and representative of two independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001; ns, not significant.
Following activation, NKT cells can facilitate CD4+ T helper (Th) cell differentiation by rapidly releasing large amounts of various cytokines. The type and quantity of cytokine release differs depending on disease or tissue context. 7 , 25 We further explored the changes in cytokine profile in the liver by RT‐PCR. Interestingly, except for TNF‐α, both Th1 (IFN‐γ) and Th2 (IL‐4 and IL‐13) cytokines were found to be significantly decreased in the liver of S. japonicum‐infected mice with AAV‐CD1d injection (Figure 4E). Additionally, immunoregulatory cytokine IL‐10 showed a trend towards increase that was not statistically significant, whereas TGF‐β level did not show any change (Figure 4E). Together, these results suggest that hepatic CD1d regulates liver NKT cell homeostasis, which may lead to impaired inflammatory responses to schistosome eggs in liver of S. japonicum‐infected mice.
Both Th1 and Th2 cell responses in livers are impaired in S. japonicum‐infected mice with CD1d overexpression
The CD4+ T cell subsets are involved in the regulation of schistosomiasis progression, and their differentiation is tightly regulated by the local cytokine profile. 26 Following S. japonicum infection, the frequencies and absolute numbers of Th1 (IFN‐γ+), Th2 (IL‐4+) and Treg (CD25+Foxp3+) cells in hepatic CD3+CD4+ T cells apparently increased (Figure 5A). In accordance with cytokine profile changes shown in Figure 4E, results further showed decreased frequencies and absolute numbers of Th1 (IFN‐γ+) and Th2 (IL‐4+) cells in hepatic CD4+ T cells of S. japonicum‐infected mice with CD1d overexpression (Figure 5B,C), whereas the frequencies and absolute numbers of Treg (CD25+Foxp3+) cells in hepatic CD4+ T cells did not show any significant change (Figure 5D). These results indicate that hepatic CD1d is important in regulating both Th1 and Th2 cell responses during S. japonicum infection.
Figure 5.
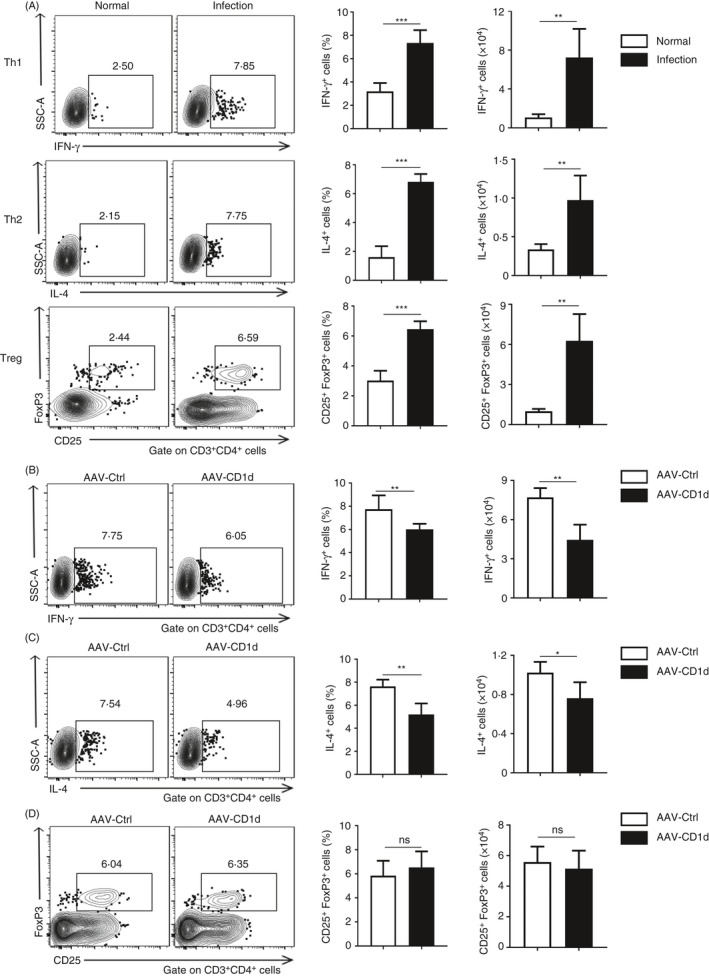
Both Th1 and Th2 cell responses are impaired in S. japonicum‐infected mice with CD1d overexpression. (A) Representative FCM dot plots (left), average percentages (middle) and absolute numbers (right) for IFN‐γ‐(Th1)‐ and IL‐4‐(Th2)‐producing CD3+CD4+ T cells, and CD3+CD4+CD25+Foxp3+ Treg cells from the livers of normal or S. japonicum‐infected mice. (B–D) Representative FCM dot plots (left), average percentages (middle) and absolute numbers (right) for IFN‐γ‐(Th1)‐ and IL‐4‐(Th2)‐producing CD3+CD4+ T cells (B and C), and CD3+CD4+CD25+Foxp3+ Treg cells (D) from the livers of S. japonicum‐infected mice injected with AAV‐CD1d or AAV‐Ctrl. Data are means ±SD from 5 mice per group and representative of two independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001; ns, not significant.
DISCUSSION
After S. mansoni or S. japonicum infection, parasite eggs are trapped in host liver and induce granulomatous inflammation, leading to liver fibrosis and severe damage. The mechanisms by which hepatic homeostasis is maintained are of central importance to inflammatory diseases. 17 However, little is known about whether tissue‐specific factor contributes to the regulation of granulomatous inflammation during schistosome infection. Here, for the first time, we have uncovered a novel role for hepatocytes of non‐haematopoietic origin in protecting against hepatic damage in mice with schistosomiasis japonica.
Hepatocytes, which constitute approximately 60% of the liver by cell number and 80% by mass, are critical for maintaining hepatic homeostasis during inflammation. 17 NKT cell subset is one of the important mediators of hepatic inflammation, 27 and CD1d constitutively expressed by hepatocyte has been reported to control local NKT cell numbers and maintain homeostasis in the liver. 14 Surprisingly, we initially observed a dramatic decrease in hepatocellular CD1d expression in S. japonicum‐infected mice, even starting before parasite egg laying. Of note, the dramatic decrease is mainly limited to hepatocytes of non‐haematopoietic origin, but not DCs, macrophages, CD3+ T cells or B cells of haematopoietic origin, which show no change or just a moderate decrease in CD1d expression after S. japonicum infection. Based on the above, our hypothesis is that loss of hepatocellular CD1d expression upon S. japonicum infection may contribute substantially to the development of granulomatous inflammation in liver.
AAV8 yields the highest transduction rate of murine hepatocytes. 24 Using AAV8‐mediated gene transfer to overexpress CD1d in hepatocytes, we show that CD1d is involved in protection against pathological damage resulted from S. japonicum infection. Coincidentally, study reported that a similar phenomenon has also been observed in inflammatory bowel disease, showing that CD1d on intestinal epithelial cells of non‐haematopoietic cell origin is critical for maintaining mucosal homeostasis and a defect of epithelial CD1d expression parallels to intestinal inflammation. 28 Taken together, our study puts forward an important notion that CD1d harbours cell type‐specific roles and parenchymal cells (non‐haematopoietic cell origin) play an important role in protecting tissue from inflammation and sustaining local homeostasis through CD1d, in contrast to CD1d on cells of haematopoietic origin (DCs, macrophages, etc.), which mainly presents phagocytosed dead cell‐ or pathogen‐derived lipid antigens to NKT cells and contributes to the pathogenesis of inflammatory diseases.
Previous studies suggest that the balance between stimulatory and inhibitory lipid antigens act as a rheostat in the regulation of NKT cell‐dependent immunity. 29 , 30 , 31 Hepatocyte metabolism‐derived self‐lipid antigens are presented by CD1d and important for maintaining local NKT homeostasis. 14 However, our results show that schistosome infection disrupts the balanced crosstalk between metabolism and immunity by reducing hepatocellular CD1d. As such, our study indicates that CD1d may be a promising therapeutic target to rescue hepatocyte‐mediated protection against liver damage in schistosomiasis.
Professional antigen‐presenting cells (APCs), notably dendritic cells, present glycolipid antigens to NKT cells by CD1d during schistosome infection, leading to the activation and expansion of NKT cells and therefore the induction of both Th1 and Th2 responses and to egg‐induced hepatic pathology. 11 , 12 , 32 Hepatocellular CD1d is reported to control local NKT cell numbers by inducing their apoptosis and therefore maintain immunological homeostasis in the liver. 14 Consistently, we observed that overexpression of hepatocellular CD1d in S. japonicum‐infected mice induced a significant decrease in absolute number of hepatic NKT cells, thereby leading to corresponding decrease in both Th1 and Th2 responses, probably due to the reduced facilitation from NKT cells. 7 , 25 All these observations suggest that CD1d plays variety, probably cell type‐based roles during schistosomiasis. However, it should be noted that CD1d knockout mice with schistosomiasis have a reduced Th2 response and are protected from egg‐induced hepatic pathology. 11 This could be explained by the fact that CD1d knockout mice without CD1d‐expressing APCs have no functional NKT cells. 33 , 34 Notably, epithelial CD1d uses a distinct mechanism to control intestinal inflammation, in a manner of inducing IL‐10 production. 35 Interestingly, our data also showed an increase in IL‐10 mRNA in CD1d‐overexpressed liver, even though it was not statistically significant. Hepatic CD1d expression in S. japonicum‐infected mice began to decrease as early as 1 week after infection. S. japonicum schistosomula arrive in the portal vein close to the liver by 3.5 ~ 6 days post‐infection. 36 , 37 Thus, the possible mediators causing the downregulation of hepatocyte CD1d in S. japonicum infection already exist from the very beginning of schistosomula stage. Additionally, in the present study we observed that several factors (SP1, PU.1 and LEF‐1) in hepatocytes related to CD1d transcriptional regulation changed accordingly during S. japonicum infection, suggesting that multiple underlying mechanisms may be involved in the dramatic decline in hepatocellular CD1d expression. However, the exact reason why hepatocellular CD1d decreases so dramatically after S. japonicum‐infected remains a mystery. Given the importance of hepatocellular CD1d in liver protection and its potential to be a tool for inflammation control, further research is needed in the future.
Taken together, our findings underscore the role of hepatocyte as a critical regulator in liver tissue inflammation and highlight the potential that hepatocellular CD1d defines a promising pathway to dampen hepatic inflammatory response during S. japonicum infection.
DISCLOSURES
The authors declare that they have no competing interests.
Supporting information
Figure S1. IL‐33 expression in hepatocyte from the liver of S. japonicum‐infected mice.
Figure S2. SP1, PU.1, and LEF‐1 mRNA levels in hepatocytes from S. japonicum‐infected mice.
Figure S3. AAV8‐mediated overexpression of CD1d does not affect NKT cells in the spleen of S. japonicum‐infected mice.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research and Development Program of China (MOST No: 2018YFA0507300) and the National Natural Science Foundation of China (NSFC No: 81871675) to Chuan Su, the Natural Science Foundation of Jiangsu Province (No. BK20170105) and the National Natural Science Foundation of China (NSFC No. 81971963) to Sha Zhou.
Zhigang Lei and Rui Tang authors contributed equally to this work.
Contributor Information
Sha Zhou, Email: shazhou@njmu.edu.cn.
Chuan Su, Email: chuansu@njmu.edu.cn.
DATA AVAILABILITY STATEMENT
Data used for the analysis are available from the corresponding author upon reasonable request.
REFERENCES
- 1. McManus DP, Bergquist R, Cai P, Ranasinghe S, Tebeje BM, You H. Schistosomiasis‐from immunopathology to vaccines. Semin Immunopathol 2020; 42:355–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou XN. Schistosomiasis. Nat Rev Dis Primers 2018; 4:13. [DOI] [PubMed] [Google Scholar]
- 3. Pearce EJ, MacDonald AS. The immunobiology of schistosomiasis. Nat Rev Immunol 2002; 2:499–511. [DOI] [PubMed] [Google Scholar]
- 4. Wynn TA, Thompson RW, Cheever AW, Mentink‐Kane MM. Immunopathogenesis of schistosomiasis. Immunol Rev 2004; 201:156–67. [DOI] [PubMed] [Google Scholar]
- 5. Moody DB, Cotton RN. Four pathways of CD1 antigen presentation to T cells. Curr Opin Immunol 2017; 46:127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lynch L. Adipose invariant natural killer T cells. Immunology 2014; 142:337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol 2013; 13:101–17. [DOI] [PubMed] [Google Scholar]
- 8. Girardi E, Zajonc DM. Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunol Rev 2012; 250:167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1‐ and MR1‐restricted T cells. Annu Rev Immunol 2014; 32:323–66. [DOI] [PubMed] [Google Scholar]
- 10. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d‐restricted antigens by natural killer T cells. Nat Rev Immunol 2012; 12:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faveeuw C, Angeli V, Fontaine J, Maliszewski C, Capron A, Van Kaer L, et al Antigen presentation by CD1d contributes to the amplification of Th2 responses to Schistosoma mansoni glycoconjugates in mice. J Immunol 2002; 169:906–12. [DOI] [PubMed] [Google Scholar]
- 12. Mallevaey T, Zanetta JP, Faveeuw C, Fontaine J, Maes E, Platt F, et al Activation of invariant NKT cells by the helminth parasite schistosoma mansoni. J Immunol 2006; 176:2476–85. [DOI] [PubMed] [Google Scholar]
- 13. Geissmann F, Cameron TO, Sidobre S, Manlongat N, Kronenberg M, Briskin MJ, et al Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol 2005; 3:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeissig S, Peuker K, Iyer S, Gensollen T, Dougan SK, Olszak T, et al CD1d‐Restricted pathways in hepatocytes control local natural killer T cell homeostasis and hepatic inflammation. Proc Natl Acad Sci U S A 2017; 114:10449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eberl G, MacDonald HR. Rapid death and regeneration of NKT cells in anti‐CD3epsilon‐ or IL‐12‐treated mice: a major role for bone marrow in NKT cell homeostasis. Immunity 1998; 9:345–53. [DOI] [PubMed] [Google Scholar]
- 16. Zeissig S, Murata K, Sweet L, Publicover J, Hu Z, Kaser A, et al Hepatitis B virus‐induced lipid alterations contribute to natural killer T cell‐dependent protective immunity. Nat Med 2012; 18:1060–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stanger BZ. Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol 2015; 77:179–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He X, Xie J, Wang Y, Fan X, Su Q, Sun Y, et al Down‐regulation of microRNA‐203‐3p initiates type 2 pathology during schistosome infection via elevation of interleukin‐33. PLoS Pathog 2018; 14:e1006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen QY, Jackson N. Human CD1D gene has TATA boxless dual promoters: an SP1‐binding element determines the function of the proximal promoter. J Immunol 2004; 172:5512–21. [DOI] [PubMed] [Google Scholar]
- 20. Geng Y, Laslo P, Barton K, Wang CR. Transcriptional regulation of CD1D1 by Ets family transcription factors. J Immunol 2005; 175:1022–9. [DOI] [PubMed] [Google Scholar]
- 21. Chen QY, Zhang T, Pincus SH, Wu S, Ricks D, Liu D, et al Human CD1D gene expression is regulated by LEF‐1 through distal promoter regulatory elements. J Immunol 2010; 184:5047–54. [DOI] [PubMed] [Google Scholar]
- 22. Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno‐associated virus serotype 8 vectors in mice. J Virol 2005; 79:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sands MS. AAV‐mediated liver‐directed gene therapy. Methods Mol Biol 2011; 807:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vercauteren K, Hoffman BE, Zolotukhin I, Keeler GD, Xiao JW, Basner‐Tschakarjan E, et al Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol Ther 2016; 24:1042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu L, Van Kaer L. Natural killer T cells in health and disease. Front Biosci (Schol Ed) 2011; 3:236–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chuah C, Jones MK, Burke ML, McManus DP, Gobert GN. Cellular and chemokine‐mediated regulation in schistosome‐induced hepatic pathology. Trends Parasitol 2014; 30:141–50. [DOI] [PubMed] [Google Scholar]
- 27. Kumar V. NKT‐cell subsets: promoters and protectors in inflammatory liver disease. J Hepatol 2013; 59:618–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perera L, Shao L, Patel A, Evans K, Meresse B, Blumberg R, et al Expression of nonclassical class I molecules by intestinal epithelial cells. Inflamm Bowel Dis 2007; 13:298–307. [DOI] [PubMed] [Google Scholar]
- 29. Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, et al Recognition of lyso‐phospholipids by human natural killer T lymphocytes. PLoS Biol 2009; 7:e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk‐Hasdemir D, et al Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014; 156:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mallevaey T, Fontaine J, Breuilh L, Paget C, Castro‐Keller A, Vendeville C, et al Invariant and noninvariant natural killer T cells exert opposite regulatory functions on the immune response during murine schistosomiasis. Infect Immun 2007; 75:2171–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL‐4 production in CD1‐deficient mice. Immunity 1997; 6:459–67. [DOI] [PubMed] [Google Scholar]
- 34. Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL‐4. Immunity 1997; 6:469–77. [DOI] [PubMed] [Google Scholar]
- 35. Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO, et al Protective mucosal immunity mediated by epithelial CD1d and IL‐10. Nature 2014; 509:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang CC, Chung‐Ti T, Chao T. Studies on the migratory route of Schistosoma japonicum in its final host. Acta Zoologica Sinica 1973; 19. [Google Scholar]
- 37. Gobert GN, Chai M, McManus DP. Biology of the schistosome lung‐stage schistosomulum. Parasitology 2007; 134:453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. IL‐33 expression in hepatocyte from the liver of S. japonicum‐infected mice.
Figure S2. SP1, PU.1, and LEF‐1 mRNA levels in hepatocytes from S. japonicum‐infected mice.
Figure S3. AAV8‐mediated overexpression of CD1d does not affect NKT cells in the spleen of S. japonicum‐infected mice.
Data Availability Statement
Data used for the analysis are available from the corresponding author upon reasonable request.


