In this review, we summarize recent understandings on the assembly, activation and regulation patterns of the NLRP6 inflammasome, as well as its diverse impacts on tissue homeostasis and disease in multiple organs.
Keywords: caspase, inflammasome, innate immune receptors, microbiome, NLRP6, pattern recognition receptors
Summary
The NOD‐like receptor family pyrin domain containing 6 (NLRP6), a member of the NOD‐like receptor (NLR) family, acts as a cytosolic innate immune sensor that recognizes microbe‐associated molecular patterns. In some circumstances upon activation, NLRP6 recruits the adaptor apoptosis‐associated speck‐like protein (ASC) and the inflammatory caspase‐1 or caspase‐11 to form an inflammasome, which mediates the maturation and secretion of the pro‐inflammatory cytokines IL‐18 and IL‐1β. In other contexts, NLRP6 can exert its function in an inflammasome‐independent manner. Tight regulation of the NLRP6 inflammasome is critical in maintaining tissue homeostasis, while improper inflammasome activation may contribute to the development of multiple diseases. In intestinal epithelial cells, the NLRP6 inflammasome is suggested to play a role in regulating gut microbiome composition, goblet cell function and related susceptibility to gastrointestinal inflammatory, infectious and neoplastic diseases. Additionally, NLRP6 may regulate extra‐intestinal diseases. In this review, we summarize current knowledge on the NLRP6 inflammasome and its activation and regulation patterns, as well as its effector functions contributing to disease modulation. We discuss current challenges in NLRP6 research and future prospects in harnessing its function into potential human interventions.
Abbreviations
- AMP
Antimicrobial peptide
- ASC
Apoptosis‐associated speck‐like protein
- Asp‐Glu‐Ala‐His
RNA helicase DEAH
- ATP
Adenosine triphosphate
- CRH
Corticotropin‐releasing hormone
- DAMPs
Damage‐associated molecular patterns
- Dhx15
DEAH (Asp‐Glu‐Ala‐His) box helicase 15
- DSS
Dextran sulphate sodium
- HSCT
Haematopoietic stem cell transplantation
- IEC
Mammalian intestinal epithelial cell
- IL
Interleukin
- KO
Knockout
- LPS
Lipopolysaccharide
- LRR
Leucine‐rich repeat
- LTA
Lipoteichoic acid
- MAMPs
Microbe‐associated molecular patterns
- MAPK
Mitogen‐activated protein kinase
- NAFLD
Non‐alcoholic fatty liver disease
- NBD
Nucleotide‐binding domain
- NF‐κB
'kappa‐light‐chain‐enhancer' of activated B cells
- NLR
NOD‐like receptor
- NLRP6
NOD‐like receptor family pyrin domain containing 6
- NLRs
Nucleotide oligomerization domain (NOD)‐like receptors
- PPAR‐γ
Peroxisome proliferator‐activated receptor γ
- PRRs
Pattern recognition receptors
- PYD
Pyrin domain
- TLR
Toll‐like receptors
- TNF‐α
Tumour necrosis factor α
- WT
Wild type
INTRODUCTION
Viruses and bacteria infecting or colonizing mammalian organisms induce host signalling, mediated by physical contact, or by toxin, microbial nucleic acid, metabolite or foreign protein section, which result in a variety of host responses to such infection. These so‐called microbe‐associated molecular patterns (MAMPs) can be sensed by host pattern recognition receptors (PRRs), which initiate innate immune responses. 1 , 2 , 3 , 4 , 5 Apart from MAMPs, sterile stressors associated with host cell damage such as ATP or heat‐shock proteins, termed damage‐associated molecular patterns (DAMPs), are likewise sensed by PRRs. 6 PRRs are localized either extracellularly like the Toll‐like receptors (TLR) or intracellularly like the nucleotide oligomerization domain (NOD)‐like receptors (NLRs), enabling the cell to sense both extra‐ and intracellular stressors and infections. A subset of these NLRs is able to build multiprotein complexes with apoptosis‐associated speck‐like protein (ASC) and caspase‐1 or caspase‐11 7 , 8 upon DAMP or MAMP‐mediated activation, termed inflammasomes. Through inflammasome assembly, the associated caspase gets activated and cleaves the inactive precursors of interleukin (IL)‐1β or IL‐18, respectively, into the mature cytokine, thereby inducing an inflammatory immune response in reacting to infection or cellular damage. 8 , 9 Alternatively, the activated caspase processes gasdermin D, consequently resulting in pore formation in the membrane of affected cells, coupled with a cytokine release, collectively driving pyroptosis, a form of programmed cell death. 10 , 11
NLRP6 (originally termed PYPAF5) belongs to the NLR family and together with NLRP1, NLRP3, NLRP7 and NLRC4 constitutes a capacity to build a fully operational inflammasome. 9 Consequently, conventional Nlrp6 deficiency in mice is accompanied by reduced serum IL‐18 levels in steady‐state conditions and decreased IL‐18 levels in colon and serum in mice upon induction of chemical‐induced colitis and colitis‐associated tumorigenesis. 12 , 13 While in vitro experiments demonstrate an activation of IL‐1β via the NLRP6 inflammasome, 14 mature IL‐1β protein level remains unaltered or even increased in Nlrp6 KO mice. 13 Of note, hepatic NLRP3 is upregulated in Nlrp6‐deficient mice, 15 suggesting a compensatory mechanism of NLRP3 activation upon NLRP6 deficiency, potentially explaining the unexpected surge in IL‐1β levels in this setting. Like other NLR family members, NLRP6 is composed of three domains. The N‐terminus consists of a pyrin domain (PYD) and is considered the essential element for inflammasome assembly as it interacts with ASC. 16 The nucleotide‐binding domain (NBD) builds the central module of NLPR6 and is followed by the C‐terminal leucine‐rich repeat (LRR) domain, which senses DAMPs and MAMPs. 9 , 17 The NLRP6 protein is mainly expressed in the lung and liver but is highest at the intestine, 12 , 18 making the gastrointestinal tract the most widely studied organ for the investigation of the NLRP6 inflammasome in health and disease. Interestingly, in human periodontium and gingiva, NLPR6 seems to also impact on the homeostasis of the oral cavity. 19 , 20
A tight regulation of inflammasomes in general is of special organismal importance, as its chronic and unbalanced activation manifests in several inflammatory and metabolic diseases, for example familial Mediterranean fever, which is caused by mutations in the pyrin‐coding gene MEFV, or cryopyrin‐associated periodic syndrome, which is ascribed to point mutations in Nlrp3 gene. 21 Unlike the NLRP3 inflammasome, the cell‐specific role of its close relative NLRP6 in health and disease is not fully elucidated to date. In this review, we summarize recent insights related to assembly and activation mechanisms of NLRP6 and the NLRP6 inflammasome, as well its pleiotropic roles in contributing to tissue homeostasis and disease in multiple organs.
NLRP6 INFLAMMASOME ASSEMBLY, REGULATION AND EFFECTOR FUNCTION
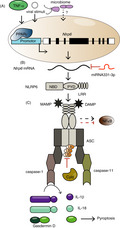
Nlrp6 expression is reported to be regulated at the transcriptional and post‐translational level, which, in turn, is mainly controlled by upstream microbial and metabolic stimuli (Figure 1). In vitro studies on Caco‐2 cells demonstrate that the Nlrp6 promoter region contains binding sites for the transcription factor peroxisome proliferator‐activated receptor γ (PPAR‐γ), commonly known to be involved in metabolic regulation. 22 Furthermore, expression of Nlrp6 is induced by the PPAR‐γ agonist rosiglitazone. 23 In addition, an increase in PPAR‐γ protein levels during embryonic development is accompanied by an increase in Nlrp6 mRNA transcription. 23 Moreover, inflammatory signals such as tumour necrosis factor‐α (TNF‐α) or viral stimuli are able to induce the transcription of Nlrp6, at least in mouse embryonic fibroblasts. 24 Beyond transcription, Nlrp6 was shown to be regulated by miRNA‐331‐3p in haemin‐treated BV2 cells. 25 Once translated, NLRP6 activity is tightly regulated, which requires specific triggers to induce an assembly of a bona fide NLRP6 inflammasome. Upon activation, NLRP6 assembles with ASC, leading to caspase‐1 and nuclear factor 'kappa‐light‐chain‐enhancer' of activated B cell (NF‐κB) activation in vitro. 14 A recent cryo‐electron microscopy study revealed the structural mechanism of NLRP6 inflammasome assembly. 16 The PYD domain of NLRP6 forms filamentous structures through self‐assembly, followed by conformational changes, which enable the recruitment of the adaptor ASC via PYD–PYD binding. This assembling process is strengthened by the NBD domain of NLRP6.
FIGURE 1.
Regulation of Nlrp6 expression and NLRP6 inflammasome assembly. (A) Tumour necrosis factor‐α (TNF‐α), viral stimuli and elusive microbiome‐mediated signalling can induce the transcription of Nlrp6. The transcription factor peroxisome proliferator‐activated receptor‐γ (PPARγ) binds to the promoter region of Nlrp6 and induces its transcription. (B) The miRNA 331‐3p inhibits translation of the Nlrp6 transcript. (C) Microbe‐associated molecular patterns (MAMPs) and damage‐associated molecular patterns (DAMPs) bind to the C‐terminal leucine‐rich repeats (LRR) of the NLRP6 protein to enable a bona fide inflammasome assembly. However, these MAMPs and DAMPs remain mostly unidentified to date. The NLRP6 protein further contains a nucleotide‐binding domain (NBD) and a pyrin domain (PYD), which interact with the ASC adaptor. The NLPR6 inflammasome activates caspase‐1 or caspase‐11 and cleaves the proIL‐1β and proIL‐18 precursors into the mature cytokines. Gasdermin D is cleaved and drives pyroptosis. NLRP6 inhibits or increases NFκb activity, probably in a cell‐ and context‐specific manner. The deubiquitinase Cyld inhibits NLRP6 inflammasome formation
The activation of NLRP6 inflammasome can be regulated by different activators or inhibitors (Figure 1). As very recently described, the deubiquitinase Cyld regulates the function of the NLRP6 inflammasome and prevents excessive inflammation via the production of IL‐18. Cyld cleaves the K63‐linked ubiquitination of NLRP6, thereby inhibiting NLRP6’s ability to interact with ASC. 26 Among others, bacteria of different taxa were shown to regulate NLPR6 inflammasome formation. 27 , 28 , 29 In line, the microbial ligands lipopolysaccharide (LPS), MDP, iE‐DAP and Pam3CSK4 have been reported to induce NLPR6 activation in human periodontal ligament cells. 19 Additionally, lipoteichoic acid (LTA), a surface‐associated adhesion amphiphile of Gram‐positive bacteria, induces the formation of NLRP6 inflammasome, and interestingly, in this case NLRP6 activates the non‐canonical pathway involving caspase‐11 in vivo. 27 Furthermore, the microbial metabolites taurine, histamine and spermine have been shown to modulate the immune responses by regulating NLRP6 inflammasome assembly. 29 Beyond bacterial products, NLRP6 also functions as a viral sensor by interacting with the adenosine triphosphate (ATP)‐dependent RNA helicase DEAH (Asp‐Glu‐Ala‐His) box helicase 15 (Dhx15). 24 Full‐length NLRP6, but not NLRP3, is required for binding the viral protein Dhx15. 24
Upon NLRP6 inflammasome assembly, it induces inflammatory responses via the cleavage of proIL‐1β and proIL‐18 into their mature forms. Other downstream signalling pathways can also be induced by the NLRP6 inflammasome. Chen et al show that alkali‐burn injury induces NLRP3 and NLRP6 inflammasome formation, whereby NLRP3 is activated by NF‐κB, while NLRP6 dampens NF‐κB activity, demonstrating the opposing roles of these two inflammasomes in this context. 30 In human periodontal ligament cells, NLRP6 inhibits NF‐κB and ERK signalling, thereby dampening the release of IL‐6 and TNF‐α. 19 Additionally, NLRP6 inhibits the activation of NF‐κB signalling pathway and p38 mitogen‐activated protein kinase (MAPK) signalling pathway after allogeneic haematopoietic stem cell transplantation‐induced liver damage in mice. Following haematopoietic stem cell transplantation in Nlrp6 knockout (KO) mice, an overcompensatory NLRP3 expression is accompanied by exacerbated liver damage, suggesting an opposing role of the NLRP6 and NLRP3 inflammasome in this context. 15 Interestingly, Nlrp6‐deficient mice are highly resistant to infections with the facultative intracellular pathogens Listeria monocytogenes and Salmonella typhimurium and the extracellular pathogen Escherichia coli, indicating that NLRP6 might dampen the inflammatory response and acts as a counterpart to the other PRRs by downregulating the TLR‐mediated NF‐κB and MAPK signalling. 31 Collectively, several independent studies demonstrate an interaction of the NLRP6 inflammasome with NF‐κB signalling pathways, driving inflammatory or anti‐inflammatory functions at different physiological contexts. To better delineate these partly opposing results, a cell‐specific role of NLRP6 needs to be further investigated in future studies.
ROLE OF NLRP6 IN HEALTH AND DISEASE
NLRP6 in the gut
The mammalian intestinal mucosa and lumen contain trillions of microorganisms, including mostly not only bacteria but also fungi, virus and parasites, which have coevolved with the host and are collectively termed the microbiome. Regulating host–microbe symbiosis is critically important in enabling critical functions conferred by the microbes, while avoiding pathogenic invasion into the sterile host or elicitation of uncontrolled inflammation against non‐invading resident microbes. Such a fine and tightly regulated balance is achieved by an induction of host immune tolerance to innocuous stimuli during steady state, coupled with an ability to induce a potent immune response against invading pathogens or pathobionts. 32 The high expression of Nlrp6 in the mammalian intestinal epithelial cells (IECs), including both enterocytes and goblet cells, 12 , 33 positions it as a potential culprit regulator of these interactions (Figure 2). Indeed, in one vivarium, Nlrp6 deletion in mice led to faecal microbiota shifts towards higher abundance of the bacterial family Prevotellaceae and phyla TM7. 12 This altered microbiome was transferrable to wild‐type (WT) mice and associated with higher susceptibility to dextran sulphate sodium (DSS)‐induced colitis. 12 In another vivarium, NLRP6 was important in controlling the abundance of the colitis‐promoting pathobiont Akkermansia muciniphila, thus protecting IL‐10 KO mice from developing spontaneous colitis. 34 The aberrant gut microbial community in Nlrp6 inflammasome‐deficient mice also promotes the susceptibility to colitis‐induced carcinogenesis through the activation of IL‐6 signalling in IEC. 35 Importantly, the impact of NLRP6 on gut microbiota composition is community‐dependent and manifests itself upon exposure to a diverse enough microbiome configuration. As such, microbiome‐deficient Nlrp6‐deficient mice transferred with microbiome configurations from a high barrier facility, in which community diversity was highly limited and pathobionts that were absent failed to induce a state of dysbiosis in recipient Nlrp6‐deficient mice. In contrast, transfer of a diverse microbiome configuration led to development of a marked dysbiosis, suggesting that the absence of Nlrp6, coupled with environmental microbial diversity, mutually engage in promoting dysbiosis. 36 , 37 , 38 These observations provide a framework for vivarium specificity of microbiome alterations that spans beyond that of NLRP6, and mirror human interindividual microbiome variability driving distinct disease features even in the presence of identical host genetic susceptibilities. 39 From a research perspective, this highlights the importance of combining littermate‐controlled experimental design together with faecal microbiome transfer into germ‐free mice (or microbiome‐depleted mice) in studying microbiome modulatory roles of this and other NLRs. 40
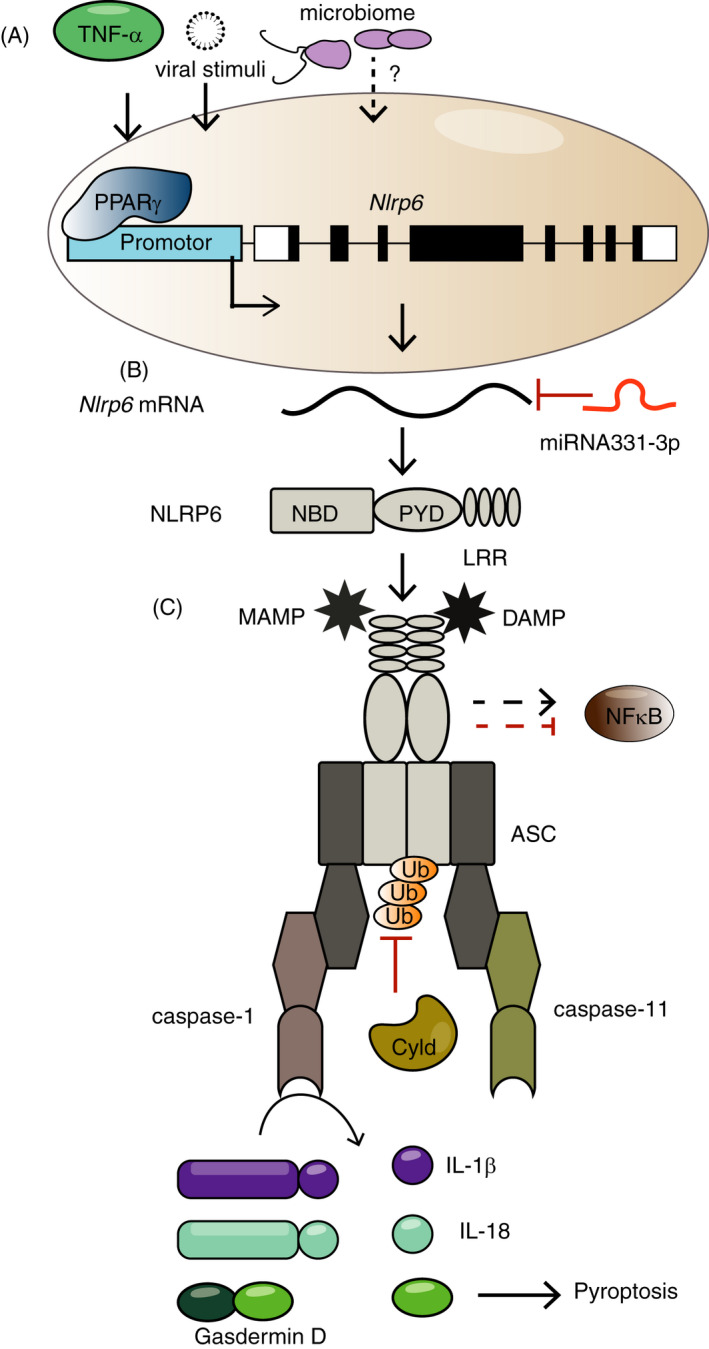
FIGURE 2.
NLRP6 in the gut. In intestinal enterocytes, NLRP6 inflammasome can be modulated by commensal‐derived metabolites (eg taurine, spermine and histamine), resulting in regulation of mature IL‐18 production and of gut microbiome composition in a vivarium‐specific and community‐dependent manner (A), as well as the generation of unique antimicrobial peptide repertoires (B). The shaping of gut microbiome is suggested, in turn, to contribute to the susceptibility to dextran sulphate sodium (DSS)‐induced intestinal inflammation (C). Upon DSS‐induced intestinal injury, the upregulated Nlrp6 expression in infiltrating inflammatory monocytes also reduces susceptibility to DSS colitis, mediated by IL‐18‐dependent tumour necrosis factor‐α (TNF‐α) production. In goblet cells, NLRP6 inflammasome activation impacts the secretion of mucus (D) through two suggested mechanisms. The first involves regulation of autophagy, which controls exocytosis of goblet cell mucin granule, while the second involves sensing of Toll‐like receptor (TLR) ligands by TLRs, triggering Myd88‐dependent reactive oxygen species (ROS) production, which induces Muc2 exocytosis
The molecular mechanism by which NLRP6‐associated signalling modulates the gut microbiome is just in the beginning to be revealed. As mentioned above, commensal‐derived metabolites (including taurine, histamine and spermine) modulate the gut epithelial NLRP6 inflammasome and IL‐18 secretion, and subsequently impact the expression of downstream antimicrobial peptide (AMP) genes in the gut, thus shaping the host–microbiome interface and determining susceptibility to DSS‐induced colitis. 29 A mechanism for this modulation may involve metabolite‐induced morphological alterations of IEC intercellular connections and cellular morphology. 41 In addition to commensal‐associated modulators, NLRP6 can directly bind and be activated by LTA derived from Gram‐positive bacteria, that is Listeria monocytogenes. 27 Sensing of cytosolic LTA by NLRP6 in macrophages results in the activation of caspase‐11 and caspase‐1 via the adaptor ASC, which in turn leads to IL‐1β and IL‐18 maturation and enhances Gram‐positive pathogen infection in vivo. Additionally, the Gram‐negative bacterial ligand LPS binds directly to NLRP6 monomer, and triggers its oligomerization and recruitment of ASC, thus initiating the assembly of NLRP6 inflammasome, suggesting that NLPR6 might also act as a potential LPS sensor. 42 Moreover, the dietary component flavone is suggested to be another modulator of NLRP6 signalling pathway, and exerts its anti‐inflammatory effect by alteration of gut microbiome downstream of the inflammasome‐independent NLRP6 signalling. 43 As mentioned above, the deubiquitinase Cyld inhibits NLRP6 protein interactions, which tightly regulates NLRP6‐mediated IL‐18 secretion and limits severe intestinal inflammation in mice upon Citrobacter Rodentium infection, 26 providing insights into possible mechanisms promoting negative regulation of the NLRP6 inflammasome. Corticotropin‐releasing hormone (CRH), a stress hormone released during a water‐avoidance stress model in mice, also exerts inhibiting effects on Nlrp6 expression and induces gut dysbiosis, which consequently mediates the development of enteritis in mice. 44
The NLRP6 inflammasome is also expressed in intestinal goblet cells and thus impacts on mucus secretion by goblet cells. One suggested mechanism to this effect involves autophagy, a critical process for exocytosis of goblet cell mucin granule. 33 Nlrp6‐deficient mice feature impaired autophagy leading to altered secretion of mucus granules in goblet cells, thereby contributing to susceptibility to Citrobacter Rodentium infection. 33 A second mechanism involves sentinel goblet cells situated at the entrance to the colonic crypt, which may sense non‐specific TLR ligands and trigger intracellular NLRP6 activation, subsequently inducing mucin exocytosis to expel bacteria. 45 As above, in some vivaria, only excess administration of LPS enables this NLRP6 goblet cell function. 46
It should be noted that, in addition to IEC, NLRP6 may also play a role in regulating intestinal homeostasis through its functions in other intestinal cell types. For example, upon DSS‐induced intestinal injury, an upregulated expression of Nlrp6 in infiltrating Ly6Chi inflammatory monocytes is critical in controlling bacteria‐driven inflammation, mediated through induction of TNF‐α production and IL‐18 signalling. 47 Likewise, the activity of NLRP6 in colonic myofibroblast 48 and haematopoietic cells 13 is suggested to be important in controlling the epithelial proliferation and self‐renewal upon chronic injury. In experimental mouse models of allogeneic bone marrow transplantation, host NLRP6 exacerbates the severity and mortality of gastrointestinal graft‐versus‐host disease, independent of microbial dysbiosis. This phenotype relies on the expression of Nlrp6 in host non‐haematopoietic compartment, and in part stems from signalling through the intestinal metabolite taurine. 49 Furthermore, NLRP6 can modulate host response to bacterial infection in other inflammasome‐ and microbiome‐independent manners and physiological contexts. For example, Nlrp6‐deficient mice are less susceptible to several Gram‐positive and Gram‐negative bacterial pathogens, along with enhanced recruitment of monocytes and neutrophils. 31 In this context, NLRP6 plays the detrimental role by negatively regulating the activation of NF‐κB and MAPK signalling. 31 In addition to the aforementioned antibacterial immunity, NLRP6 is critical in controlling the enteric virus infection when mice are challenged with encephalomyocarditis virus or murine norovirus 1. 24 Furthermore, upregulation of Nlrp6 expression (and also Pycard, Caspase‐1 and IL18) in rat intestine at late fetal development 23 may point towards a potential role in intestinal organogenesis, which merits further mechanistic studies. In summary, NLRP6 emerges as an important player in gut homeostasis and interactions with commensal bacteria, viruses and pathogens. This highlights NLRP6 as a potential future exploitable checkpoint, whose manipulations may be harnessed to control infectious, inflammatory and potentially metabolic disease. However, the role of NLRP6 at the cellular and molecular level needs to be investigated in more depth to develop such adequate therapeutic strategies.
NLRP6 in the liver
At the hepatic sinusoids, the oxygen‐rich blood of the hepatic artery is mixed with the less oxygenated but nutrient‐rich portal blood returning from the intestines. The portal venous blood also contains ample antigens and microbial products of digested food and the intestinal microbiome, thereby constantly challenging the liver as the ‘first‐pass organ’. 50 This influx bears pathophysiological relevance. In obesity, for example, an altered release of DAMPs and MAMPs by a dysbiotic gut microbiome composition 51 is delivered to the liver through the portal circulation, where it creates a state of chronic low‐grade inflammation contributing to the development of non‐alcoholic fatty liver disease (NAFLD). 52 , 53 NLRP6 modulates these interactions in mice, through regulation of gut microbiota leading to enhanced influx of TLR4 and TLR9 ligands from the intestine into the liver upon Nlrp6 deficiency. 52 Other NLRP6 hepatic effects may be independent of its impact on the gut microbiome, as is described below (Figure 3).
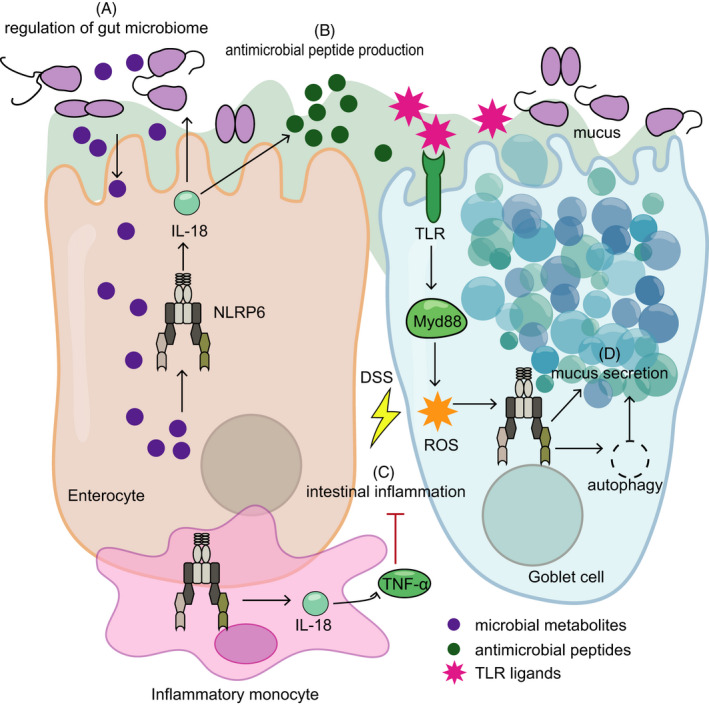
FIGURE 3.
NLRP6 in the liver. (A) Stimulation of hepatocytes with LPS or palmitic acid is accompanied by increased Nlrp6 expression. (B) In hepatic stellate cells, NLRP6 interacts with PPM1A to inhibit transforming growth factor‐β (TGF‐β). This, in turn, leads to reduced phosphorylation of Smad2/3 and reduced proliferation and collagen expression. (C) NLRP6 prevents the infiltration of macrophages into the liver by reducing C‐C chemokine ligand 20 (CCL20) levels. (D) Whole‐body Nlrp6 deletion leads to a dysregulated microbiota, which increases the influx of TLR ligands in the circulation, thereby promoting fibrosis. (E) In fibrotic livers, Nlrp6 expression is decreased, the functional implications of which remain unknown to date
Indeed, when investigating the expression patters of PRRs in human and murine solid organs, a relatively high expression level of Nlrp6 mRNA was found not only in the murine intestine but also in the liver when compared to the spleen. 12 , 18 Likewise, NLRP6 protein expression is most dominant in the intestine, followed by the liver. 24 Importantly, Nlrp6 expression is decreased in human biopsies from fibrotic and cirrhotic livers when compared to healthy controls, 54 suggesting a putative role of NLRP6 in human liver disease. Indeed, Zhu and colleges 54 identified a protective in vitro function of NLRP6 in the hepatic stellate cell line LX‐2, in which it inhibited cell proliferation and reduced collagen expression. On a molecular level, NLRP6 inhibited TGF‐β‐mediated phosphorylation of Smad2/3 in a PPM1A‐dependent manner, suggesting that it builds a complex with PPM1A to exert its inhibitory effect on TGF‐β. 54 Interestingly, overexpression of Nlrp6 in gingival fibroblasts induces pyroptosis, 20 suggesting a potential pan‐fibroblast activity across organs. Hepatocytes treated with either palmitic acid or LPS featured an increase in Nlrp3, Nlrp10 and Nlrp6 mRNA levels, 55 suggesting an additional hepatocyte‐specific role of NLPR6 in these conditions. Intriguingly, deep RNA sequencing and single‐molecule transcript imaging demonstrated that Nlrp6 mRNA level is more abundant in the nucleus than the cytoplasm in murine liver, which might play a role in buffering the cytosolic Nlrp6 gene expression noise. 56
Following the induction of liver damage secondary to allogenic haematopoietic stem cell transplantation (HSCT), hepatic Nlrp6 expression is increased in mice, while Nlrp6‐deficient mice manifest more severe liver damage after HSCT compared to controls. This exacerbated liver damage is coupled with higher levels of TGF‐β, suggesting a protective role of NLRP6 in the liver via regulation of TGF‐β. 15 However, the lack of a conditional Nlrp6 KO models precludes the identification of a cell‐specific role of NLRP6 in this model. In an alcoholic hepatitis model in mice, Nlrp6 expression is downregulated compared with controls, while NLPR6 is suggested to prevent the infiltration of macrophages into the liver by reducing CCL20 levels, thereby putatively impacts disease severity. In agreement, a viral overexpression of Nlrp6 repressed the development of fibrosis in an alcoholic hepatitis model. 57 Taken together, a protective role of NLPR6 in the liver is suggested across different disease models. In contrast, parasitic‐induced liver fibrosis by Schistosoma mansoni infection in whole body of Nlrp6‐deficient mice is associated with a reduced macrophage and neutrophil infiltration, coupled with abrogated cytokine release (eg CCL2, CCL3, IL‐10 and IL‐5) and collagen deposition. 58 Cell‐specific depletion of Nlrp6 may clarify these apparent contradictory NLRP6‐driven innate immune responses noted in different liver disease models. Of note, in whole body of Nlrp6‐depleted mice, secondary effects induced by Nlrp6‐deficiency, such as compositional changes of the gut microbiome, cannot be excluded at present as potential indirect contributors to the observed phenotypes and may impact the discrepancies noted between different models induced at different vivaria.
NLRP6 in other organs
The function of the NLRP6 inflammasome, and implications on organ‐specific diseases, is increasingly investigated in tissues beyond the gut and liver, including, but not limited to, the central nervous system, lung and kidney. For example, in a rat model of middle cerebral artery occlusion/reperfusion, an upregulated brain expression of Nlrp6 aggravates cerebral ischaemic and reperfusion injury and worsens neurological functions, indicating a potential neuroprotective role of NLRP6. 59 In this regard, a recent in vitro study showed that upon oxygen–glucose deprivation, NLRP6 inflammasome assembly in astrocytes induces the production of the pro‐inflammatory cytokines IL‐1β and IL‐8, coupled with promotion of pyroptosis. 60 Likewise in another mouse model of intracerebral haemorrhage, Nlrp6 downregulation targeted by a microRNA miR‐331‐3p dampens the inflammatory responses and restores the neurological function after cerebral haemorrhage. 25 In contrast, NLRP6 plays a protective role in promoting the recovery after peripheral nerve injury, suggestively via dampening the MAPK signalling pathway in an inflammasome‐independent manner. 61
In the lung, Nlrp6‐deficient mice are more resistant to pulmonary Staphylococcus aureus infection compared with WT counterparts, and this protective effect is mediated by enhanced neutrophil recruitment and neutrophil‐associated bacterial clearance following infection. 28 Nlrp6 is also expressed in the tubular epithelium of healthy murine and human kidneys, and its expression is reduced upon acute kidney injury induced by folate acid or cisplatin overdose, in both human or mouse models. 62 Furthermore, deficiency of Nlrp6 in mice aggravates the severity of nephrotoxic acute kidney injury, characterized by enhanced apoptotic and pro‐inflammatory responses in the tubular cells, suggesting a nephroprotective role of NLRP6 in acute kidney injury. 62 Taken together, the complicated and multifaceted functions of NLRP6 in regulating disease associated with a multitude of organs warrant a continued thorough investigation.
CHALLENGES AND UNKNOWNS IN NLRP6 INFLAMMASOME RESEARCH
The above‐mentioned pleiotropic and multifaceted functions of the NLRP6 inflammasome make it an enigmatic but intriguing NLR, along with many unknowns and challenges remaining to be investigated. First, the sterile and non‐sterile ligands of NLRP6, and their mechanisms of activity, have only partially been deciphered to date. Despite the revealed NLRP6 activators such as commensal‐associated metabolites, bacterial ligands LTA, LPS and viral RNA, 24 , 27 , 29 , 42 the manners and mechanisms by which other pathogenic stimuli derived from fungi, parasites and host‐derived activating signals (eg DAMPs) bind and trigger NLRP6 signalling remain to be explored. This would likely necessitate unravelling of the Cryo‐EM atomic‐resolution structure of NLRP6 ligand binding. Second, identification of negative regulators of NLRP6 inflammasome in the context of different diseases is an equally important and fascinating topic. Given the intimate interaction between NLRP6 and the gut microbiome, 12 it is yet to be determined how various microbial activators and inhibitors counteract to fine‐tune the activation of the NLRP6 inflammasome and therefore maintain gut homeostasis, especially at the human setting. Third, more researches are warranted in identifying the downstream effectors of NLRP6 inflammasome activation beyond secretion of pro‐inflammatory cytokines, including but not limited to induction of pyroptosis, regulation of intestinal barrier function, modulation of adaptive immunity and orchestration of cell metabolism. Moreover, given inconsistent results regarding the role of NLRP6 in regulating gut microbiome composition across different vivaria, future studies investigating its role in modulating gastrointestinal and extra‐gastrointestinal diseases will require proper littermate‐controlled and microbial‐transfer strategies. However, in all the above‐mentioned strategies, deciphering a cell‐specific role of NLRP6 is of essential importance as it could help to delineate opposing and complementary cellular functions, while decoding the roles of elusive NLRP6‐modulated pathways. Last but not least, the role of NLRP6 inflammasome in the context of human health and disease is still poorly understood, as the majority of studies to date are conducted in murine models. To date, several inhibitors of NLRP3 inflammasome activation (eg MCC950, CY‐09) have been recognized and applied as promising interventions in different disease models, 63 while similar therapeutic applications have not been identified to date for the NLRP6 inflammasome. It is imperative that future research will focus on translating the NLRP6 inflammasome biology towards such identification and application of potential therapeutic targets as disease treatment in human NLRP6‐driven disease.
CONCLUDING REMARKS
Recent advances in revealing the biology and functions of NLRP6, in the context of inflammasome formation or non‐inflammasome functions, and its connection to health and disease have greatly enhanced our understandings of how NLRP6 impacts the mammalian host. However, conflicting observations in different studies suggest that NLRP6 can harness a context‐specific pro‐inflammatory or anti‐inflammatory innate immune activation. Variable microbiome configurations can further impact NLRP6‐mediated modulation of mammalian health and disease traits. We strongly believe that utilization of such inherent variability may facilitate achieving a mechanistic understanding of NLRP6 interactions with the host and its microbiome, thereby enabling deciphering molecular and cell‐specific mechanism of NLRP6 regulation and effector function. Unveiling of such personalized structural and molecular considerations governing the modulation of NLRP6 may enable to identify new regulatory checkpoints, to be further developed as potential therapeutics of NLRP6‐associated disease.
FUNDING STATEMENT
Open Access funding enabled and organized by ProjektDEAL.
CONFLICT OF INTEREST
EE is a salaried scientific consultant for DayTwo and BiomX. DZ and LK have nothing to declare.
AUTHOR CONTRIBUTION
All authors performed an extensive literature research, contributed substantially to discussion of the content, and wrote and edited the manuscript.
ACKNOWLEDGMENTS
We thank the members of the Elinav Lab for discussions and apologize for authors whose work was not cited because of space constraints. DZ is the recipient of the European Crohn's and Colitis Organization (ECCO) Fellowship, and is supported by the Ke Lin Program of the First Affiliated Hospital, Sun Yat‐sen University. LK is funded by a postdoctoral fellowship by the Walter Benjamin fellowship from Deutsche Forschungsgemeinschaft (DFG). EE is the incumbent of the Sir Marc and Lady Tania Feldmann Professorial Chair, a senior fellow at the Canadian Institute of Advanced Research and an international scholar at the Bill & Melinda Gates Foundation and the Howard Hughes Medical Institute (HHMI).
Danping Zheng and Lara Kern are contributed equally.
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
Enigmatic inflammasomes. Immunology 2021, 162: 249‐251.
MHC class I transactivator NLRC5 in host immunity, cancer and beyond. Immunology 2021, 162: 252‐261.
NLRP9 in innate immunity and inflammation. Immunology 2021, 162: 262‐267.
NLR in eXile: Emerging roles of NLRX1 in immunity and human disease. Immunology 2021, 162: 268‐280.
DATA AVAILABILITY STATEMENT
There are no raw data generated in this manuscript.
REFERENCES
- 1. Kanneganti T‐D, Body‐Malapel M, Amer A, Park J‐H, Whitfield J, Franchi L, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase‐1 in response to viral infection and double‐stranded RNA. J Biol Chem. 2006;281:36560–8. [DOI] [PubMed] [Google Scholar]
- 2. Kanneganti T‐D, Özören N, Body‐Malapel M, Amer A, Park J‐H, Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase‐1 through cryopyrin/Nalp3. Nature. 2006;440:233–6. [DOI] [PubMed] [Google Scholar]
- 3. Muruve DA, Pétrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–7. [DOI] [PubMed] [Google Scholar]
- 4. Amer A, Franchi L, Kanneganti TD, Body‐Malapel M, Özören N, Brady G, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–23. [DOI] [PubMed] [Google Scholar]
- 5. Miao EA, Alpuche‐Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, et al. Cytoplasmic flagellin activates caspase‐1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–75. [DOI] [PubMed] [Google Scholar]
- 6. Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012;249:158–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kayagaki N, Warming S, Lamkanfi M, Walle LV, Louie S, Dong J, et al. Non‐canonical inflammasome activation targets caspase‐11. Nature. 2011;479:117–21. [DOI] [PubMed] [Google Scholar]
- 8. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL‐beta. Mol Cell. 2002;10:417–26. [DOI] [PubMed] [Google Scholar]
- 9. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–22. [DOI] [PubMed] [Google Scholar]
- 10. Ding J, Wang K, Liu W, She Y, Sun QI, Shi J, et al. Pore‐forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–6. [DOI] [PubMed] [Google Scholar]
- 11. Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome‐activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elinav E, Strowig T, Kau A, Henao‐Mejia J, Thaiss C, Booth C, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen GY, Liu M, Wang F, Bertin J, Núñez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grenier JM, Wang L, Manji GA, Huang WJ, Al‐Garawi A, Kelly R, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF‐kappaB and caspase‐1. FEBS Lett. 2002;530(1–3):73–8. [DOI] [PubMed] [Google Scholar]
- 15. Li M, Chen Y, Shi J, Ju W, Qi K, Fu C, et al. NLRP6 deficiency aggravates liver injury after allogeneic hematopoietic stem cell transplantation. Int Immunopharmacol. 2019;74:105740. [DOI] [PubMed] [Google Scholar]
- 16. Shen C, Lu A, Xie WJ, Ruan J, Negro R, Egelman EH, et al. Molecular mechanism for NLRP6 inflammasome assembly and activation. Proc Natl Acad Sci USA. 2019;116:2052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol Rev. 2015;265:35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lech M, Avila‐Ferrufino A, Skuginna V, Susanti HE, Anders HJ. Quantitative expression of RIG‐like helicase, NOD‐like receptor and inflammasome‐related mRNAs in humans and mice. Int Immunol. 2010;22:717–28. [DOI] [PubMed] [Google Scholar]
- 19. Lu WL, Zhang L, Song DZ, Yi XW, Xu WZ, Ye L, et al. NLRP6 suppresses the inflammatory response of human periodontal ligament cells by inhibiting NF‐κB and ERK signal pathways. Int Endod J. 2019;52:999–1009. [DOI] [PubMed] [Google Scholar]
- 20. Liu W, Liu J, Wang W, Wang Y, Ouyang X. NLRP6 induces pyroptosis by activation of caspase‐1 in gingival fibroblasts. J Dent Res. 2018;97:1391–8. [DOI] [PubMed] [Google Scholar]
- 21. Broderick L, De Nardo D, Franklin BS, Hoffman HM, Latz E. The inflammasomes and autoinflammatory syndromes. Annu Rev Pathol. 2015;10:395–424. [DOI] [PubMed] [Google Scholar]
- 22. Janani C, Ranjitha Kumari BD. PPAR gamma gene–a review. Diabetes Metab Syndr. 2015;9:46–50. [DOI] [PubMed] [Google Scholar]
- 23. Kempster SL, Belteki G, Forhead AJ, Fowden AL, Catalano RD, Lam BY, et al. Developmental control of the Nlrp6 inflammasome and a substrate, IL‐18, in mammalian intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300:G253–G263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nie H, Hu Y, Guo W, Wang W, Yang Q, Dong Q, et al. miR‐331‐3p inhibits inflammatory response after intracerebral hemorrhage by directly targeting NLRP6. Biomed Res Int. 2020;2020:6182464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukherjee S, Kumar R, Tsakem Lenou E, Basrur V, Kontoyiannis DL, Ioakeimidis F, et al. Deubiquitination of NLRP6 inflammasome by Cyld critically regulates intestinal inflammation. Nat Immunol. 2020;21:626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, et al. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram‐positive pathogen infection. Cell. 2018;175:1651–64. e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghimire L, Paudel S, Jin L, Baral P, Cai S, Jeyaseelan S. NLRP6 negatively regulates pulmonary host defense in Gram‐positive bacterial infection through modulating neutrophil recruitment and function. PLoS Pathog. 2018;14:e1007308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levy M, Thaiss C, Zeevi D, Dohnalová L, Zilberman‐Schapira G, Mahdi J, et al. Microbiota‐modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen H, Li Y, Gu J, Yin L, Bian F, Su L, et al. TLR4‐MyD88 pathway promotes the imbalanced activation of NLRP3/NLRP6 via caspase‐8 stimulation after alkali burn injury. Exp Eye Res. 2018;176:59–68. [DOI] [PubMed] [Google Scholar]
- 31. Anand PK, Malireddi RKS, Lukens JR, Vogel P, Bertin J, Lamkanfi M, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host‐microbial symbiosis. Nat Immunol. 2013;14:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wlodarska M, Thaiss C, Nowarski R, Henao‐Mejia J, Zhang J‐P, Brown E, et al. NLRP6 inflammasome orchestrates the colonic host‐microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, et al. NLRP6 protects Il10(‐/‐) mice from colitis by limiting colonization of akkermansia muciniphila. Cell Rep. 2017;19:733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al. Microbiota‐induced activation of epithelial IL‐6 signaling links inflammasome‐driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A. 2013;110:9862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gálvez EJC, Iljazovic A, Gronow A, Flavell R, Strowig T. Shaping of intestinal microbiota in Nlrp6‐ and Rag2‐deficient mice depends on community structure. Cell Rep. 2017;21:3914–26. [DOI] [PubMed] [Google Scholar]
- 37. Mamantopoulos M, Ronchi F, Van Hauwermeiren F, Vieira‐Silva S, Yilmaz B, Martens L, et al. Nlrp6‐ and ASC‐dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity. 2017;47:339–48. e4. [DOI] [PubMed] [Google Scholar]
- 38. Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, et al. The NLR protein NLRP6 does not impact gut microbiota composition. Cell Rep. 2017;21:3653–61. [DOI] [PubMed] [Google Scholar]
- 39. Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, et al. A specific gene‐microbe interaction drives the development of Crohn's disease‐like colitis in mice. Sci Immunol. 2019;4:eaaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elinav E, Henao‐Mejia J, Strowig T, Flavell R. NLRP6 and dysbiosis: avoiding the luring attraction of over‐simplification. Immunity. 2018;48:603–4. [DOI] [PubMed] [Google Scholar]
- 41. Grosheva I, Zheng D, Levy M, Polansky O, Lichtenstein A, Golani O, et al. High‐throughput screen identifies host and microbiota regulators of intestinal barrier function. Gastroenterology. 2020;159:1807–23. [DOI] [PubMed] [Google Scholar]
- 42. Leng F, Yin H, Qin S, Zhang K, Guan Y, Fang R, et al. NLRP6 self‐assembles into a linear molecular platform following LPS binding and ATP stimulation. Sci Rep. 2020;10:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radulovic K, Normand S, Rehman A, Delanoye‐Crespin A, Chatagnon J, Delacre M, et al. A dietary flavone confers communicable protection against colitis through NLRP6 signaling independently of inflammasome activation. Mucosal Immunol. 2018;11:811–9. [DOI] [PubMed] [Google Scholar]
- 44. Sun Y, Zhang M, Chen CC, Gillilland M III, Sun X, El‐Zaatari M,, et al. Stress‐induced corticotropin‐releasing hormone‐mediated NLRP6 inflammasome inhibition and transmissible enteritis in mice. Gastroenterology. 2013;144:1478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Birchenough GM, Nyström EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6‐dependent Muc2 secretion. Science. 2016;352:1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Volk JK, Nyström EEL, van der Post S, Abad BM, Schroeder BO, Johansson Å, et al. The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function. J Exp Med. 2019;216:2602–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seregin SS, Golovchenko N, Schaf B, Chen J, Eaton KA, Chen GY. NLRP6 function in inflammatory monocytes reduces susceptibility to chemically induced intestinal injury. Mucosal Immunol. 2017;10:434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Normand S, Delanoye‐Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin‐Biroulet L, et al. Nod‐like receptor pyrin domain‐containing protein 6 (NLRP6) controls epithelial self‐renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci USA. 2011;108:9601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Toubai T, Fujiwara H, Rossi C, Riwes M, Tamaki H, Zajac C, et al. Host NLRP6 exacerbates graft‐versus‐host disease independent of gut microbial composition. Nat Microbiol. 2019;4:800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–63. [DOI] [PubMed] [Google Scholar]
- 51. Sarwar R, Pierce N, Koppe S. Obesity and nonalcoholic fatty liver disease: current perspectives. Diabetes Metab Syndr Obes. 2018;11:533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Henao‐Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome‐mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. 2011;54:133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu Y, Ni T, Deng W, Lin J, Zheng L, Zhang C, et al. Effects of NLRP6 on the proliferation and activation of human hepatic stellate cells. Exp Cell Res. 2018;370:383–8. [DOI] [PubMed] [Google Scholar]
- 55. Lee HJ, Yeon JE, Ko EJ, Yoon EL, Suh SJ, Kang K, et al. Peroxisome proliferator‐activated receptor‐delta agonist ameliorated inflammasome activation in nonalcoholic fatty liver disease. World J Gastroenterol. 2015;21:12787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bahar Halpern K, Caspi I, Lemze D, Levy M, Landen S, Elinav E, et al. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 2015;13:2653–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ji X, Li L, Lu P, Li X, Tian D, Liu M. NLRP6 exerts a protective role via NF‐kB with involvement of CCL20 in a mouse model of alcoholic hepatitis. Biochem Biophys Res Commun. 2020;528:485–92. [DOI] [PubMed] [Google Scholar]
- 58. Sanches RCO, Souza C, Marinho FV, Mambelli FS, Morais SB, Guimarães ES, et al. NLRP6 plays an important role in early hepatic immunopathology caused by schistosoma mansoni infection. Front Immunol. 2020;11:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meng C, Zhang J, Zhang L, Wang Y, Li Z, Zhao J. Effects of NLRP6 in cerebral ischemia/reperfusion (I/R) injury in rats. J Mol Neurosci. 2019;69:411–8. [DOI] [PubMed] [Google Scholar]
- 60. Zhang J, Jiang N, Zhang L, Meng C, Zhao J, Wu J. NLRP6 expressed in astrocytes aggravates neurons injury after OGD/R through activating the inflammasome and inducing pyroptosis. Int Immunopharmacol. 2020;80:106183. [DOI] [PubMed] [Google Scholar]
- 61. Ydens E, Demon D, Lornet G, De Winter V, Timmerman V, Lamkanfi M, et al. Nlrp6 promotes recovery after peripheral nerve injury independently of inflammasomes. J Neuroinflammation. 2015;12:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Valiño‐Rivas L, Cuarental L, Nuñez G, Sanz AB, Ortiz A, Sanchez‐Niño MD. Loss of NLRP6 expression increases the severity of acute kidney injury. Nephrol Dial Transplant. 2020;35:587–98. [DOI] [PubMed] [Google Scholar]
- 63. Zheng D, Liwinski T, Elinav E. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discov. 2020;6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no raw data generated in this manuscript.


