Abstract
Background
SLC16A1-AS1 has been characterized as an oncogenic long non-coding (lncRNA) in breast cancer and bladder cancer, while its role in cervical squamous cell carcinoma (CSCC) is unknown.
Methods
CSCC and non-tumor tissue samples were collected from 60 female patients, and qPCR was performed to detect the expression of SLC16A1-AS1, miR-194 and SOCS2. Luciferase reporter assay was performed to detect the interaction between SLC16A1-AS1 and miR-194. Colony formation assay was used to detect cell proliferation.
Results
SLC16A1-AS1 was down-regulated in CSCC and correlated with poor survival. Overexpression of SLC16A1-AS1 could inhibit the proliferation of cervical cancer cells. In addition, SLC16A1-AS1 could sponge miR-194 and increase the expression levels of SOCS2, ultimately inhibiting the proliferation of cervical cancer cells.
Conclusion
SLC16A1-AS1 was downregulated in CSCC and suppressed cell proliferation in cervical squamous cell carcinoma (CSCC) through the miR-194/SOCS2 axis.
Keywords: cervical squamous cell carcinoma, SLC16A1-AS1, miR-194, SOCS2
Introduction
As a type of malignancy starts in the cervix, cervical cancer is among the most commonly diagnosed female cancers and affects millions of females each year.1 It is reported that cervical cancer accounted for 3.2% of all new cancer cases and 3.3% of all cancer deaths in 2018.2 Human papillomavirus (HPV) infection is the major cause of cervical cancer.3,4 With the increased vaccination rate of HPV and popularization of HPV infection screen, the incidence of cervical cancer has significantly dropped.5 However, HPV infection rate is still high across the world,6 and HPV vaccination is not beneficial for HPV-positive cases.3 In addition, HPV-negative cervical cancer also exists.7 Therefore, better understanding of the molecular pathogenesis of cervical cancer is of great importance to facilitate the development of novel anti-cervical cancer therapy.
HPV infection is not sufficient for the occurrence and development of cervical cancer.8 It has been well established that the progression of cervical cancer also requires the involvement of molecular players.9 Functional studies of the molecular players involved in cervical cancer provided novel insights into the development of targeted therapy.9 Long non-coding RNAs (lncRNAs) encode no proteins but regulate the expression of protein-coding gene or interact with other non-coding RNAs, such as miRNAs, to participate in cancer biology.10,11 Therefore, lncRNAs may serve as novel therapeutic targets for cancer treatment. However, the functions of most lncRNAs in cancer biology remain unclear. LncRNA SLC16A1-AS1 has been reported to be downregulated in lung cancer,12 indicating its potential involvement in cancer biology. However, its functions in cancer development and progression remain unclear. Our preliminary bioinformatics analysis found that SLC16A1-AS1 may interact with miR-194, which can promote cancer development. This study was therefore carried out to explore the interaction between SLC16A1-AS1 and miR-194 in cervical squamous cell carcinoma (CSCC), which is a major subtype of cervical cancer.
Materials and Methods
CSCC Patients and Tissue Collection
A total of 60 female patients (44 to 68 years old, mean age 56.8 ± 4.6 years old) diagnosed as CSCC between March 2013 and March 2015 at Heping Hospital Affiliated to Changzhi Medical College hospital were enrolled in this study. The Ethics Committee of this hospital approved the study before admission of the patients. All 60 patients had complete medical records. Based on their medical records, 53 cases were HPV-positive and 7 cases were HPV-negative. All CSCC patients were diagnosed for the first time. Recurrent CSCC cases were excluded from this study. Patients complicated with other clinical disorders or with initiated therapy were also excluded. All patients were diagnosed through histopathological biopsy, during which CSCC and paired non-tumor tissue samples were collected from each patient. Tissues were stored in liquid nitrogen before use. All patients signed the written informed consent. Based on the AJCC staging criteria, the 60 patients included 12, 15, 12 and 21 cases at I–IV stages, respectively.
A 5-Year Follow-Up
All patients were followed-up for 5 years after the admission of patients. Follow-up was performed every month through telephone to record their survival. All patients completed the follow-up.
CSCC Cells and Cell Culture
Human CSCC cell line SiHa (ATCC, USA) was used. Cells were cultured with DMEM medium containing 10% FBS in cell culture incubator at 37°C with 5% CO2. Cells were collected when the confluence was about 85% for subsequent experiments.
Cell Transfections
The pcDNA3.1 vector (Sangon, Shanghai, China) was used as backbone to construct SLC16A1-AS1 and SOCS2 expression vector (The sequences are shown in Table 1). Mimic of miR-194 and negative control (NC) miRNA were purchased from Sigma-Aldrich (USA). SiHa cells were collected and counted, and 106 cells were transfected with either 10 nM vector and/or 40 nM miRNA using lipofectamine 2000 (Invitrogen). Untransfected cells were the control (Ctrl.) cells. NC cells were miRNA-NC or empty vector-transfected cells. Cells were harvested at 48 h post-transfection to perform the following experiments.
Table 1.
Vectors Sequences Applied
| PcDNA3.1-SLC16A1-AS1-F | CCGCTCGAGGCGCGCTGCGCCCCTGCTGA |
| PcDNA3.1-SLC16A1-AS1-R | CGGGATCCAAAAAAATTTTATTATTGTTACAC |
| PcDNA3.1-SOCS2-F | CCGCTCGAGATGACCCTGCGGTGCCTTGAG |
| PcDNA3.1-SOCS2-R | CGGGATCCTTATACCTGGAATTTATATTCTTCCAAG |
| pGL3-SLC16A1-AS1-F | GGGGTACCCCTGTTTCCTCCCTCTGAAGTG |
| pGL3-SLC16A1-AS1-R | GAAGATCTTACCATAGCAGCATCCAGTCTATC |
Luciferase Activity Assay
The pGL3 (Promega Corporation) backbone vector was used to construct Luciferase vector of SLC16A1-AS1 (The sequences are shown in Table 1). To explore the interactions between SLC16A1-AS1 and miR-194, SiHa cells were co-transfected with SLC16A1-AS1 luciferase vector + miR-194 mimic (miR-194 group) or SLC16A1-AS1 luciferase vector + NC miRNA (NC group) using lipofectamine 2000. Luciferase activity was measured and compared 48 h later.
RNA Extraction
Trizol reagent (Sigma-Aldrich) was used to extract total RNAs from CSCC and paired non-tumor tissue samples as well as SiHa cells. Genomic DNA was removed using DNase I (Sigma-Aldrich).
RT-qPCR
RNA samples were subjected to reverse transcriptions (RTs) using QuantiTect Reverse Transcription Kit (QIAGEN). All qPCRs reactions were prepared using QuantiFast SYBR Green PCR Kit (QIAGEN). The expression levels of SLC16A1-AS1 and SOCS2 were determined with 18S rRNA as the internal control. To measure the expression levels of miR-194, addition of poly (A), miRNA RTs, and qPCRs were performed using All-in-OneTM miRNA qRT-PCR Detection Kit (GeneCopoeia). U6 was used as the internal control for the expression levels of miR-194. Each experiment was performed in triplicate. Ct values were processed using 2−ΔΔCq method to calculate the relative gene expression levels. The primers used in this study are shown in Table 2.
Table 2.
q-PCR Primer Sequences Applied
| Primer | 5ʹ-3’ |
|---|---|
| SLC16A1-AS1-F | GGGAGACTTAGGCACAAATTAACC |
| SLC16A1-AS1-R | ATGTTGGTGTGCTTGAAATCTTCC |
| U6-F | CTCGCTTCGGCAGCACATATAC |
| U6-R | AACGCTTCACGAATTTGCGTGTC |
| MiR-194-S | GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACTCCACAT |
| MiR-194-F | TGTAACAGCAACTCCATGTG |
| MiR-194-R | AGAGCAGGGTCCGAGGT |
| SOCS2-F | AGGTACAGGTGAACAGTCCCATT |
| SOCS2-R | TCCAGATGTGCAAGGATAAACG |
Western Blot Analysis
His-Spin Protein Miniprep (Zymo Research) was used to isolate total proteins from SiHa cells. Protein concentrations were measured using BCA assay (Sigma-Aldrich). Following protein denaturation in boiling water for 10 min, proteins were separated using SDS-PAGE gel (8%). Proteins were then transferred to PVDF membranes, and blocking was performed in PBS containing 5% non-fat milk. Membranes were then incubated with SOCS2 (ab3692, Abcam) and GAPDH (ab9485, Abcam) rabbit primary antibodies at 4°C for 14 h, followed by incubation with lgG-HRP secondary antibody (ab6721, Abcam) at room temperature for another 2 h. ECL (Sigma-Aldrich) was used to develop signals. Image J v1.48 software was used for data normalizations.
Colony Formation Assay
Colony formation assay was performed to detect cell proliferation. SiHa cells were plated in 6-well plate. Colonies (>50 cells per colony) were stained with crystal violet, counted, and photographed.
CCK-8 Assay
Cell Counting Kit 8 (ab228554, Abcam) was used to evaluate the effects of transfections on the proliferation of SiHa cells. Each well of a 96-well cell culture plate was filled with 0.1 mL cell suspension containing 5000 cells. Cell culture was performed at 37°C. Measurement of OD values was performed at 450 nm every 24 h for a total of 96 h. CCK-8 solution was added into each well to 10% at 4 h before measuring the OD values.
Statistical Analyses
Three replicates were included in each experiment and data were expressed as mean ± stand deviation (SD) values. Differences between paired tissue samples were compared by paired t test. Two independent groups were compared by unpaired t test. Multiple groups were compared by ANOVA Tukey’s test. The 60 patients were divided into high and low SLC16A1-AS1 level groups (n = 30, with the median expression levels of SLC16A1-AS1 in CSCC tissues as cutoff value) and survival curves were plotted. Survival curves were compared by Log rank test. P < 0.05 was considered as statistically significant.
Results
Downregulation of SLC16A1-AS1 Predicted Poor Survival of CSCC Patients
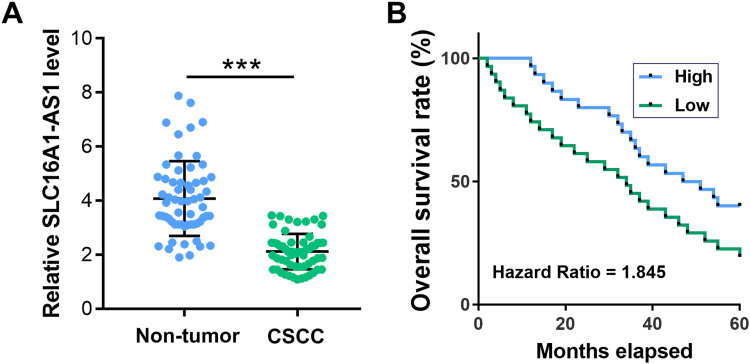
The expression levels of SLC16A1-AS1 in CSCC and paired non-tumor tissues from CSCC patients (n = 60) were determined by RT-qPCR. Compared with non-tumor tissues, the expression levels of SLC16A1-AS1 were significantly lower in CSCC tissues (Figure 1A, p < 0.05). Survival curve analysis showed that the overall survival of CSCC patients with lower expression levels of SLC16A1-AS1 was inferior to those with higher expression levels of SLC16A1-AS1 (Figure 1B).
Figure 1.
Downregulation of SLC16A1-AS1 predicted poor survival of CSCC patients. (A) The expression of SLC16A1-AS1 in CSCC and paired non-tumor tissues from CSCC patients (n = 60) were determined by RT-qPCR. PCRs were performed in triplicate and mean values were presented and compared. ***p < 0.05. (B) The 60 patients were divided into high and low SLC16A1-AS1 level groups (n = 30, with median expression level of SLC16A1-AS1 in CSCC tissues as cutoff value) and survival curves were plotted.
SLC16A1-AS1 Interacted with miR-194
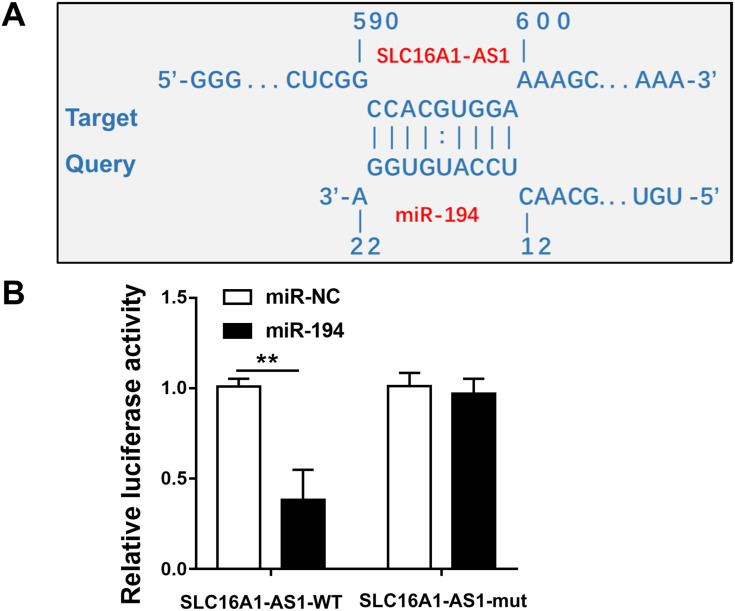
IntaRNA 2.0 was used to predict the interaction between SLC16A1-AS1 and miR-194. It was observed that SLC16A1-AS1 and miR-194 could form strong base pairing (Figure 2A). To validate the interaction between SLC16A1-AS1 and miR-194, SLC16A1-AS1-luciferase reporter and miR-194 mimic were used for luciferase reporter assay. It showed that miR-194 mimic significantly reduced luciferase activities, while mutating the miR-194 binding sites restored the luciferase activity (Figure 2B, p < 0.05).
Figure 2.
SLC16A1-AS1 interacted with miR-194. (A) Schematic model of interaction between SLC16A1-AS1 and miR-194. (B) Luciferase reporter assay for luciferase activity of SLC16A1-AS1-reporter with mutated miR-194 binding sites in SiHa cells co-transfected with miR-194 mimics. n = 3. **p < 0.01.
Overexpression of SLC16A1-AS1 and miR-194 Did Not Affect the Expression of Each Other
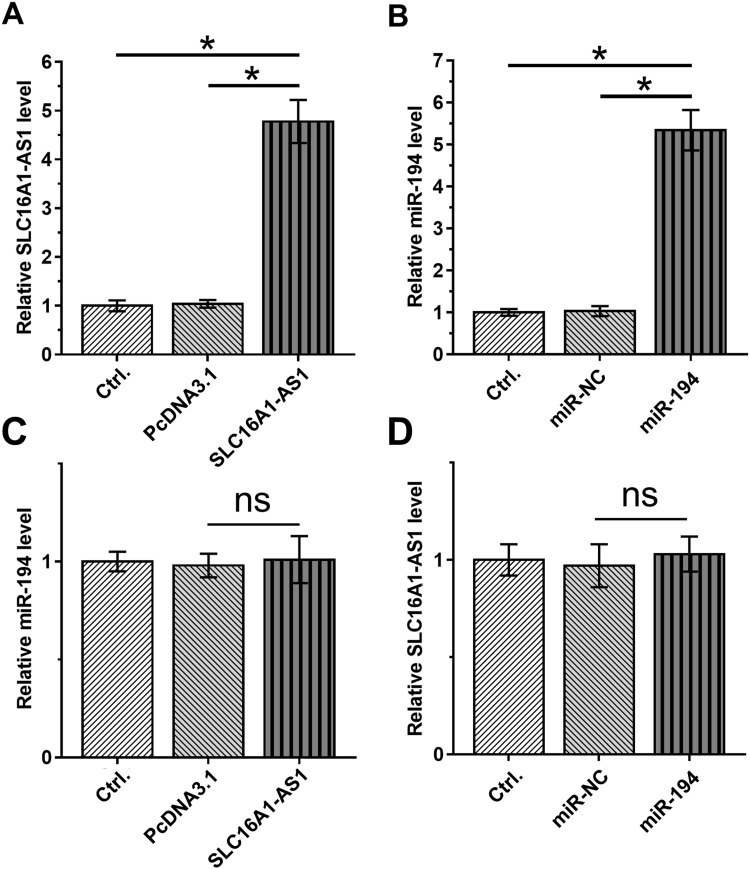
To further explore the interaction between SLC16A1-AS1 and miR-194, SiHa cells were transfected with either SLC16A1-AS1 expression vector or miR-194 mimic, and the overexpression of SLC16A1-AS1 and miR-194 were confirmed 48 h later by RT-qPCR (Figure 3A and B). Compared with the Control and NC groups, overexpression of SLC16A1-AS1 and miR-194 did not affect the expression of each other (Figure 3C and D).
Figure 3.
Overexpression of SLC16A1-AS1 and miR-194 did not affect the expression of each other. (A and B) The effects of overexpression of SLC16A1-AS1 and miR-194 mimic transfection on SLC16A1-AS1 and miR-194 expression in siHa cells. n = 3. (C and D) The effects of SLC16A1-AS1 overexpression and miR-194 mimic transfection on miR-194 and SLC16A1-AS1 expression in SiHa cells. n = 3. Ns means the difference is not statistically significant. *p < 0.05.
Overexpression of SLC16A1-AS1 Led to Upregulated SOCS2 Through miR-194
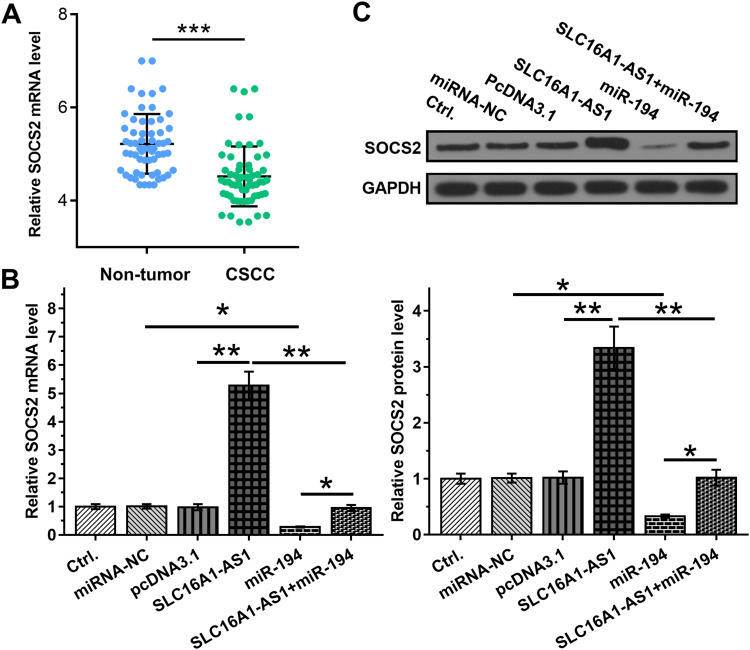
Studies have shown that miR-194 binds to the 3ʹ-UTR of SOCS2,13 so the expression of SOCS2 was detected in CSCC. The results showed that SOCS2 is downregulated in CSCC (Figure 4A). SLC16A1-AS1 may sponge miR-194 to affect the expression of SOCS2. To verify this hypothesis, the effects of SLC16A1-AS1 and miR-194 on SOCS2 by RT-qPCR (Figure 4B) and Western blot (Figure 4C). We found that overexpression of miR-194 inhibited the expression of SOCS2, which could be abrogated by simultaneous upregulation of SLC16A1-AS1 (Figure 4B and C).
Figure 4.
Overexpression of SLC16A1-AS1 led to upregulated SOCS2 through miR-194. (A) The expression of SOCS2 in CSCC and paired non-tumor tissues from CSCC patients were determined by RT-qPCR. (B and C) The effects of overexpression of SLC16A1-AS1 and miR-194 on the expression of SOCS2, which is a target of miR-194, were analyzed by RT-qPCR (B) and Western blot (C). n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
SLC16A1-AS1 Suppressed SiHa Cells Proliferation Through the miR-194/SOCS2 Axis
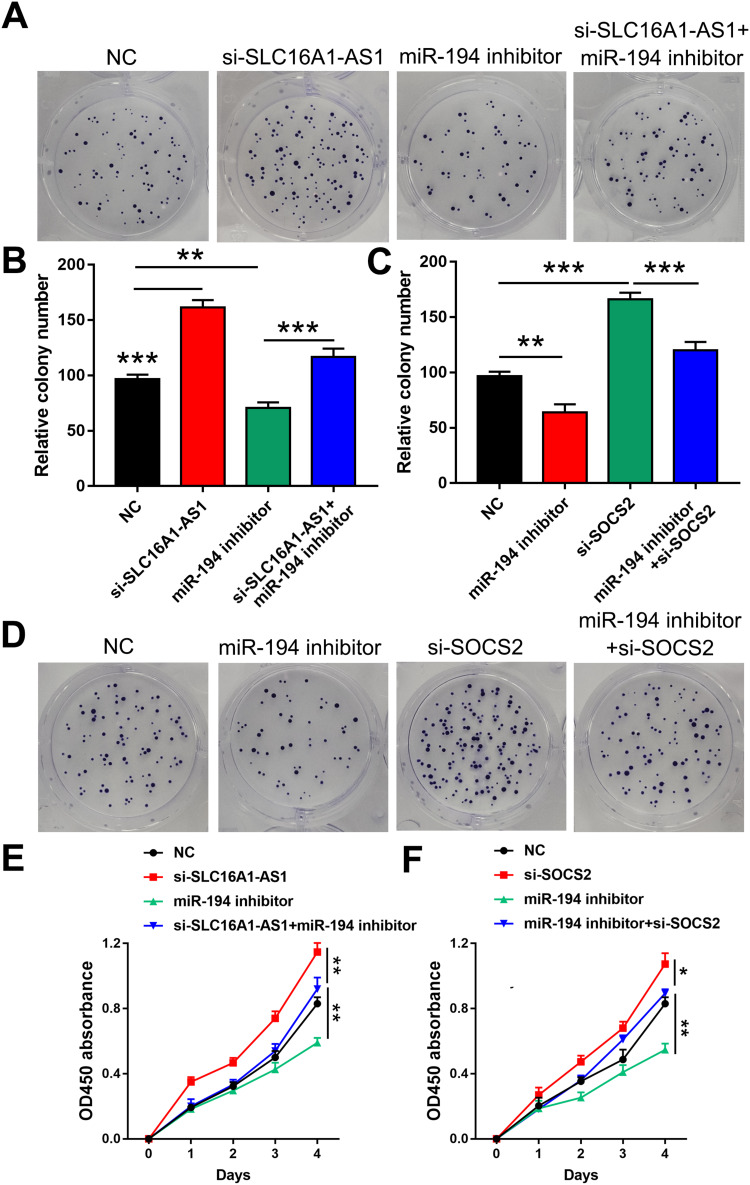
To further explore whether SLC16A1-AS1 promoted malignant phenotype of CSCC via the miR-194/SOCS2 axis, colony formation assay (Figure 5A–D) and CCK-8 assay (Figure 5E and F) were performed. The results showed that knockdown of SLC16A1-AS1 and SOCS2 (si-SOCS2 and si-control sequences are shown in Table 3.) promoted cell proliferation. In addition, cell growth decreased after miR-194 inhibitor treated cells, which could be reversed by knockdown of SLC16A1-AS1 (Figure 5A, B and E). Similarly, inhibition of miR-194 could also reverse the pro-proliferation effect caused by knockdown of SOCS2 (Figure 5C, D and F).
Figure 5.
SLC16A1-AS1 suppressed SiHa cell proliferation through the miR-194/SOCS2 axis. (A–D) Colony formation assay was performed to evaluate the effects of si-SLC16A1-AS1, miR-194 inhibitor and si-SOCS2 on the proliferation of SiHa cells. n = 3. (E and F) CCK-8 assay was used to detect the effects of si-SLC16A1-AS1, miR-194 inhibitor and si-SOCS2 on the proliferation of SiHa cells. n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
Table 3.
siRNAs Sequences Applied
| siRNA | 5ʹ-3’ |
|---|---|
| Si-SOCS2 #1 | ATGCAGCTATGTGAAAGAGAA |
| Si-SOCS2 #2 | ATGTGTCAAGTCCAAGCTTAA |
| Si-Control | CAGGGTATCGACGATTACAAA |
Discussion
The study explored the interactions among SLC16A1-AS1, miR-194 and SOCS2 in CSCC. We found that SLC16A1-AS1 was downregulated in CSCC, and it may sponge miR-194 to upregulate SOCS2, thereby promoting cancer cell proliferation.
The involvement of SLC16A1-AS1 in cancer biology has only been investigated in non-small cell lung cancer (NSCLC). In NSCLC, SLC16A1-AS1 is downregulated and overexpression of SLC16A1-AS1 inhibits the proliferation of NSCLC cells.12 In addition, downregulation of SLC16A1-AS1 is closely correlated with the poor survival of NSCLC patients. Based on our knowledge, the involvement of SLC16A1-AS1 in other cancers remains unclear. This study is the first to report the downregulation of SLC16A1-AS1 in CSCC. In addition, overexpression of SLC16A1-AS1 resulted in decreased proliferation rate of CSCC cells. Therefore, SLC16A1-AS1 is likely a tumor suppressor in CSCC.
The 5-year overall survival of early-stage CSCC can reach 90%, while only about 40% of patients are diagnosed at early stages.14 Once tumor metastasis occurred, the overall 5-year survival rate of cervical cancer drops to 30%.15 Therefore, early diagnosis is still the key for the treatment of CSCC. However, due to the lack of effective markers, the early diagnosis of cervical cancer is unlikely significantly improved in the near future. This study showed that downregulation of SLC16A1-AS1 in CSCC tissues is closely correlated with poor survival of patients. Therefore, measuring the expression levels of SLC16A1-AS1 before therapy may assist the prognosis of CSCC and guide the determination of therapeutic approaches, thereby promoting the survival of patients.
MiR-194 plays different roles in different cancers.13,16 MiR-194 is downregulated in glioma and targets Bmi1 to suppress epithelial-to-mesenchymal transition, indicating its tumor-suppressive function.16 In contrast, miR-194 is upregulated in prostate cancer and downregulates SOCS2 to promote cancer development.13 In this study, we showed that miR-194 can also target SOCS2 in CSCC to promote cancer cell proliferation, indicating its oncogenic role in CSCC.
It has been well established that lncRNAs may sponge miRNAs to participate in cancer biology. For instance, lncRNA FEZF1-AS1 sponges miR-34a to upregulate Notch-1 and promote cancer development in glioblastoma.17 Interestingly, SLC16A1-AS1 may sponge miR-194 to suppress its function in regulating the expression of SOCS2 and CSCC cell proliferation. However, other mechanisms may exist and future studies are still needed.
Conclusion
In conclusion, SLC16A1-AS1, expressed in low abundance in CSCC, functioned as miR-194 sponge to inhibit miR-194 activity and increased the expression levels of SOCS2, ultimately inhibiting the proliferation of cervical cancer cells. Taken together, our results revealed the key role of SLC16A1-AS1 in breast cancer cell proliferation.
Disclosure
The authors report conflicts of interest in this work.
References
- 1.Cohen P, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi: 10.1016/S0140-6736(18)32470-X [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Stanley M. Pathology and epidemiology of HPV infection in females. Gynecol Oncol. 2010;117(2 Suppl):S5–S10. doi: 10.1016/j.ygyno.2010.01.024 [DOI] [PubMed] [Google Scholar]
- 4.Phianmongkhol Y, Suwan N, Srisomboon J, Kietpeerakool C. Knowledge about human papillomavirus infection and cervical cancer prevention among nurses in Chiang Mai University Hospital, Thailand. Asian Pac J Cancer Prev. 2011;12(3):823–825. [PubMed] [Google Scholar]
- 5.Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383(9916):524–532. doi: 10.1016/S0140-6736(13)62218-7 [DOI] [PubMed] [Google Scholar]
- 6.de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141(4):664–670. doi: 10.1002/ijc.30716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez-Carunchio L, Soveral I, Steenbergen RD, et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. BJOG. 2015;122(1):119–127. doi: 10.1111/1471-0528.13071 [DOI] [PubMed] [Google Scholar]
- 8.Ibeanu OA. Molecular pathogenesis of cervical cancer. Cancer Biol Ther. 2011;11(3):295–306. doi: 10.4161/cbt.11.3.14686 [DOI] [PubMed] [Google Scholar]
- 9.Crafton SM, Salani R. Beyond chemotherapy: an overview and review of targeted therapy in cervical cancer. Clin Ther. 2016;38(3):449–458. doi: 10.1016/j.clinthera.2016.02.007 [DOI] [PubMed] [Google Scholar]
- 10.Peng L, Yuan X, Jiang B, Tang Z, Li G-C. LncRNAs: key players and novel insights into cervical cancer. Tumor Biol. 2016;37(3):2779–2788. doi: 10.1007/s13277-015-4663-9 [DOI] [PubMed] [Google Scholar]
- 11.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Lu S, Guo Z, et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J Investig Med. 2019;68:jim–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das R, Gregory PA, Fernandes RC, et al. MicroRNA-194 promotes prostate cancer metastasis by inhibiting SOCS2. Cancer Res. 2017;77(4):1021–1034. doi: 10.1158/0008-5472.CAN-16-2529 [DOI] [PubMed] [Google Scholar]
- 14.Biewenga P, van der Velden J, Mol BW, et al. Prognostic model for survival in patients with early stage cervical cancer. Cancer. 2011;117(4):768–776. doi: 10.1002/cncr.25658 [DOI] [PubMed] [Google Scholar]
- 15.Li H, Wu X, Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. J Gynecol Oncol. 2016;27(4):e43–e43. doi: 10.3802/jgo.2016.27.e43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Wei C, Li J, Liu J, Qu J. MicroRNA-194 represses glioma cell epithelial‑to‑mesenchymal transition by targeting Bmi1. Oncol Rep. 2017;37(3):1593–1600. doi: 10.3892/or.2017.5376 [DOI] [PubMed] [Google Scholar]
- 17.Luo L, Zhang Y, He H, Chen C, Zhang B, Cai M. LncRNA FEZF1-AS1 sponges miR-34a to upregulate Notch-1 in glioblastoma. Cancer Manag Res. 2020;12:1827–1833. doi: 10.2147/CMAR.S240531 [DOI] [PMC free article] [PubMed] [Google Scholar]