Abstract
Background
Disturbance in the oropharyngeal microbiota is common in hospitalized patients and contributes to the development of nosocomial pneumonia. Lactiplantibacillus plantarum 299 and 299v (Lp299 and Lp299v) are probiotic bacteria with beneficial effects on the human microbiome.
Aim
To investigate how Lp299 and Lp299v affect the growth of nosocomial oropharyngeal pathogens in vitro and to evaluate the efficacy in vivo when these probiotics are administered prophylactically in hospitalized patients.
Methods
The in vitro effect of Lp299 and Lp299v on nosocomial respiratory tract pathogens was evaluated using two methods, the co‐culture and agar overlay. In the clinical study, patients were randomized to orally receive either probiotics or placebo twice daily during their hospital stay. Oropharyngeal swabs were analyzed at inclusion and every fourth day throughout hospitalization.
Findings
All tested pathogens were completely inhibited by both Lp299 and Lp299v using the agar‐overlay method. In the co‐culture experiment, Lp299 and Lp299v significantly (p < 0.05) reduced the growth of all pathogens except for Enterococcus faecalis co‐incubated with Lp299. In the clinical study, daily oral treatment with Lp299 and Lp299v did not influence the development of disturbed oropharyngeal microbiota or nosocomial infection. Proton pump inhibitors, antibiotics, and steroid treatment were identified as risk factors for developing disturbed oropharyngeal microbiota.
Conclusions
Lp299 and Lp299v inhibited pathogen growth in vitro but did not affect the oropharyngeal microbiota in vivo. The ClinicalTrials.gov Identifier for this study is NCT02303301.
Keywords: hospitalization, nosocomial infection, nosocomial pathogens, oropharyngeal microbiota, probiotics
This combined laboratory and clinical study showed that Lactiplantibacillus plantarum 299 and 299v significantly inhibited in vitro growth of nosocomial pathogens often found in the oropharynx of hospitalized patients. In the randomized controlled trial, hospitalized patients were randomized to receive either oral twice‐daily treatment with lactobacilli or placebo, and oropharyngeal swabs were performed regularly throughout hospitalization. No differences could be shown between the Lactobacilli and placebo groups regarding changes in oropharyngeal microbiota or the occurrence of nosocomial infections.

1. INTRODUCTION
Probiotics are defined by the International Scientific Association for Probiotics and Prebiotics (ISAPP) as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host (Hill et al., 2014). Different probiotics display a variety of antimicrobial properties, for example, the production of ammonia, lactic acid, free fatty chains, hydrogen peroxide, and bacteriocins (de Vrese & Schrezenmeir, 2008). Lactiplantibacillus plantarum 299 and 299v (Lp299 and Lp299v) are probiotic bacteria within the lactic acid bacteria group. L. plantarum can secrete bacteriocins, also called plantaricins (Adebayo et al., 2014; Prabhurajeshwar & Chandrakanth, 2017; Seddik et al., 2017), with inhibitory effects on, for example, oral Streptococcus mutans (Hasslöf et al., 2010). A recent review (Simons et al., 2020) presents the possible role of bacteriocins as a part of future antibiotic treatment.
A disturbance in the microbiome of the oropharynx, defined as an overgrowth of normally existing species and/or establishment of new potential pathogens, has been shown to indicate the degree of sickness in the host, and to be associated with increased mortality in the intensive care unit (ICU) and non‐ICU patients (Dickson et al., 2020; Johanson et al., 1969). The microbiome of the oropharynx and that of the lower respiratory tract resemble each other, probably due to the microaspiration of oropharyngeal microbiota (Bassis et al., 2015). Microaspiration of disturbed oropharyngeal microbiota has been shown to play a part in the complex pathogenesis behind the development of pneumonia (Bahrani‐Mougeot et al., 2007; Huffnagle et al., 2016) Therefore, a large number of studies have investigated the effects of decontamination of the oropharynx (using chlorhexidine or local antibiotics) or of administration of probiotics, with varying results (Bo et al., 2020; Gu et al., 2012; Karacaer et al., 2017; Klarin et al., 2008; Morrow et al., 2010; Wang et al., 2013; Weng et al., 2017). These studies have been carried out in ICU settings or pediatric populations, diminishing the occurrence of respiratory disease as well as antibiotic‐associated diarrhea (Hatakka et al., 2001; Ling et al., 2019; Niveen et al., 2016). Because of the heterogeneity in the populations studied, generalized conclusions and recommendations about probiotic benefits are difficult to present.
In this study, we investigated whether Lp299 and Lp299v can reduce or inhibit the growth of nosocomial pathogens in vitro and in vivo. Seven pathogens were selected for the in vitro study, due to their frequent appearance in oropharyngeal cultures in ICU and non‐ICU patients according to previous studies (Klarin et al., 2018; Tranberg et al., 2018). The randomized controlled trial aimed to study whether oral administration with lactobacilli could prevent or delay the occurrence of disturbed oropharyngeal microbiota in non‐ICU hospitalized patients.
2. MATERIALS AND METHODS
2.1. In vitro study
2.1.1. Bacterial strains
Lp299 and Lp299v were provided by Probi AB, Lund, Sweden. Reference strains of the pathogens Escherichia coli (CCUG 24), Staphylococcus aureus (CCUG 1800), Enterococcus faecalis (CCUG 19916) and E. faecium (CCUG 542), Klebsiella pneumoniae (CCUG 225), Pseudomonas aeruginosa (CCUG 551) and Enterobacter cloacae (CCUG 6323) were purchased from Culture Collection, University of Gothenburg, Sweden. Clinical isolates came from the Department of Clinical Microbiology at Skåne University Hospital, Sweden.
2.1.2. Growth conditions
Lp299 and Lp299v were grown in De Man‐Rogosa‐Sharpe (MRS) broth (Merck, Darmstadt, Germany) and on MRS agar. E. faecalis, E. faecium, and S. aureus were cultured in Todd Hewitt (TH) broth (Becton Dickinson) and agar, whereas E. coli, K. pneumoniae, P. aeruginosa, and E. cloacae were grown in lysogeny broth (LB) (Sigma‐Aldrich, St. Louis, MO, USA) and agar. All strains were cultured at 37°C under aerobic conditions (21% oxygen, 5% CO2).
2.1.3. Agar overlay
Overnight cultures of Lp299 and Lp299v were washed in Phosphate‐buffered saline (PBS) and adjusted to final concentrations of approximately 2 × 109 colony‐forming units (CFU)/ml. Varying amounts (4 × 104, 4 × 105, and 4 × 106 CFU, respectively) of Lp299 or Lp299v were added to 8 ml of warm (42–45°C) MRS agar and poured into Petri dishes. Control plates contained no lactobacilli. After solidification, this bottom agar was incubated at 37°C overnight. The second layer of agar (24 ml), suited for the pathogen, was then cast on top of the MRS agar. Overnight cultures of the pathogens were diluted 1:1000, 1:10,000, and 1:100,000 in PBS, and 10 μl drops of the dilutions were seeded on the top agar. After overnight incubation at 37°C, the growth of the pathogen was assessed. Experiments were repeated twice using reference strains and once with clinical isolates of the pathogen.
2.1.4. Inhibitory activity of Lp299 and Lp229v
Co‐cultures of lactobacilli and pathogens were grown in a mixed broth consisting of 25% (v/v) MRS and 75% (v/v) TH or LB broth. These proportions provide good growth conditions for both lactobacilli and pathogens. Overnight cultures of Lp299, Lp299v, and pathogen strains were washed and adjusted to bacterial suspensions of 2 × 109 CFU/ml, and 50 μl of the pathogen and 500 μl of Lp229 or Lp299v were added to 10 ml of mixed broth. As a control, 50 μl of the pathogen suspension was grown in mixed broth in the absence of lactobacilli. The co‐cultures were incubated for 5 hours at 37°C. Before and after incubation, a small aliquot of each sample was diluted in PBS and plated on 15–20 ml agar suitable for the pathogen. After incubation overnight at 37°C, colonies of the pathogens were counted, and the growth of lactobacilli was ensured. Experiments were performed in triplicate.
2.1.5. Antibacterial activity of Lp299 and Lp299v supernatants
Overnight cultures of Lp229 or Lp299v were pelleted by centrifugation, and the supernatants were sterile filtered through a 0.22 μm Millex®‐ GP, Millipore Express® PES Membrane Filter (Merck Millipore Ltd). The cell‐free supernatants were then either pH‐neutralized with 1 M NaOH to a pH of 5.4 (corresponding to the natural pH of MRS broth); heat‐treated at 99°C for 5 minutes, or incubated with pepsin (Sigma‐Aldrich), proteinase K (Thermo Scientific), or trypsin (Sigma‐Aldrich) at a final concentration of 1 mg/ml for 2 hours at 37°C. After that, the samples were heated to 99°C for 5 minutes to eliminate the protease activity. The inhibitory effect of the supernatants was tested against E. cloacae and S. aureus. These two were chosen as they were inhibited by Lp299 and 299v, and they differ in Gram staining and natural habitat. 2.5 ml of pH‐neutralized, heat‐treated, or protease‐treated supernatant was added to 7.5 ml of TH or LB to obtain a mixed broth. Untreated supernatant, sterile MRS broth, and MRS adjusted to pH 4.2 with acetic acid were included for comparison. Pathogens were washed and diluted as described in the co‐culture experiment, and 50 μl of pathogen solution was added to the mixed broths. As a control, pathogens were incubated with Lp299 or Lp299v in a mixed broth with sterile MRS. Samples were plated on agar before and after incubation for 5 hours as described above. After incubation overnight at 37°C, colonies of the pathogens were counted, and the growth of lactobacilli was ensured when relevant. Experiments were performed in triplicate.
2.2. Randomized controlled trial
2.2.1. Study population
Patients were enrolled in the study between 2014 and 2019 at the University Hospital in Lund using the following inclusion criteria: age ≥18 years, obtaining the first oropharyngeal swabs (OPS) within 24 hours of hospital admission, and an expected length of stay of more than 72 hours. Exclusion criteria were respiratory infection and prior hospitalization within two weeks. The patients were enrolled by the investigators, research nurses, or medical students. The patients were admitted to medical, surgical, or orthopedic wards. A standardized case report form was used to record patient data.
We based our approximative power calculation on a study from 1969, where oropharyngeal cultures had been analyzed throughout the hospitalization in ward patients (Johanson et al., 1969). From that study, we estimated that a sample size of 75 patients in each group would be sufficient to show a significant difference between the groups in the occurrence of disturbed microbiota in the oropharynx. The patients were younger but much sicker in the study from 1969, and patients that would now be admitted to the ICU were treated in general wards. The study from 1969 was observational, and the above factors put together made it difficult to make an exact power calculation.
2.2.2. Randomization
The randomization was performed directly after inclusion via sealed envelopes at a 1:1 ratio. The randomization was blinded to recruiters, staff, and patients.
2.2.3. Intervention
Patients received either a combination of 1010 CFU Lp299 and 1010 CFU Lp299v with 3 grams of maltodextrin or a placebo consisting of only 3 grams of maltodextrin. Both lactobacilli and placebo were manufactured and generously provided by Probi AB, Lund, Sweden, and delivered in identical freeze‐dried sachets labeled “A” and “B,” respectively. The sachets were kept in a −80°C freezer until use. In the ward, the sachets were kept at 4°C for a maximum of five days. Viability controls of the lactobacilli in the sachets were performed yearly throughout the study period, analyzing sachets stored at both −80°C and 4°C. Before administration to the patient, the contents of the sachets were resuspended in 15 ml of sterile water, allowing the revival of the potential lactobacilli for 20–40 minutes before administering the mixture to the patient. Patients received the assigned mixture twice daily throughout the hospital stay, with instructions to gargle the mixture as long as possible and then swallow. OPSs were taken at inclusion (day 1), on day 3, and after that approximately every fourth day. In all other respects, the patients received standard care.
2.2.4. Microbiological procedures and definitions
The OPSs were processed by extended microbiological procedures at the Department of Clinical Microbiology, Skane University Hospital in Lund. The laboratory is accredited by the accreditation body (SWEDAC) designated by the Swedish government and is formally recognized as competent according to European and international standards.
For bacteria cultivation, sampling media were inoculated on five types of agar plates (three selective, one differentiating, and one nonselective). All plates were produced in‐house, sometimes using commercially available media components (5% horse blood, hematin agar, and UriSelect 4 agar), as listed below:
Agar with 5% horse blood (LabM, Heywood) supplemented with 10 mg/L colistin and 15 mg/L nalidixic acid with an optochin disk (selective);
Agar with 5% horse blood supplemented with 2 mg/L gentamicin and 25 mg/L nalidixic acid for Gram‐positive cocci including Streptococcus pneumoniae (selective);
Hematin agar (Oxoid™, Thermo Science) supplemented with 300 mg/L bacitracin for fastidious Gram‐negative rods including Haemophilus influenzae (selective)
UriSelect 4 agar (Bio‐Rad Laboratories) supplemented with 10 mg/L vancomycin for non‐fastidious Gram‐negative rods (differentiating)
Hematin agar with a colistin disk (nonselective).
The plates were inspected for growth after 16 and 40 hours of aerobic, anaerobic, or CO2 incubation at 35–37°C. If an inspection result was ambiguous at 40 hours, the plate was incubated for an additional 24 hours to obtain a more definite result. Species identification of bacteria was performed using matrix‐assisted laser desorption/ionization time‐of‐flight (MALDI‐TOF) mass spectrometry (MALDI Biotyper Microbial Identification System, Bruker), using software FlexControl 3.4 and MALDI Biotyper (MBT) Compass 4.1, with MBT Compass Library, DB‐7854 MSP (Bruker, Bremen, Germany).
Cultivation and differentiation of Candida spp. were based on colony appearance on CHROM Candida agar (CHROMagar, Hägersten, Sweden) after 48 hours of incubation at 35°C.
For a sample to be considered representative of “oropharyngeal microbiota,” bacterial species normally found in the oropharynx were required to grow on the nonselective hematin plate as determined by visual inspection by an experienced senior microbiologist and following standard practice (Retchless et al., 2020). Samples were classified as disturbed oropharyngeal microbiota when there was a growth of species not normally found in the oral cavity and/or overgrowth of normal oropharyngeal microbiota on selective and differentiating plates. Samples with disturbed oropharyngeal microbiota were divided into three subclasses (see Figure 4): gut pathogens, respiratory tract pathogens, and yeast.
FIGURE 4.
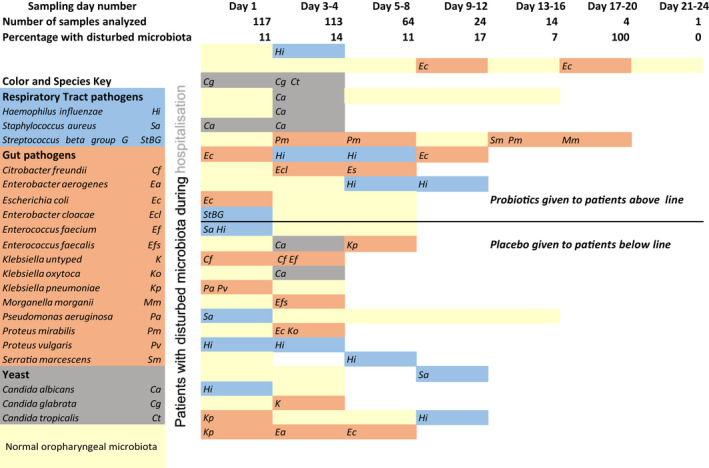
Oropharyngeal swab (OPS) culture results for the 27 patients who had at least one OPS sample with disturbed microbiota during their hospitalization. Each horizontal bar represents the patient's observation time, and the colored bars indicate an OPS culture result for each sampling time: yellow = normal, blue = respiratory pathogens, terracotta = gut microbiota, gray = yeast species. The top row describes the time frames within which the OPS was obtained. The second and third row shows the number of OPSs collected/analyzed and the percentage of OPSs with disturbed microbiota for the total cohort at each sampling time point. The patients are divided according to whether they received probiotics or placebo during hospitalization
2.2.5. Statistical analyses
Inhibitory effects in vitro were analyzed using Student's paired t‐test. In the randomized controlled trial, continuous variables were presented as median, minimum, and maximum values. Dichotomous variables were presented as numbers and as a percentage of the total number. For subjects with a normal oropharyngeal microbiota at inclusion, a univariate Poisson regression was used to analyze the association between the patients’ characteristics (predicting variables) and the intervention they were randomized to (dependent variable). Thereafter, a multivariate Poisson regression model using the two strongest predicting variables from the univariate analysis was constructed, in which one additional potential explanatory variable was added to determine whether the model improved or did not improve by including a third variable. A Kaplan–Meyer analysis was performed to test for differences between the placebo and the lactobacilli group regarding “time to first disturbed oropharyngeal swab.” Fisher's exact test was used to assess the relationship between the intervention group and nosocomial infection rate.
Statistical analyses were performed using IBM SPSS Statistics 26 for Windows (IBM Corp., Armonk, NY, USA). Odds ratios (ORs) are presented with a 95% confidence interval. p < 0.05 was considered significant, and all statistical tests were two‐tailed.
3. RESULTS
3.1. Lp299 and Lp299v inhibit the growth of bacterial pathogens in vitro
The inhibitory effect of Lp299 and Lp299v on other bacteria was first tested in an agar‐overlay assay, where varying concentrations of lactobacilli were grown in a bottom MRS agar, and the pathogens were seeded on a top agar. Under these conditions, all experiments showed a complete absence of pathogen growth compared to control plates without lactobacilli. Both clinical isolates and the corresponding reference strains were tested and gave the same clear results. See Table A1.
Next, pathogens and lactobacilli were co‐cultured in broth to further characterize the inhibitory effect. In this experimental set‐up, both Lp299 and Lp299v significantly inhibited the growth of S. aureus, E. cloacae, K. pneumoniae, E. coli, E. faecium, and P. aeruginosa. For two pathogens, E. cloacae and K. pneumoniae, almost complete eradication of the pathogens was seen, as the number of colonies was close to zero after incubation. E. faecalis was significantly inhibited by Lp299v, but in co‐culture, with Lp299 the inhibition did not reach statistical significance (p = 0.12). See Figure 1.
FIGURE 1.
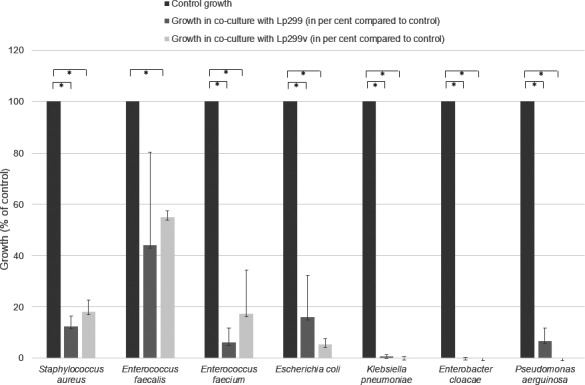
Co‐culture of different pathogens with Lp299 or Lp299v resulted in significant growth inhibition for all pathogens except Enterococcus faecalis co‐incubated with Lp299. The growth of the pathogen alone in the absence of lactobacilli served as control. *p < 0.05
3.2. The antibacterial activity of Lp299 AND Lp299v IS pH‐dependent
To investigate the mechanism behind the growth‐inhibitory effects of lactobacilli, S. aureus was incubated in a mixed broth containing cell‐free supernatants from overnight cultures of Lp299 and Lp299v. The pH of the supernatants was 4.1 and 4.0, respectively. Figure 2 shows that the supernatants significantly inhibited the growth of S. aureus to the same extent as co‐incubation with live bacteria. When the pH of the supernatants was elevated to 5.4, corresponding to the pH of MRS broth, the inhibitory effect was abolished, and S. aureus grew equally well as in the control. Further, MRS broth made acidic to pH 4.0 (same pH as the supernatants) significantly inhibited S. aureus growth to the same extent as the supernatants from both Lp299 and Lp299v (see Figure 2). The same results were obtained when the experiment was performed with E. cloacae (see Figure A1).
FIGURE 2.
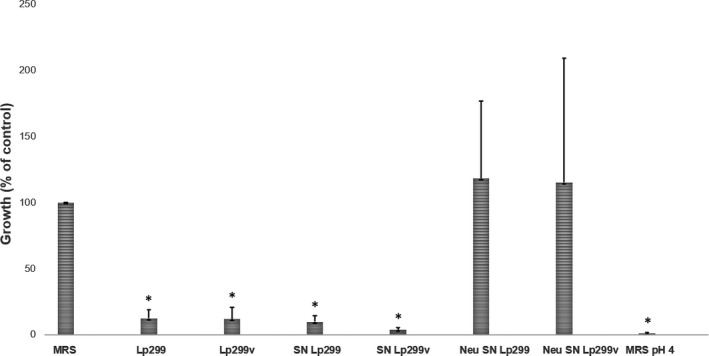
The inhibitory effect of Lp299 and Lp299v on S. aureus is pH‐dependent. S. aureus was incubated with Lp299, Lp299v, or cell‐free supernatants from overnight cultures of the lactobacilli. To explore the role of pH, S. aureus was also incubated with pH‐neutralized supernatants and with acidified MRS broth. The bars with an asterisk above them indicate a significant growth inhibition compared to the control with S. aureus grown in a mixed broth without lactobacilli. SN =supernatant. Neu =neutralized. *p < 0.05
To examine the possible role of plantaricins secreted by Lp299 and Lp299v, E. Cloacae was incubated with Lp299 and Lp299v supernatants that had been heat‐treated to denature the protein content, or pre‐incubated with the proteinases pepsin, proteinase K or trypsin to digest proteins in the supernatants. All of the pre‐treated supernatants showed the same clear growth inhibition as untreated supernatants, whereas the controls incubated with MRS broth showed expected growth of the pathogen during the incubation time (see Figure A2).
Taken together, the overall conclusion of these experiments is that the inhibitory effect was mainly pH‐dependent.
3.3. Randomized controlled trial
Between the 18th of September 2014 to 1st of May 2019, 135 patients were included and randomized. Eighteen patients were excluded due to non‐adherence to protocol. Thus, 117 patients met all inclusion criteria and contributed a total of 337 OPSs. (See Figure 3 for the CONSORT diagram). The median number of OPSs per patient in both groups was 3. All OPSs were representative of oropharyngeal microbiota. The baseline patient characteristics and hospitalization characteristics of the placebo and lactobacilli groups were similar and are presented in Table 1.
FIGURE 3.
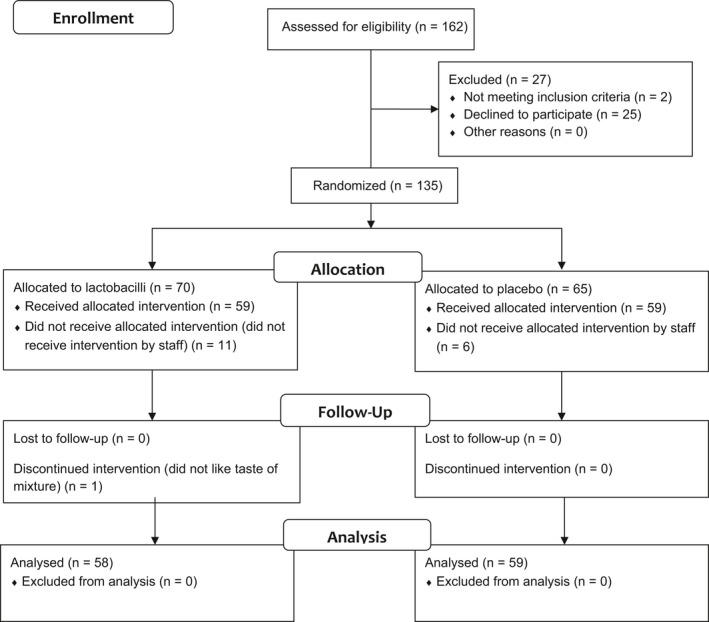
The CONSORT flow diagram. CONSORT (Consolidated Standards of Reporting Trials) diagram demonstrating the progress through the phases of the randomized trial of two groups
TABLE 1.
Descriptive statistics of patients (n = 117)
| Variable | Lactobacilli (n = 58) | Placebo (n = 59) | p |
|---|---|---|---|
| Age, years | 76 (22‒96) | 76 (36‒97) | 0.926 b |
| Gender, male | 28 (48%) | 26 (44%) | 0.712 a |
| Body mass index | 26 (18‒40) | 26 (17‒40) | 0.311 b |
| Current or ex‐smoker | 23 (40%) | 24 (41%) | 1.000 a |
| Diabetes | 15 (26%) | 8 (14%) | 0,108 a |
| Alcohol intake >2 times/week | 19 (33%) | 21 (37%) | 1.000 a |
| Proton pump inhibitor | 12 (21%) | 19 (32%) | 0.345 a |
| Able to walk two flights of stairs | 33 (57%) | 33 (57%) | 0.490 a |
| Cortisone medication | 3 (5%) | 6 (10%) | 0.490 a |
| Unplanned admission | 45 (78%) | 42 (71%) | 0.526 a |
| Antibiotics >24 hours before hospitalization | 2 (3%) | 1 (1.7%) | 0.619 a |
| Antibiotics >24 hours during hospitalization | 14(24%) | 20 (34%) | 0.309 a |
| Prophylactic antibiotics perioperatively | 36 (62%) | 31 (53%) | 0.352 a |
| Oropharyngeal swabs | 3 (2‒8) | 3 (2‒6) | 0.236 c |
| Length of hospital stay (days) | 8 (3‒32) | 7 (3‒32) | 0.260 c |
| Nosocomial infection | 4 (6.9%) | 10 (17%) | 0.153 a |
Data are presented as median (range) or number (percentage).
Fisher's exact test, exact sig. (2‐sided).
Independent samples t‐test, sig. (2‐tailed).
Mann–Whitney U test, exact sig. (2‐tailed).
Figure 4 presents the microbiological results for the 27 patients showing any type of disturbed oropharyngeal microbiota on any sampling occasion. Each horizontal row corresponds to a patient's observation time, and by following the row from left to right it is possible to see OPS changes over time. Using the color and species key, the figure shows the subclass and pathogen for each disturbed OPS. The upper part of the figure shows the patients receiving placebo and the lower part shows the patients receiving lactobacilli.
In 104 patients (89%), the first OPS at admission was normal. We analyzed results from these patients using univariate and multivariate Poisson analyses. The univariate analyses showed that treatment with lactobacilli yielded an RR of 0.96 (CI 0.36–2.55, p = 0.94) for acquiring disturbed oropharyngeal microbiota during hospitalization. Both univariate and multivariate analyses showed that treatment with proton pump inhibitor (PPI), cortisone, or antibiotics during hospitalization could be associated with an added risk of developing disturbed microbiota during hospitalization (Table 2).
TABLE 2.
Poisson regression analysis for developing disturbed oropharyngeal microbiota during hospitalization in 104 subjects with a normal microbiota at admission, (yes n = 14, no n = 90). The number of patients receiving Lactobacilli in this analysis was 53, the control group consisted of 51 patients
|
Univariate RR (95% CI) |
p |
Multivariate RR (95% CI) |
p | |
|---|---|---|---|---|
| Lactobacilli group | 0.96 (0.36‒2.55) | 0.938 | ||
| Lactobacilli group | 0.92 (0.33‒2.51) | 0.864 | ||
| Diabetes | 1.41 (0.49‒4.08) | 0.528 | 1.43 (0.48‒4.31) | 0.523 |
| Lactobacilli | 1.10 (0.41‒2.96) a | 0.849 a | ||
| PPI | 2.85 (1.10‒7.39) | 0.031 | 2.89 (1.08‒7.76) a | 0.035 a |
| Lactobacilli | 1.13 (0.44‒2.90) | 0.805 | ||
| Cortisone | 4.22 (1.65‒10.8) | 0.0026 | 4.32 (1.68‒11.1) | 0.0025 |
| Lactobacilli | 0.94 (0.36‒2.46) | 0.893 | ||
| Antibiotics before hospitalization | 2.59 (0.48‒13.9) | 0.267 | 2.62 (0.48‒14.2) | 0.264 |
| Lactobacilli | 1.17 (0.44‒3.13) | 0.753 | ||
| Antibiotics during hospitalization | 3.14 (1.19‒8.30) | 0.021 | 3.23 (1.18‒8.84) | 0.023 |
| Lactobacilli | 0.97 (0.37‒2.55) | 0.952 | ||
| Unplanned admission | 0.83 (0.29‒2.43) | 0.739 | 0.83 (0.29‒2.41) | 0.738 |
the maximum number of step‐halvings was reached but the log‐likelihood value cannot be further improved. Output for the last iteration is displayed. RR =risk ratio, PPI =proton pump inhibitor.
Kaplan–Meyer analyses were performed to determine whether treatment with Lp299 and Lp299v could delay the development of disturbed microbiota. There was a slight tendency to later development of disturbed microbiota in the treatment group, but the difference was not significant.
Concerning the risk of developing a nosocomial infection during hospitalization, the difference between the two groups did not reach significance. In the treatment group, 4/58 patients (7%) developed a nosocomial infection, while the incidence in the placebo group was 10/59 (17%, p = 0.153), with no obvious difference in the severity of infection between the groups (Table 1). The causes of nosocomial infection were urinary tract infections, wound infections, and pneumonia.
4. DISCUSSION
In this combined laboratory and clinical study, we found that Lp299 and Lp299v significantly inhibited in vitro growth of nosocomial pathogens commonly found in the oropharyngeal tract of hospitalized patients. In the randomized controlled trial, no difference between the intervention group and the placebo group could be found regarding changes in the oropharyngeal microbiota or the occurrence of nosocomial infections. The study confirmed the already known risk factors for the development of disturbed oropharyngeal microbiota (Frandah et al., 2013).
The ability of lactobacilli to inhibit pathogen growth has been shown before (Annuk et al., 2003). This has also been specifically shown for Lp299v when Hutt et al. demonstrated its antagonistic effect on Salmonella enterica and Helicobacter pylori (Hutt et al., 2006). Furthermore, a study on oral care in ICU patients showed that Lp299 could be identified in the oropharynx in all patients given the study product (Klarin et al., 2008), indicating that the lactobacilli remain in the oropharynx after oral administration. In this study, we show for the first time that Lp299 and Lp299v inhibit in vitro growth of pathogens known to cause nosocomial respiratory tract infections. Notably, 60% of ICU patients are colonized with at least one of the seven investigated pathogens as early as 24 hours after admission to the ICU (Tranberg et al., 2018). The L. plantarum species have a high production of lactic acid compared to others in the lactic acid bacteria group, and a relatively small production of, for example, hydrogen peroxide and carbon dioxide, which is typical for this group of facultatively heterofermentative lactobacilli (Annuk et al., 2003). In our study, the acidic environment produced by the lactobacilli was essential for inhibiting the in vitro growth of the pathogens under study. However, other factors may also be involved. For example, it has been shown that an acidic pH is necessary for other inhibitory mechanisms to be activated. Several studies on plantaricins show that they are activated at a pH <5 (Lin & Pan, 2019; Song et al., 2014). Although we were unable to demonstrate a plantaricin effect in our study, we cannot rule out a possible role of plantaricins in our strains due to the overwhelming effect of acidic pH. Further experiments are required to determine the possible presence and requisites for the activity of bacteriocins in Lp299 and Lp299v.
In the clinical trial, we could not show that the oral administration of Lp299 and Lp299v prevents or delays the occurrence of disturbed oropharyngeal microbiota in non‐ICU hospitalized patients compared to placebo. In agreement with earlier findings (Tranberg et al., 2018), we found that 11% of the patients had disturbed oropharyngeal microbiota at admission and that an increasing proportion of the patients developed disturbed oropharyngeal microbiota during their hospitalization (see Figure 3). We also confirmed previously reported findings that treatment with PPI and antibiotics were risk factors for disturbed oropharyngeal microbiota (Frandah et al., 2013; Tranberg et al., 2018). In this study, oral cortisone was strongly associated with disturbed oropharyngeal microbiota (p = 0.0025 in the multivariate Poisson regression analyses shown in Table 2, and p = 0.0026 in the univariate Poisson regression), which has not been shown before. The contribution of steroid inhalation treatment to the risk of developing oral candidiasis is well known. In our study, none of the patients who were on cortisone treatment developed disturbed microbiota consisting of Candida species, and only the third was simultaneously on PPI medication.
This study is unique, as it focuses on ward patients. Most previous studies on disturbed oropharyngeal microbiota and its possible contribution to nosocomial pneumonia have focused on intensive care patients. The idea of giving hospitalized patients probiotics is tempting, in many ways. Probiotics are harmless, inexpensive, and may reduce antibiotic use by restoring the patient's microbiota toward being healthier and more normal. When swallowed and digested, the probiotics also influence the intestinal tract immune system. New connections between the composition of the gut microbiota and a wide range of diseases such as irritable bowel syndrome and depression have emerged in the last decade (Didari et al., 2015; Wallace & Milev, 2017).
An important weakness of the clinical trial is that our power calculation was based on older studies with longer hospital stays (Johanson et al., 1969). Consequently, the study was underpowered, and we can therefore unfairly rule out our hypothesis that treatment with lactobacilli can decrease or delay the incidence of disturbed oropharyngeal microbiota during hospitalization. An additional explanation for the lack of effect of probiotic treatment is that changes in the oropharyngeal microbiota take time and a potential contribution to the development of pneumonia even longer. Even if Lp299 and Lp299v showed clear growth inhibition on pathogens in vitro, we might need longer treatment times and longer local exposure to clinically be able to influence the oropharyngeal microbiota. Thus, it cannot be excluded that a study involving a larger patient population and more intense administration of a combination of probiotics would show an effect on a clinically meaningful level.
In conclusion, this study shows that Lp299 and Lp299v inhibit in vitro growth of commonly found nosocomial pathogens in the oropharynx. Oral administration of Lp299 and Lp299v to non‐ICU patients did not reduce the risk of disturbed oropharyngeal microbiota or nosocomial infection.
ETHICS STATEMENT
The study protocol was approved by the Regional Ethical Review Board, Lund, Sweden (no. 2013/764). Written informed consent, including permission to collect and publish anonymous data, was obtained from the patients at inclusion.
CONFLICTS OF INTEREST
None declared.
AUTHOR CONTRIBUTION
Anna Tranberg: Conceptualization (equal); Data curation (equal); Formal analysis (lead); Funding acquisition (lead); Investigation (lead); Methodology (supporting); Project administration (equal); Visualization (lead); Writing‐original draft (lead); Writing‐review & editing (equal). Bengt Klarin: Conceptualization (equal); Data curation (equal); Formal analysis (supporting); Funding acquisition (supporting); Investigation (supporting); Methodology (supporting); Project administration (equal); Visualization (supporting); Writing‐original draft (supporting); Writing‐review & editing (equal). Julia Johansson: Conceptualization (supporting); Data curation (equal); Formal analysis (equal); Funding acquisition (supporting); Investigation (equal); Methodology (supporting); Project administration (equal); Visualization (supporting); Writing‐original draft (supporting); Writing‐review & editing (equal). Lisa I. Påhlman: Conceptualization (supporting); Investigation (supporting); Methodology (equal); Project administration (equal); Supervision (equal); Visualization (supporting); Writing‐original draft (equal); Writing‐review & editing (equal).
ACKNOWLEDGMENTS
We wish to thank research nurses Anne Adolfsson and Susann Schrey for invaluable help, and Gisela Hovold for excellent technical assistance. We also wish to thank Probi AB for generously producing both the probiotic and placebo sachets. This work was funded by research grants from Swedish Region Skane (Doktorand‐2019‐0144 and Doktorand‐2020‐0459).
APPENDIX 1.
FIGURE A1.
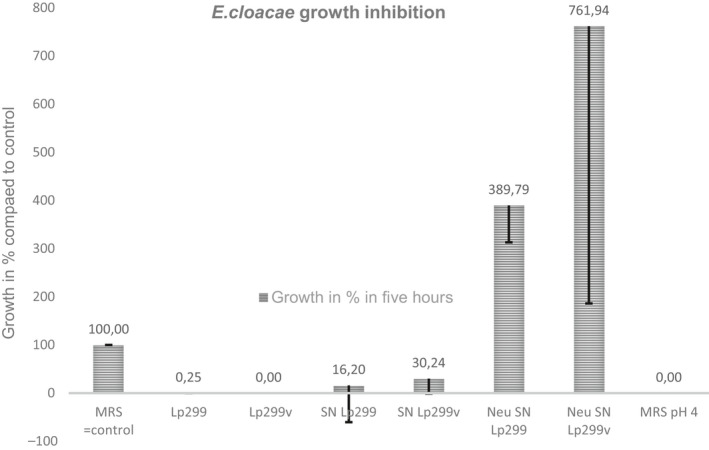
The inhibitory effect of Lp299 and Lp299v on E. cloacae is pH‐dependent. E. cloacae was incubated with Lp299, Lp299v, or cell‐free supernatants from overnight cultures of the lactobacilli. To explore the role of pH, E. cloacae was also incubated with pH‐neutralized supernatants and with acidified MRS broth. There is a significant difference in five‐hour growth between the control (E. cloacae grown in 25% MRS broth) and all other groups (p < 0.05). There is also a significant difference in the five‐hour growth between neutralized supernatants and all other groups (p < 0.05). SN =supernatant. Neu =neutralized
FIGURE A2.
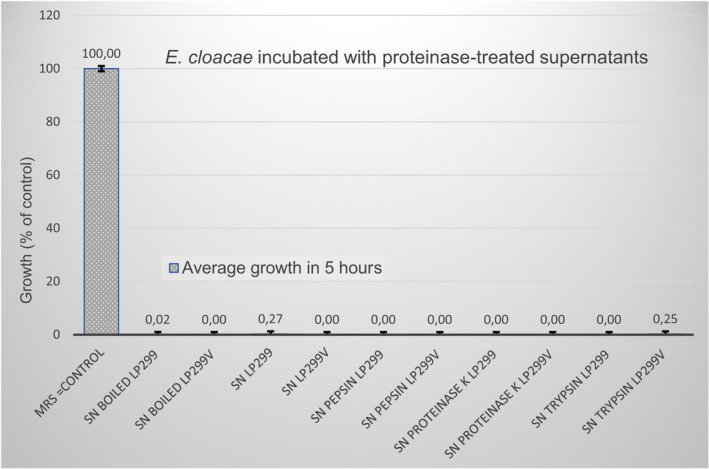
Overnight cultures of E. cloacae were allowed to incubate with pure supernatant (SN); boiled SN; and SN treated with the proteinases pepsin, proteinase K, and trypsin. Both Lp299 and Lp299v SN were included in the experiment. The results show that all of the pre‐treated supernatants showed the same clear growth inhibition as untreated supernatants, whereas the control (E. cloacae incubated with MRS broth) showed expected growth of the pathogen during the incubation time
TABLE A1.
Results from the agar overlay experiment
| Lp299 | Lp299v | |||||
|---|---|---|---|---|---|---|
| 4 × 104 CFU | 4 × 105 CFU | 4 × 106 CFU | 4 × 104 CFU | 4 × 105 CFU | 4 × 106 CFU | |
| S. aureus (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| S. aureus (CI) | 0 | ‐ | ‐ | 0 | ‐ | ‐ |
| E. faecalis (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| E. faecalis (CI) | 0 | ‐ | ‐ | 0 | ‐ | ‐ |
| E. faecium (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| E. faecium (CI) | 0 | ‐ | ‐ | 0 | ‐ | ‐ |
| E. coli (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| E. coli (CI) | 0 | ‐ | ‐ | 0 | ‐ | ‐ |
| K. pneumoniae (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| K. pneumoniae (CI) | 0 | ‐ | ‐ | 0 | ‐ | ‐ |
| E. cloacae (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| P. aeruginosa (RS) | 0 | 0 | 0 | 0 | 0 | 0 |
| P. aeruginosa (CI) | 0 | ‐ | ‐ | 0 | ‐ | ‐ |
Abbreviations: CI, Clinical isolate; RS, Reference strain.
Growth described as 0 = no growth, or 1= growth ‐ = experiment not performed.
DATA AVAILABILITY STATEMENT
The datasets generated and/or analyzed during the in vitro part of the study are available in the figshare repository at https://doi.org/10.6084/m9.figshare.13312118.v1 and https://doi.org/10.6084/m9.figshare.13311929.v1. The datasets generated and/or analyzed during the RCT part of the study are not publicly available due to patient confidentiality but are available from the corresponding author on reasonable request. The study is registered at ClinicalTrials.gov, with the clinical trial number: NCT02303301: https://clinicaltrials.gov/ct2/show/NCT02303301
REFERENCES
- Adebayo, F. A. , Afolabi, O. R. , & Akintokun, A. K. (2014). Antimicrobial properties of purified bacteriocins produced from Lactobacillus casei and Lactobacillus fermentum against selected pathogenic microorganisms. British Journal of Medicine and Medical Research, 4(18), 3415–3431. 10.9734/bjmmr/2014/8584 [DOI] [Google Scholar]
- Annuk, H. , Shchepetova, J. , Kullisaar, T. , Songisepp, E. , Zilmer, M. , & Mikelsaar, M. (2003). Characterization of intestinal lactobacilli as putative probiotic candidates. Journal of Applied Microbiology, 94(3), 403–412. 10.1046/j.1365-2672.2003.01847.x [DOI] [PubMed] [Google Scholar]
- Bahrani‐Mougeot, F. , Paster, B. , Coleman, S. , Barbuto, S. , Brennan, M. , Noll, J. , Kennedy, T. , Fox, P. , & Lockhart, P. (2007). Molecular analysis of oral and respiratory bacterial species associated with ventilator‐associated pneumonia. Journal of Clinical Microbiology, 45(5), 1588–1593. 10.1128/jcm.01963-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis, C. , Erb‐Downward, J. , Dickson, R. , Freeman, C. , Schmidt, T. , Young, V. , Beck, J. , Curtis, J. , & Huffnagle, G. (2015). Analysis of the upper respiratory tract Microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio, 6(2), 10.1128/mBio.00037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo, L. , Li, J. , Tao, T. , Bai, Y. , Ye, X. , Hotchkiss, R. , Kollef, M. , Crooks, N. , & Deng, X. (2020). Probiotics for preventing ventilator‐associated pneumonia. The Cochrane Database of Systematic Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrese, M. , & Schrezenmeir, J. (2008). Probiotics, prebiotics, and synbiotics. Food Biotechnology, 1–66. 10.1007/10_2008_097 [DOI] [PubMed] [Google Scholar]
- Dickson, R. , Schultz, M. , van der Poll, T. , Schouten, L. , Falkowski, N. , Luth, J. , Sjoding, M. , Brown, C. , Chanderraj, R. , Huffnagle, G. , Bos, L. , de Beer, F. , Bos, L. , Claushuis, T. , Glas, G. , Horn, J. , Hoogendijk, A. , van Hooijdonk, R. , Huson, M. , … Witteveen, E. (2020). Lung microbiota predict clinical outcomes in critically ill patients. American Journal of Respiratory and Critical Care Medicine, 201(5), 555–563. 10.1164/rccm.201907-1487oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didari, T. , Mozaffari, S. , Nikfar, S. , & Abdollahi, M. (2015). Effectiveness of probiotics in irritable bowel syndrome: Updated systematic review with meta‐analysis. World Journal of Gastroenterology, 21(10), 3072 10.3748/wjg.v21.i10.3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandah, W. , Colmer‐Hamood, J. , Mojazi Amiri, H. , Raj, R. , & Nugent, K. (2013). Oropharyngeal flora in patients admitted to the medical intensive care unit: clinical factors and acid suppressive therapy. Journal of Medical Microbiology, 62(5), 778–784. 10.1099/jmm.0.053066-0 [DOI] [PubMed] [Google Scholar]
- Gu, W. , Wei, C. , & Yin, R. (2012). Lack of Efficacy of Probiotics in Preventing Ventilator‐Associated Pneumonia. Chest, 142(4), 859–868. 10.1378/chest.12-0679 [DOI] [PubMed] [Google Scholar]
- Hasslöf, P. , Hedberg, M. , Twetman, S. , & Stecksén‐Blicks, C. (2010). Growth inhibition of oral mutans streptococci and candida by commercial probiotic lactobacilli ‐ an in vitro study. BMC Oral Health, 10(1), 10.1186/1472-6831-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakka, K. , Savilahti, E. , Pönkä, A. , Meurman, J. H. , Poussa, T. , Näse, L. , Saxelin, M. , & Korpela, R. (2001). Effect of long‐term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised. BMJ, 322(7298), 1327 10.1136/bmj.322.7298.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C. , Guarner, F. , Reid, G. , Gibson, G. , Merenstein, D. , Pot, B. , Morelli, L. , Canani, R. , Flint, H. , Salminen, S. , Calder, P. , & Sanders, M. (2014). The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Reviews Gastroenterology & Hepatology, 11(8), 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Huffnagle, G. , Dickson, R. , & Lukacs, N. (2016). The respiratory tract microbiome and lung inflammation: A two‐way street. Mucosal Immunology, 10(2), 299–306. 10.1038/mi.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt, P. , Shchepetova, J. , Loivukene, K. , Kullisaar, T. , & Mikelsaar, M. (2006). Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero‐ and uropathogens. Journal of Applied Microbiology, 100(6), 1324–1332. 10.1111/j.1365-2672.2006.02857.x [DOI] [PubMed] [Google Scholar]
- Johanson, W. , Pierce, A. , & Sanford, J. (1969). Changing Pharyngeal Bacterial Flora of Hospitalized Patients. New England Journal of Medicine, 281(21), 1137–1140. 10.1056/NEJM196911202812101 [DOI] [PubMed] [Google Scholar]
- Karacaer, F. , Hamed, I. , Özogul, F. , Glew, R. , & Özcengiz, D. (2017). The function of probiotics on the treatment of ventilator‐associated pneumonia (VAP): facts and gaps. Journal of Medical Microbiology, 66(9), 1275–1285. 10.1099/jmm.0.000579 [DOI] [PubMed] [Google Scholar]
- Klarin, B. , Adolfsson, A. , Torstensson, A. , & Larsson, A. (2018). Can probiotics be an alternative to chlorhexidine for oral care in the mechanically ventilated patient? A multicentre, prospective, randomised controlled open trial. Critical Care, 22(1), 10.1186/s13054-018-2209-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarin, B. , Molin, G. , Jeppsson, B. , & Larsson, A. (2008). Use of the probiotic Lactobacillus plantarum 299 to reduce pathogenic bacteria in the oropharynx of intubated patients: a randomised controlled open pilot study. Critical Care, 12(6), R136 10.1186/cc7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, T. , & Pan, T. (2019). Characterization of an antimicrobial substance produced by Lactobacillus plantarum NTU 102. Journal of Microbiology, Immunology and Infection, 52(3), 409–417. 10.1016/j.jmii.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Ling, Z. , Liu, X. , Guo, S. , Cheng, Y. , Shao, L. , Guan, D. , Cui, X. , Yang, M. , & Xu, X. (2019). Role of probiotics in Mycoplasma pneumoniae pneumonia in children: A short‐term pilot project. Frontiers in Microbiology, 9, 10.3389/fmicb.2018.03261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, L. , Kollef, M. , & Casale, T. (2010). Probiotic Prophylaxis of Ventilator‐associated Pneumonia. American Journal of Respiratory and Critical Care Medicine, 182(8), 1058–1064. 10.1164/rccm.200912-1853OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niveen, E. , Angi, E. , Mohamed, E. , Ghada, B. , & Wafaa, S. (2016). Role of probiotics in prevention of hospital acquired pneumonia in Egyptian children admitted to Pediatric Intensive Care Unit of Mansoura University Children’s Hospital. African Journal of Microbiology Research, 10(31), 1240–1247. 10.5897/ajmr2016.8085 [DOI] [Google Scholar]
- Prabhurajeshwar, C. , & Chandrakanth, R. (2017). Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomedical Journal, 40(5), 270–283. 10.1016/j.bj.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retchless, A. , Kretz, C. , Rodriguez‐Rivera, L. , Chen, A. , Soeters, H. , Whaley, M. , & Wang, X. (2020). Oropharyngeal microbiome of a college population following a meningococcal disease outbreak. Scientific Reports, 10(1), 10.1038/s41598-020-57450-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddik, H. , Bendali, F. , Gancel, F. , Fliss, I. , Spano, G. , & Drider, D. (2017). Lactobacillus plantarum and Its Probiotic and Food Potentialities. Probiotics and Antimicrobial Proteins, 9(2), 111–122. 10.1007/s12602-017-9264-z [DOI] [PubMed] [Google Scholar]
- Simons, A. , Alhanout, K. , & Duval, R. (2020). Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug‐resistant bacteria. Microorganisms, 8(5), 639 10.3390/microorganisms8050639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, D. , Zhu, M. , & Gu, Q. (2014). Purification and characterization of plantaricin ZJ5, a new bacteriocin produced by Lactobacillus plantarum ZJ5. PLoS One, 9(8), e105549 10.1371/journal.pone.0105549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranberg, A. , Thorarinsdottir, H. , Holmberg, A. , Schött, U. , & Klarin, B. (2018). Proton pump inhibitor medication is associated with colonisation of gut flora in the oropharynx. Acta Anaesthesiologica Scandinavica, 62(6), 791–800. 10.1111/aas.13094 [DOI] [PubMed] [Google Scholar]
- Wallace, C. , & Milev, R. (2017). The effects of probiotics on depressive symptoms in humans: A systematic review. Annals of General Psychiatry, 16(1), 10.1186/s12991-017-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Liu, K. , Ariani, F. , Tao, L. , Zhang, J. , & Qu, J. (2013). Probiotics for preventing ventilator‐associated pneumonia: a systematic review and meta‐analysis of high‐quality randomized controlled trials. PLoS One, 8(12), e83934 10.1371/journal.pone.0083934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, H. , Li, J. , Mao, Z. , Feng, Y. , Wang, C. , Ren, X. , & Zeng, X. (2017). Probiotics for preventing ventilator‐associated pneumonia in mechanically ventilated patients: a meta‐analysis with trial sequential analysis. Frontiers in Pharmacology, 8, 10.3389/fphar.2017.00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the in vitro part of the study are available in the figshare repository at https://doi.org/10.6084/m9.figshare.13312118.v1 and https://doi.org/10.6084/m9.figshare.13311929.v1. The datasets generated and/or analyzed during the RCT part of the study are not publicly available due to patient confidentiality but are available from the corresponding author on reasonable request. The study is registered at ClinicalTrials.gov, with the clinical trial number: NCT02303301: https://clinicaltrials.gov/ct2/show/NCT02303301


