Abstract
Cranioectodermal dysplasia (CED) or Sensenbrenner syndrome is a very rare autosomal-recessive disease that is characterized by craniofacial, skeletal and ectodermal abnormalities. The proteins encoded by six CED-associated genes are members of the intraflagelline transport (IFT) system, which serves an essential role in the assembly, maintenance and function of primary cilia. The current study identified compound novel heterozygous IFT122 (NM_052985.3) variants in a male Chinese infant with CED. The latter variant changes the length of the protein and may result in the partial loss-of-function of IFT122. With the simultaneous presence of frameshift and stop-loss variants, the patient manifested typical CED with fine and sparse hair, macrocephaly, dysmorphic facial features and upper limb phocomelia. A number of unusual phenotypic characteristics were additionally observed and included postaxial polydactyly of both hands and feet. The molecular confirmation of CED in this patient expands the CED-associated variant spectrum of IFT122 in CED, while the manifestation of CED in this patient provides additional clinical information regarding this syndrome. Moreover, the two variants identified in the proband provide a novel perspective into the phenotypes caused by different combinations of variants.
Keywords: cranioectodermal dysplasia, ciliopathy, combinations of mutations, loss-of-function, IFT122
Introduction
Cranioectodermal dysplasia (CED), which is also known as Sensenbrenner syndrome, is a rare autosomal recessive disorder. To date, IFT122 (WDR10), WDR35 (IFT121), IFT43 (C14orf179), WDR19 (IFT144), IFT52 (C20orf9) and IFT140 variants have been linked to CED (1-6). The products of these genes are members of the intraflagellar transport (IFT) system, which serves an essential role in the assembly, maintenance and functioning of primary cilia (7). Mutations in the ciliary protein genes cause defects in the structure and function of cilia, which can lead to ‘ciliopathies’ (8). Ciliopathies are a collection of complex developmental disorders involving multiple organ systems (7-10).
Clinically, CED is highly heterogeneous and is characterized by craniofacial, skeletal and ectodermal abnormalities (8). Facial dysmorphic features with frontal bossing and low set ears, hepatic fibrosis, stunted growth and retinal dysfunction have also been noted to vary across patients (11-13). While there is currently no clear association between molecular differences and clinical phenotypes, >60 patients diagnosed with CED have been reported (11,14-16). Additional reports on these gene variants and their phenotypes will therefore be critical to the understanding of this condition. The current study reports the compound heterozygous IFT122 (NM_052985.3) variants, c.366_376delAGGCCAAGGTG(p.Gly123Glufs*3) and c.3879A>G (p.Ter1293Trpext), identified in a male Chinese child with CED and additionally describes the patient's associated clinical profile.
Case report
Participants
From the CED family, the proband, his parents and two healthy siblings were recruited at Guangxi Maternal and Child Health Hospital (Guangxi, China) on November 22, 2018. Physical and imaging examinations of the proband were performed by an experienced orthopedic surgeon. Detailed written informed consent was obtained from the parents, with all study participants agreeing to the genetic analysis of collected samples. All procedures in the present study were approved by the Guangxi Maternal and Child Health Hospital Ethics Committee.
Genetic analysis Whole-exome and Sanger sequencing
Genomic DNA was extracted from 2 ml peripheral blood of the proband and his family members using a Lab-Aid DNA kit (Xiamen Zeesan Biotech Co., Ltd.) by standard methods, according to the manufacturer's protocol. Whole-exome sequencing using proband-derived genomic DNA was performed using an Agilent SureSelect Human All Exon V5 kit (Agilent Technologies, Inc.) on a Hiseq2500 platform (Illumina, Inc.), according to the manufacturers' protocol. FASTQ files were generated and reads were compared with the reference human genome (hg19) using Burrows-Wheeler Aligner software (version 0.7.17; http://bio-bwa.sourceforge.net/bwa.shtml). A custom pipeline, built predominantly using the Genome Analysis Tool Kit GATK(https://wiki.rc.usf.edu/index.php/Genome_Analysis_ToolKit_), was used for sequence data analysis and annotation. Identification of causal variants by annotating the selected single-nucleotide variants (SNVs) and indels was performed using TGex software (LifeMap Sciences; Biotime, Inc.). The variants were defined as a 'rare deleterious' mutation if: i) The GnomAD database showed that it had an allele frequency ≤0.5%; and ii) a stop-gain, stop-loss, non-synonymous, frameshift or splice-site mutation was present. When a potential novel mutation was considered following alignment of the patient's genome sequence against the ClinVar (www.ncbi.nlm.nih.gov), HGMD (www.hgmd.cf.ac.uk/ac/), HPSD (liweilab.genetics.ac.cn/HPSD/) and SNP (www.ncbi.nlm.nih.gov/SNP) databases, and control databases (ExAC and gnomAD), bioinformatics softwares PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.bii.a-star.edu.sg/www/SIFT_seq_submit2.html), the Human Splicing Finder website (http://www.umd.be/HSF/) and MutationTaster (http://www.mutationtaster.org) were used to predict whether the novel variant(s) had any deleterious effects on the protein(s). All the variants were classified according to American College of Medical Genetics/Association of Molecular Pathology guidelines (17).
Specific primers for amplification and sequencing were designed using Primer3web version 4.1.0 (http://primer3.ut.ee/). PCR was performed using genomic DNA of the proband and his family members followed by Sanger sequencing using ABI3730xl Genetic analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. PCR was performed using Takara PrimeSTAR Max DNA Polymerase (Takara Biotechnology Co., Ltd.) under the following thermocycling conditions: initial denaturization at 95˚C for 5 min, followed by 35 cycles of denaturation at 95˚C for 3 sec, annealing at 60˚C for 30 sec and extension at 72˚C for 30 sec. Final extension occurred at 72˚C for 5 min. IFT122 (NM_052985.3) variants were amplified and sequenced using two pairs of IFT122-specific primers: IFT122 primer1 forward (F; 5'-AGACGCCTTGCATGGTACA-3') and primer1 reverse (R; 5'-CCAGGACAGGACATCAGGAA-3') were used for exon 5 amplification; and IFT122 primer2F (5'-GGCTTCAGCCTGGTCTTACA-3') and primer2R (5'-CAAGGCATGAAATGCCATAA-3') were used for exon 31 amplification.
Results
Patient description
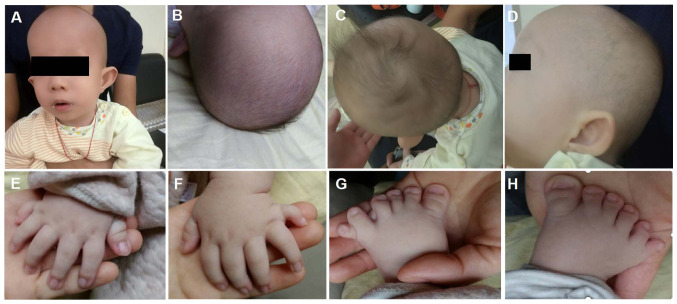
The third child born to healthy, non-consanguineous parents originating from Guangxi, China was a 2-month-old infant male proband who was admitted to Guangxi Maternal and Child Health Hospital (Guangxi, China) for genetic counseling due to macrocephaly, postaxial hand polydactyly and postaxial polysyndactyly of the feet on November 22, 2018. The clinical images were obtained when the proband was 3 months old (Fig. 1). The proband was born full term with a birth weight of 3.1 kg, a birth length of 51 cm (51st centile) and a head circumference of 37.5 cm (91st centile). Typical CED-associated craniofacial features were observed (Fig. 1). These features included: Macrocephaly, dolichocephaly, a high forehead with frontal bossing, long face, broad nasal bridge, long philtrum, telecanthus, bilateral epicanthic folds, hypertelorism, esotropia, abnormality of the pinna, everted lower lip vermilion, abnormal and low-set pinnae and micrognathia. The probands hair and eyebrows were sparse, with hair that was also fine. Upper limb phocomelia and postaxial polydactyly of both hands and feet were also observed (Fig. 1). When the proband was two-year-old, a renal ultrasound scan indicated normal-sized kidneys and a normal renal function (estimated glomerular filtration rate, 93 ml/min/1.73m2; serum creatinine level, 51 µmol/l).
Figure 1.
Images of the patient at the age of 3 months. (A and B) Macrocephaly, dolichocephaly, high forehead with frontal bossing, long face, low set ears, broad nasal bridge, long philtrum, telecanthus, bilateral epicanthic folds bilaterally, sparse eyebrows, broad philtrum and prominent vermilion of the lower lip. (C) Fine and sparse hair. (D) Abnormality of the pinnae. (E-H) Postaxial hand polydactyly and postaxial polysyndactyly of feet.
Mutation analysis
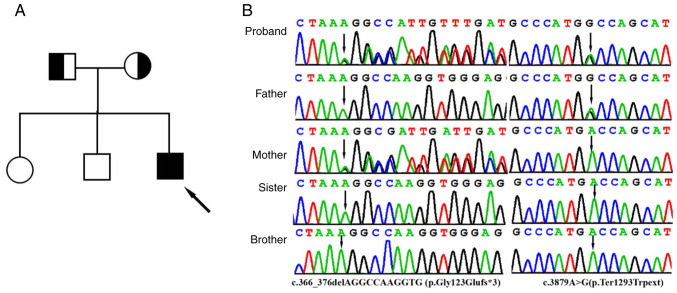
With 20X coverage for >97% of the bases, a total of >99% of reads were mapped to genomic targets. In the proband, a total of 130,330 variants were identified in the proband and included 17,560 variants (94.7%) classified with known SNP identification numbers. Collectively, 121,448 SNVs, 3,951 insertions and 4,931 deletions were identified. After data analysis using of TGex analysis software (https://tgex.genecards.cn/), two novel heterozygous IFT122 variants, c.366_376delAGGCCAAGGTG (p.Gly123Glufs*3) and c.3879A>G (p.Ter1293Trpext), were identified in the proband (Fig. 2B). Sanger sequencing further revealed that heterozygous c.366_376delAGGCCAAGGTG and c.3879A>G variants were identified in the father and mother, respectively, and that his sister and brother did not have either variant (Fig. 2B).
Figure 2.
Clinical and genetic features. (A) Pedigree chart of the family of the patient with Sensenbrenner syndrome. The proband is indicated by a black arrow. (B) Sanger sequencing DNA chromatograms of IFT122 indicating the frameshift c.366_376delAGGCCAAGGTG (p.Gly123Glufs*3) variant inherited from the mother and the stop-loss variant c.3879A>G(p.Ter1293Trpext) was transmitted by the father. Black arrows indicate the c.366_376delAGGCCAAGGTG (p.Gly123Glufs*3) and c.3879A>G(p.Ter1293Trpext) variants.
These variants were not present in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/), HPSD (http://liweilab.genetics.ac.cn/HPSD/) and the dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) databases, nor were they present in ExAC and gnomAD). The c.366_376delAGGCCAAGGTG(p.Gly123Glufs*3) variant in exon 5, which was inherited from the mother, is an 11 bp deletion that causes a frameshift alteration after codon 123 (Glycine) and leads to a premature termination codon that is located at codon 126. The c.3879A>G (p.Ter1293Trpext) variant was inherited from the father and is located in exon 31. Changing the length of the protein, this variant results in the substitution of a TGA stop codon to a missense TGG codon. It is predicted that the novel variant likely has a deleterious effect by damaging the function of the IFT122 protein. According to the ACMG standards and guidelines (17) for the interpretation of sequence variants, the novel variants are likely to be pathogenic.
Discussion
Defects in the IFT122 gene have been identified in patients with type IV short-rib polydactyly (SRP) and type 1 CED (6,18). While SRP is characterized by multiple skeletal anomalies that include short ribs, shortening of the tubular bones, acetabula, irregular metaphyses and post-axial polydactyly, CED is considered to be a mild form of SRP (19). The current study performed whole-exome sequencing and identified compound heterozygosity novel variants in the IFT122 gene of a male Chinese infant. Consistent with previously reported CED patient presentation (20-22), the child manifested fine and sparse hair, macrocephaly, dysmorphic facial features, upper limb phocomelia and postaxial polydactyly of both hands and feet. Therefore, the patient was diagnosed with CED.
In previous reports, the phenotypic severity of ciliary disorders has been indicated to be determined by a combination of missense and truncating mutations (21-23). In the patient described in the present study, the biallelic IFT122 variants identified were a combination of frameshift and stop-loss variants. The IFT122 gene contains 31 exons and encodes a member of the WD repeat protein family that is comprised of 1,292 amino acids and seven WD40 functional domains (6). These domains are involved in the formation of β-propeller structures and act as platforms for the association of binding partners (24). In the present study, the c.366_376delAGGCCAAGGTG (p.Gly123Glufs*3) variant was indicated to be a novel frameshift variant located in the third WD domain. This may subsequently result in a truncated protein, and a loss of function. It can be hypothesized that the p.Gly123Glufs*3 variant acts similarly to other truncating IFT122 variants like p.Tyr1077Valfs*10 and p.Tyr1077Valfs*11 that have been reported. It may consequently prevent the retrograde transport of cilia and affect the assembly and maintenance of eukaryotic cilia and flagella, thereby causing abnormal development (18). The c.3879A>G (p.Ter1293Trpext) variant is a novel stop-loss variant that results in a missense-derived TGA stop codon substitution to a TGG codon. The variant changes the length of the protein, which may cause alteration of the overall structure the protein. This leads to the partial loss of function of IFT122, preventing the formation of the IFT-A complex and interrupting IFT122-related intraflagellar transport. According to the ACMG standards and guidelines (17), the novel c.366_376delAGGCCAAGGTG (p.Gly123Glufs*3) variant is likely pathogenic according to PVS1, PM2 and PP4 criteria, while the novel c.3879A>G (p.Ter1293Trpext) variant is likely pathogenic according to PM2, PM3, PM4 and PP4. To date, ~16 patients with CED or lethal SRP type IV have been linked to IFT122 variants and described in the literature. It should be noted that of all mutations in these patients, none were biallelic nonsense, deletion or null mutations (6,20,21,25). Of those mutations described in living patients, the biallelic IFT122, WDR35, IFT43 and WDR19 variants were missense mutations or a combination of missense and truncated mutations (13). The combination of a missense and nonsense variant can lead to lethal phenotypes (25,26). This suggests that the phenotype is dependent on the variable degree of functional IFT122 impairment. In the current study, the patient presented with a typical CED phenotype and was identified to carry a frameshift and a stop-loss variant. It appears that p.Gly123Glufs*3 is associated with a similar truncating variant to those IFT122 variants reported, while the c.3879A>G (p.Ter1293Trpext) stop-loss variant may result in partial loss-of-function of the IFT122 protein. A future functional study of the variants would improve understanding of the disease and its mechanism, which require further study.
Although polydactyly is often a component of ciliopathies, it has infrequently been reported in patients with CED and has only described in individuals with WDR35 and IFT43 variants (18,27). Notably, polydactyly has only been identified in patients with IFT122 variants that have also been diagnosed with SRP syndrome (27). In the present study, while presenting with phenotypes typically associated with CED, to the best of our knowledge, the current study is the first reported patient with CED with IFT122 variants and with polydactyly. Polydactyly is related to the impairment of the sonic hedgehog (SHH) signaling pathway (28). Previous studies have demonstrated that the alterations of limb patterning in IFT122-null mouse embryos are associated with the inhibition of SHH signaling (29,30). IFT122 affects the expression of multiple genes in the SHH pathway, suggesting that the heterogeneity of polydactyly may be associated with the degree of IFT122 deficiency (31,32). Functional studies are necessary to demonstrate the cause of IFT122 variants and determine their associated disease phenotypes. The role of IFT122 variants in polydactyly also requires further study.
In conclusion, to the best of our knowledge, the current study is the first instance in which a combination of frameshift and stop-loss variants in a patient with CED have been identified. The current study provided new insights into phenotypes caused by different variant combinations. The molecular confirmation of CED in this patient expands the clinical profile of patients with CED as well as the CED-associated IFT122 variant spectrum.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Project of Yu-Miao (grant no. GXWCH-YMJH-2017006) and Guangxi Medical and Health Appropriate Technology Development and Promotion Application (grant no. S2020060), which was used for the patient recruitment and determining genetic variants.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QY and JL designed the study and drafted the manuscript. QZ, QY, FC and ShaY performed the data analysis and the experiments. XX analyzed the clinical data. ML, SheY and XX performed analyzed the experimental results. QY and XX revised the manuscript. QY and QZ confirmed the authenticity of all the raw data. All authorsread and approved the final version of the manuscript.
Ethics approval and consent to participate
All procedures in this study were approved by the Ethics Committee of Guangxi Maternal and Child Health Hospital. Detailed written informed consent was obtained from all participants. For the patients who are underage, written informed consent for participation in this study was obtained from the patients' parents or guardians.
Patient consent for publication
Parents of the children (under 18) provided written informed consent for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Arts HH, Bongers EM, Mans DA, van Beersum SE, Oud MM, Bolat E, Spruijt L, Cornelissen EA, Schuurs-Hoeijmakers JH, de Leeuw N, et al. C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. J Med Genet. 2011;48:390–395. doi: 10.1136/jmg.2011.088864. [DOI] [PubMed] [Google Scholar]
- 2.Bayat A, Kerr B, Douzgou S. The evolving craniofacial phenotype of a patient with Sensenbrenner syndrome caused by IFT140 compound heterozygous mutations. Clin Dysmorphol. 2017;26:247–251. doi: 10.1097/MCD.0000000000000169. DDD Study. [DOI] [PubMed] [Google Scholar]
- 3.Bredrup C, Saunier S, Oud MM, Fiskerstrand T, Hoischen A, Brackman D, Leh SM, Midtbø M, Filhol E, Bole-Feysot C, et al. Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. Am J Hum Genet. 2011;89:634–643. doi: 10.1016/j.ajhg.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilissen C, Arts HH, Hoischen A, Spruijt L, Mans DA, Arts P, van Lier B, Steehouwer M, van Reeuwijk J, Kant SG, et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet Sep. 2010;87:418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girisha KM, Shukla A, Trujillano D, Bhavani GS, Hebbar M, Kadavigere R, Rolfs A. A homozygous nonsense variant in IFT52 is associated with a human skeletal ciliopathy. Clin Genet. 2016;90:536–539. doi: 10.1111/cge.12762. [DOI] [PubMed] [Google Scholar]
- 6.Walczak-Sztulpa J, Eggenschwiler J, Osborn D, Brown DA, Emma F, Klingenberg C, Hennekam RC, Torre G, Garshasbi M, Tzschach A, et al. Cranioectodermal dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am J Hum Genet. 2010;86:949–956. doi: 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevo B, Scholey JM, Peterman EJ. Intraflagellar transport: Mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 2017;284:2905–2931. doi: 10.1111/febs.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker K, Beales PL. Making sense of cilia in disease: The human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- 9.Yuan X, Serra RA, Yang S. Function and regulation of primary cilia and intraflagellar transport proteins in the skeleton. Ann NY Acad Sci. 2015;1335:78–99. doi: 10.1111/nyas.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oud MM, Lamers IJ, Arts HH. Ciliopathies: Genetics in Pediatric Medicine. J Pediatr Genet. 2017;6:18–29. doi: 10.1055/s-0036-1593841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arts H, Knoers N. Cranioectodermal Dysplasia. GeneReviews [Internet]. Adam MP, Ardinger HH and Pagon RA (eds). University of Washington Seattle, WA, 2013. [PubMed] [Google Scholar]
- 12.Zhang W, Taylor SP, Ennis HA, Forlenza KN, Duran I, Li B, Sanchez JA, Nevarez L, Nickerson DA, Bamshad M, et al. University of Washington Center for Mendelian Genomics: Expanding the genetic architecture and phenotypic spectrum in the skeletal ciliopathies. Hum Mutat. 2018;39:152–166. doi: 10.1002/humu.23362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handa A, Voss U, Hammarsjö A, Grigelioniene G, Nishimura G. Skeletal ciliopathies: A pattern recognition approach. Jpn J Radiol. 2020;38:193–206. doi: 10.1007/s11604-020-00920-w. [DOI] [PubMed] [Google Scholar]
- 14.Lin AE, Traum AZ, Sahai I, Keppler-Noreuil K, Kukolich MK, Adam MP, Westra SJ, Arts HH. Sensenbrenner syndrome (Cranioectodermal dysplasia): Clinical and molecular analyses of 39 patients including two new patients. Am J Med Genet A. 2013;161A:2762–2776. doi: 10.1002/ajmg.a.36265. [DOI] [PubMed] [Google Scholar]
- 15.Walczak-Sztulpa J, Posmyk R, Bukowska-Olech EM, Wawrocka A, Jamsheer A, Oud MM, Schmidts M, Arts HH, Latos-Bielenska A, Wasilewska A. Compound heterozygous IFT140 variants in two Polish families with Sensenbrenner syndrome and early onset end-stage renal disease. Orphanet J Rare Dis. 2020;15(36) doi: 10.1186/s13023-020-1303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidts M. Clinical genetics and pathobiology of ciliary chondrodysplasias. J Pediatr Genet. 2014;3:46–94. doi: 10.3233/PGE-14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. ACMG Laboratory Quality Assurance Committee: Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silveira KC, Moreno CA, Cavalcanti DP. Beemer-Langer syndrome is a ciliopathy due to biallelic mutations in IFT122. Am J Med Genet A. 2017;173:1186–1189. doi: 10.1002/ajmg.a.38157. [DOI] [PubMed] [Google Scholar]
- 19.Huber C, Cormier-Daire V. Ciliary disorder of the skeleton. Am J Med Genet C Semin Med Genet. 2012;160C:165–174. doi: 10.1002/ajmg.c.31336. [DOI] [PubMed] [Google Scholar]
- 20.Alazami AM, Seidahmed MZ, Alzahrani F, Mohammed AO, Alkuraya FS. Novel IFT122 mutation associated with impaired ciliogenesis and cranioectodermal dysplasia. Mol Genet Genomic Med. 2014;2:103–106. doi: 10.1002/mgg3.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moosa S, Obregon MG, Altmüller J, Thiele H, Nürnberg P, Fano V, Wollnik B. Novel IFT122 mutations in three Argentinian patients with cranioectodermal dysplasia: Expanding the mutational spectrum. Am J Med Genet A. 2016;170A:1295–1301. doi: 10.1002/ajmg.a.37570. [DOI] [PubMed] [Google Scholar]
- 22.Cardenas-Rodriguez M, Badano JL. Ciliary biology: Understanding the cellular and genetic basis of human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:263–280. doi: 10.1002/ajmg.c.30227. [DOI] [PubMed] [Google Scholar]
- 23.Bacino CA, Dhar SU, Brunetti-Pierri N, Lee B, Bonnen PE. WDR35 mutation in siblings with Sensenbrenner syndrome: A ciliopathy with variable phenotype. Am J Med Genet A. 2012;158A:2917–2924. doi: 10.1002/ajmg.a.35608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 25.Tsurusaki Y, Yonezawa R, Furuya M, Nishimura G, Pooh RK, Nakashima M, Saitsu H, Miyake N, Saito S, Matsumoto N. Whole exome sequencing revealed biallelic IFT122 mutations in a family with CED1 and recurrent pregnancy loss. Clin Genet. 2014;85:592–594. doi: 10.1111/cge.12215. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Sun S, Li N, Yu T, Wang X, Wang J, Bao N. Identification and analysis of the genetic causes in nine unrelated probands with syndromic craniosynostosis. Gene. 2018;641:144–150. doi: 10.1016/j.gene.2017.10.041. [DOI] [PubMed] [Google Scholar]
- 27.Walczak-Sztulpa J, Wawrocka A, Sobierajewicz A, Kuszel L, Zawadzki J, Grenda R, Swiader-Lesniak A, Kocyla-Karczmarewicz B, Wnuk A, Latos-Bielenska A, et al. Intrafamilial phenotypic variability in a Polish family with Sensenbrenner syndrome and biallelic WDR35 mutations. Am J Med Genet A. 2017;173:1364–1368. doi: 10.1002/ajmg.a.38163. [DOI] [PubMed] [Google Scholar]
- 28.Bouldin CM, Gritli-Linde A, Ahn S, Harfe BD. Shh pathway activation is present and required within the vertebrate limb bud apical ectodermal ridge for normal autopod patterning. Proc Natl Acad Sci USA. 2010;107:5489–5494. doi: 10.1073/pnas.0912818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsao CC, Gorovsky MA. Tetrahymena IFT122A is not essential for cilia assembly but plays a role in returning IFT proteins from the ciliary tip to the cell body. J Cell Sci. 2008;121:428–436. doi: 10.1242/jcs.015826. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 31.Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc Natl Acad Sci USA. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks ER, Islam MT, Anderson KV, Zallen JA. Sonic hedgehog signaling directs patterned cell remodeling during cranial neural tube closure. eLife. 2020;9(e60234) doi: 10.7554/eLife.60234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.