Abstract
To characterize early cerebellum development, accurate segmentation of the cerebellum into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) tissues is one of the most pivotal steps. However, due to the weak tissue contrast, extremely folded tiny structures, and severe partial volume effect, infant cerebellum tissue segmentation is especially challenging, and the manual labels are hard to obtain and correct for learning-based methods. To the best of our knowledge, there is no work on the cerebellum segmentation for infant subjects less than 24 months of age. In this work, we develop a semi-supervised transfer learning framework guided by a confidence map for tissue segmentation of cerebellum MR images from 24-month-old to 6-month-old infants. Note that only 24-month-old subjects have reliable manual labels for training, due to their high tissue contrast. Through the proposed semi-supervised transfer learning, the labels from 24-month-old subjects are gradually propagated to the 18-, 12-, and 6-month-old subjects, which have a low tissue contrast. Comparison with the state-of-the-art methods demonstrates the superior performance of the proposed method, especially for 6-month-old subjects.
Keywords: Infant cerebellum segmentation, Confidence map, Semi-supervised learning
1. Introduction
The first 2 years of life is the most dynamic postnatal period of the human cerebellum development [1], with the cerebellum volume increasing by 240% from 2 weeks to 1 year, and by 15% from 1 to 2 years of age [2]. Cerebellum plays an important role in motor control, and is also involved in some cognitive functions as well as emotional control [3]. For instance, recent cerebellar findings in autism suggest developmental differences at multiple levels of neural structure and function, indicating that the cerebellum is an important player in the complex neural underpinnings of autism spectrum disorder, with behavioral implications beyond the motor domain [4]. To characterize early cerebellum development, accurate segmentation of the cerebellum into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) is one of the most pivotal steps. However, compared with adult cerebellum, infant cerebellum is much more challenging in tissue segmentation, due to the low tissue contrast caused by ongoing myelination, extremely folded tiny structures, and severe partial volume effect.
Most of the previous brain development studies have focused on the cerebral cortex [5–10]. For instance, Wang et al. proposed an anatomy-guided Densely-connected U-Net (ADU-Net) [11] for the cerebrum image segmentation of 6-month-old infants. However, there are very few works proposed for pediatric cerebellum tissue segmentation [12–16]. Chen et al. [17] proposed an ensemble sparse learning method for cerebellum tissue segmentation of 24-month-old subjects. Romero et al. [15] presented a patch-based multi-atlas segmentation tool called CERES, that is able to automatically parcellate the cerebellum lobules, and is the winner of a MICCAI cerebellum segmentation challenge. To the best of our knowledge, there is no work on cerebellum tissue segmentation for infant subjects less than 24 months of age. Figure 1 shows an example of T1-weighted (T1w) and T2-weighted (T2w) MR images (MRIs) of cerebellum at 6, 12, 18 and 24 months of age, and the corresponding segmentations are obtained by ADU-Net [11] and the proposed semi-supervised method in the last two rows. From 6 months to 24 months, we can observe that the cerebellum volume increases rapidly, and the tissue contrast is varying remarkably, i.e., 6-month-old cerebellum exhibits an extremely low tissue contrast, while 24-month-old cerebellum shows a much high contrast. As also confirmed from the previous work [14], 24-month-old cerebellum can be automatically or manually segmented due to its high contrast. However, for other early ages, especially for 6-month-old cerebellum, it is challenging even for manual segmentation by experienced experts. Directly applying the model trained on 24-month-old subjects to younger infants cannot derive satisfactory segmentation results. For example, we directly apply a trained model on 24-month-old subjects using ADU-Net [11] to other time points, and show the derived results in the third row of Fig. 1. It can be seen that the results are not accurate, due to distinct tissue contrast and distribution between 24-month-old subjects and other younger subjects. Therefore, in this work, we will investigate how to effectively utilize the labels from 24-month-old subjects with high contrast to the other time-point subjects with low contrast. This is a general yet challenging transfer learning task, if we consider 24-month-old subjects as a source site while the remaining time-point subjects as a target site. Note that all studied subjects in this paper are cross-sectional.
Fig. 1.
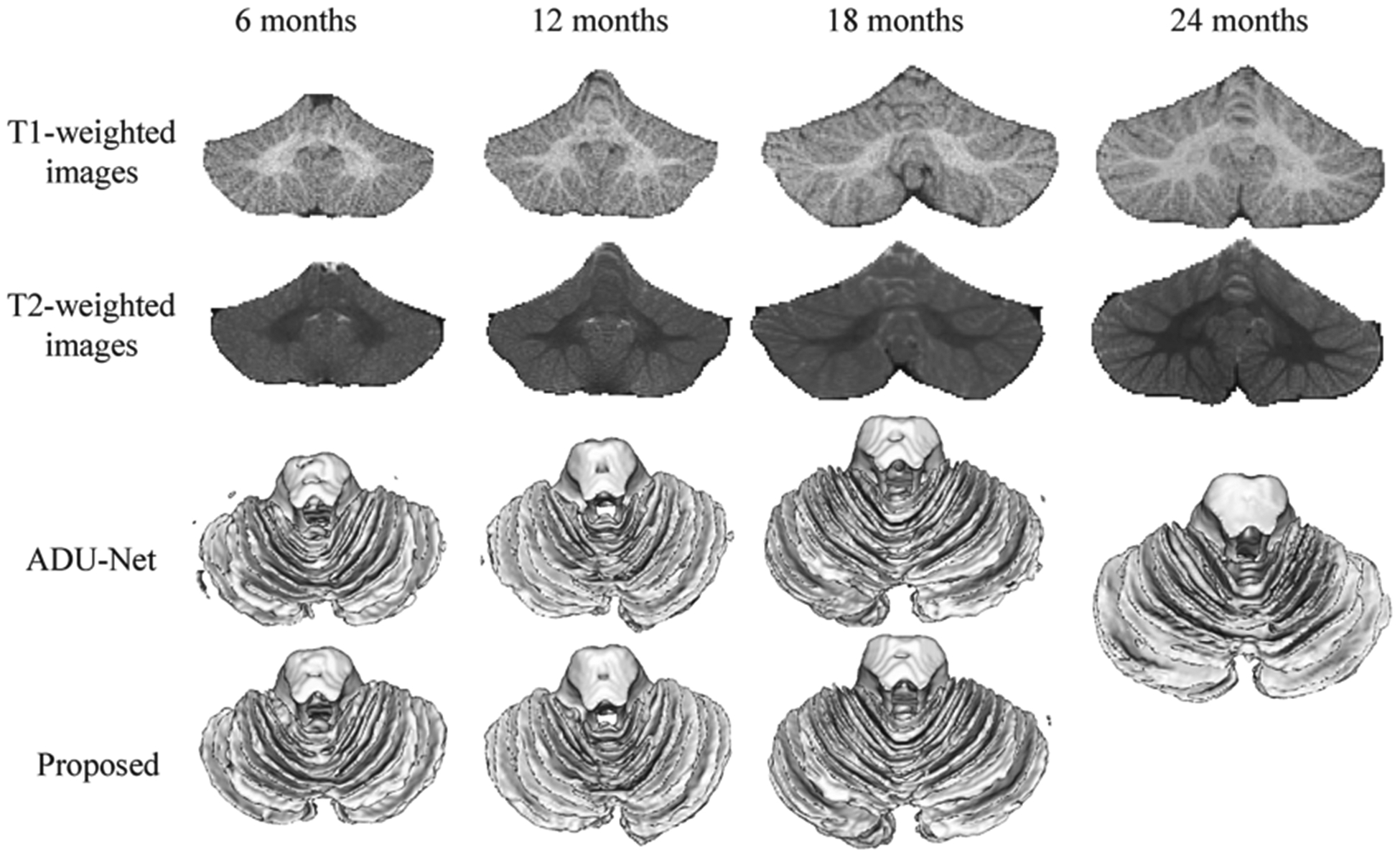
T1- and T2-weighted MRIs of the cerebellum at 6, 12, 18 and 24 months of age, with the corresponding segmentation results obtained by ADU-Net [9] and the proposed semi-supervised method.
In this paper, we propose a semi-supervised transfer learning framework guided by the a confidence map for tissue segmentation of cerebellum MRIs from 24-month-old to 6-month-old infants, where only 24-month-old subjects with high tissue contrast have manual labels for training. Then, for other younger subjects without manual labels, we retrain the segmentation models to handle the different tissue contrast and distribution guided by the confidence map. Specifically, in order to select reliable segmentations as training datasets at 18, 12, and 6 months of age, a confidence network is trained to estimate the reliability of automatic tissue segmentation results [18]. Second, the confidence map is further incorporated as a spatially-weighted loss function to alleviate the effect from these regions with unreliable segmentations. This paper is organized as follows. Section 2 introduces the dataset and related pre-processing. Then, the proposed semi-supervised framework is detailed in Sect. 3. In Sect. 4, experimental results and analyses are presented to demonstrate the superior performance of our method. Finally, Sect. 5 concludes the paper.
2. Dataset and Preprocessing
T1w and T2w infant brain MRIs used in this study were from the UNC/UMN Baby Connectome Project (BCP) [19] and were acquired at around 24, 18, 12, and 6 months of age on Siemens Prisma scanners. During scanning, infants were naturally sleeping, fitted with ear protection, and their heads were secured in a vacuum-fixation device. T1w MRIs were acquired with 160 sagittal slices using parameters: TR/TE 2400/2.2 ms and voxel resolution = 0.8 × 0.8 × 0.8 mm3. T2w MRIs were obtained with 160 sagittal slices using parameters: TR/TE = 3200/564 ms and voxel resolution = 0.8 × 0.8 × 0.8 mm3.
Accurate manual segmentation, providing labels for training and testing, is of great importance for learning-based segmentation methods. In this paper, we manually edited eighteen 24-month-old subjects to train the segmentation model. Limited number of 18-, 12-, and 6-month-old subjects are manually edited for validation. From 24- to 6-month-old subjects, the label editing becomes gradually difficult and more time-consuming due to the low tissue contrast and extremely folded tiny structures.
3. Method
Figure 2 illustrates the flowchart of the proposed semi-supervised segmentation framework guided by a confidence map. As we can see, there are infant subjects at 6, 12, 18, and 24 months of age, while only 24-month-old manual segmentations are used for training. After training the 24-month-old segmentation (shorted as 24 m-S) model, the automatic segmentations are used to generate error maps compared with ground truth, which are viewed as targets for 24-month-old confidence model (shorted as 24 m-C). However, the trained 24 m-S model cannot be directly applied to 18-month-old subjects due to different tissue contrast and data distribution, as shown in Fig. 1. To effectively utilize the labels from 24-month-old subjects to 18-month-old subjects, we apply the 24 m-C model to estimate the reliability of automatic tissue segmentation results on the 18-month-old subjects. Then, based on the confidence maps, top K-ranked subjects with good segmentations are chosen as training sets for 18-month-old subjects, and the confidence map is further incorporated as a spatially-weighted loss function to alleviate possible errors in the segmentations. The same procedure can be also applied to train 12-month-old segmentation (12 m-S) model and 6-month-old segmentation (6 m-S) model.
Fig. 2.
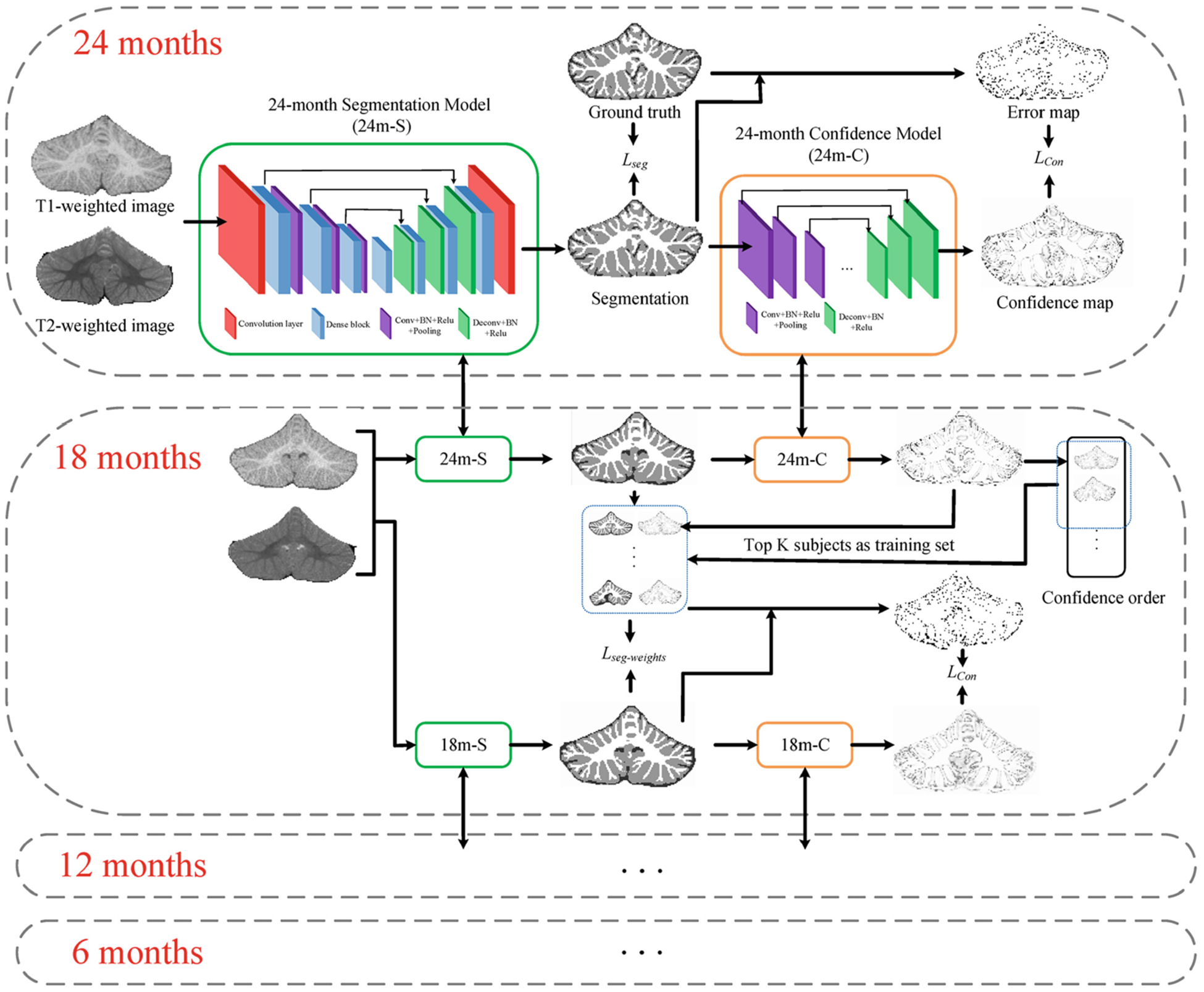
Flowchart of the proposed semi-supervised segmentation framework guided by confidence map. For 24-month-old subjects, the ground truth is edited manually, the loss function of the segmentation model Lseg is cross entropy, and the loss function of the confidence network Lcp is multi-task cross-entropy. For other months, the ground truth is chosen from automatic segmentations, the loss function of the segmentation model Lseg-weights is the proposed spatially-weighted cross entropy.
3.1. Confidence Map of Automatic Segmentations
To derive reliable 18- (12-, 6-) month cerebellum segmentations, we first employ the confidence map to evaluate the automatic segmentations generated by the trained 24 m-S (18 m-S, 12 m-S) model. Inspired by [18], we apply U-Net structure [20] with the contracting path and expansive path to achieve the confidence map. Instead of using adversarial learning, the error map (Fig. 3 (a)) generated based on the differences between manual results and automatic segmentations, is regarded as ground truth to train the confidence model. We employ a multi-task softmax loss function, which is more effective to learn whether the segmentation results are reasonable or not voxel-by-voxel, as shown in Fig. 3 (b). Note that the darker the color, the worse the segmentation. The 3D zoomed view of WM segmentation is shown in Fig. 3 (c), where the intersection of red lines denotes a missing gyrus that is the same region with Fig. 3 (b), and the corresponding ground truth is also displayed in Fig. 3 (d).
Fig. 3.
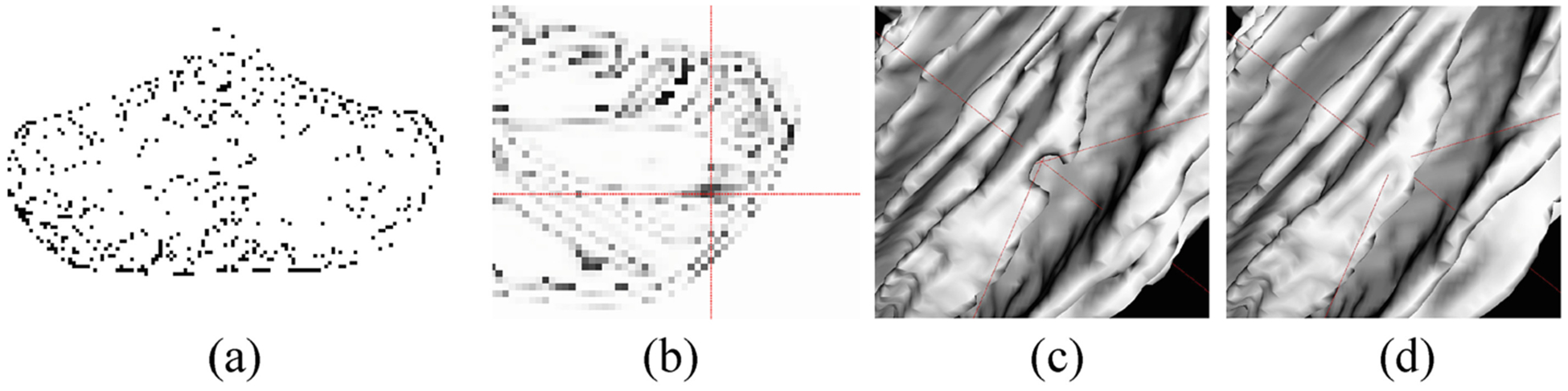
(a) is the error map generated based on the differences between ground truth and automatic segmentations. (b) shows the confidence map (a probability map), where the intersection of red lines points out a low confidence region, which means the segmentation is relatively unreasonable. (c) is the corresponding 3D surface rendering results, i.e., the missing gyrus as indicated in (b), and (d) is the ground truth.
3.2. Semi-supervised Learning
After training the 24 m-S model with manual labels, we retrain the 18 m-S, 12 m-S and 6 m-S models based on 18-, 12-, and 6-month-old subjects, respectively. In particular, the training labels are automatically generated from the automatic segmentations, and a spatially-weighted cross-entropy loss is proposed to learn from reasonable segmentations guided by the confidence map.
Training Segmentation Model for 24-month-old Subjects:
We employ the ADU-Net architecture [11] as the segmentation model, which combines the advantages of U-Net and Dense block, and demonstrates outstanding performance on infant brain segmentation. As shown in Fig. 2, the ADU-Net includes a down-sampling path and an up-sampling path, going through seven dense blocks. Then, eighteen paired T1w and T2w images with their corresponding manual segmentations are as inputs of ADU-Net to train the 24 m-S model. We evaluate the performance of the 24 m-S model in terms of Dice ratio on five 24-month-old testing subjects, with the accuracy of 90.46 ± 1.56%, 91.83 ± 0.62% and 94.11 ± 0.38% for CSF, GM and WM, respectively.
Automatic Generation of Training Labels for 18-, 12-, 6-month-old Subjects:
With the guidance of the confidence map, we select K top-ranked subjects with good segmentations as training sets for each month. The rank is based on the average confidence values of each confidence map. Figure 4 shows a set of automatic segmentation results on 12-month-old subjects, ranked by the average confidence values. Specifically, the 12-month-old cerebellum segmentations and confidence maps are obtained by 18 m-S and 18 m-C models, respectively. From Fig. 4, we can observe that the confidence order is consistent with the accuracy of segmentations, which also proves the effectiveness of the confidence network. Therefore, according to the confidence order, the chosen K top-ranked subjects are reliable for training the segmentation model of other months.
Fig. 4.
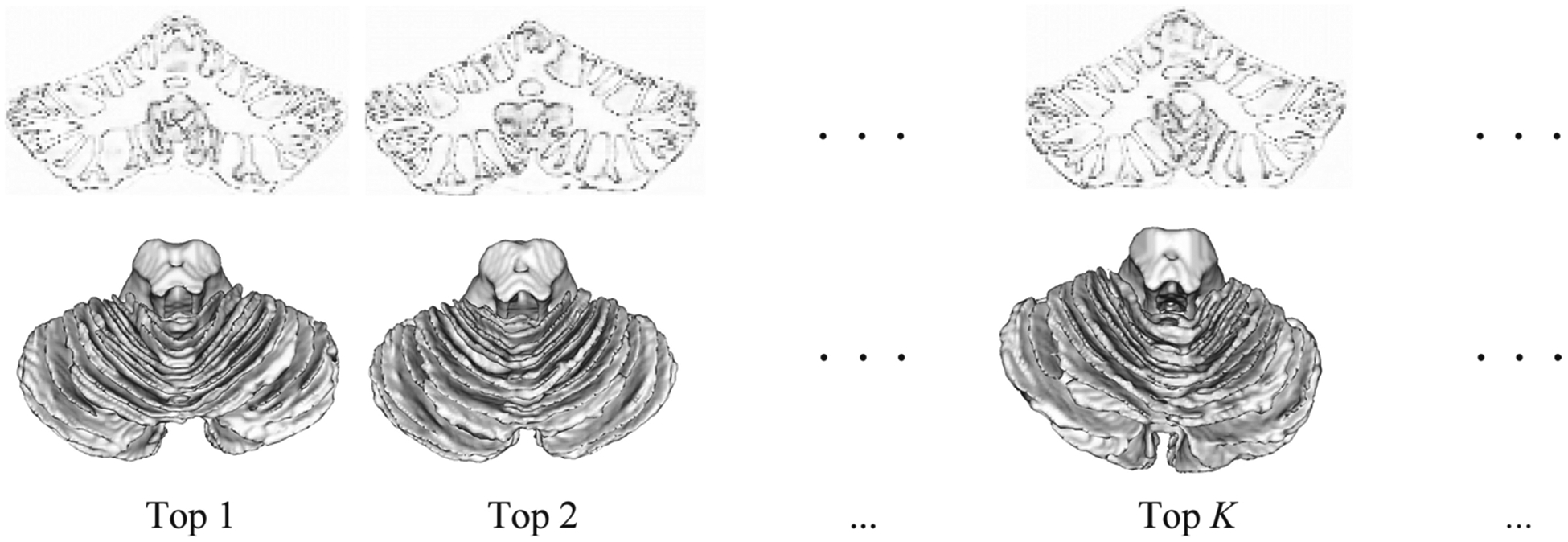
A set of automatic segmentation results on 12-month-old subjects, ranked by the average confidence values. The first row is the confidence maps, and the second row is the automatic 3D WM segmentations, which are obtained from 18 m-S and 18 m-C model, respectively.
Spatially-Weighted Cross-entropy Loss with Confidence Map:
Although we select reliable segmentation results as the training labels for 18-, 12- and 6-month-old subjects based on the confidence map, for each selected subject, there are still many locations with unreliable labels. Considering this issue, we further incorporate the confidence map into the loss function, therefore, the segmentation model would pay more attention to reliable labels, while less attention to unreliable labels. The spatially-weighted cross-entropy loss function is written as,
where C is the class number, xi represents the predicted probability map, yi is the ground truth, and w denotes the weights from the confidence map.
Implementation Details:
We randomly extract 1,000 32 × 32 × 32 3D patches from each training subject. The loss Lcp for the confidence network is multi-task cross-entropy. The kernels are initialized by Xavier [38]. We use SGD optimization strategy. The learning rate is 0.005 and multiplies by 0.1 after each epoch.
4. Experimental Results
In this section, we first investigate the optimal choice of number K of training subjects, then make a comparison of semi-supervised learning and supervised learning to demonstrate the effectiveness of the proposed method. Later, we perform an ablation study of confidence weights. Finally, the performance of our method is compared with volBrain [21] and ADU-Net method [11] on five 18-month-old subjects, five 12-month-old subjects, and five 6-month-old subjects with manual labels.
Selection of the Number K of Training Subjects:
According to the confidence map, we select the K top-ranked subjects as training images for the 18 m-S (12 m-S and 6 m-S) models. Considering that the reliability of ordered segmentations gradually decreases, we compare the performance associated with the different number K to choose the best one, as shown in Fig. 5. It is expected with the increase of K (<10), the Dice ratio is gradually improved in terms of CSF, GM and WM results, whereas the value begins to drop after K > 10, due to introducing too many unreliable labels into the training. Therefore, we set K = 10 in all experiments.
Fig. 5.
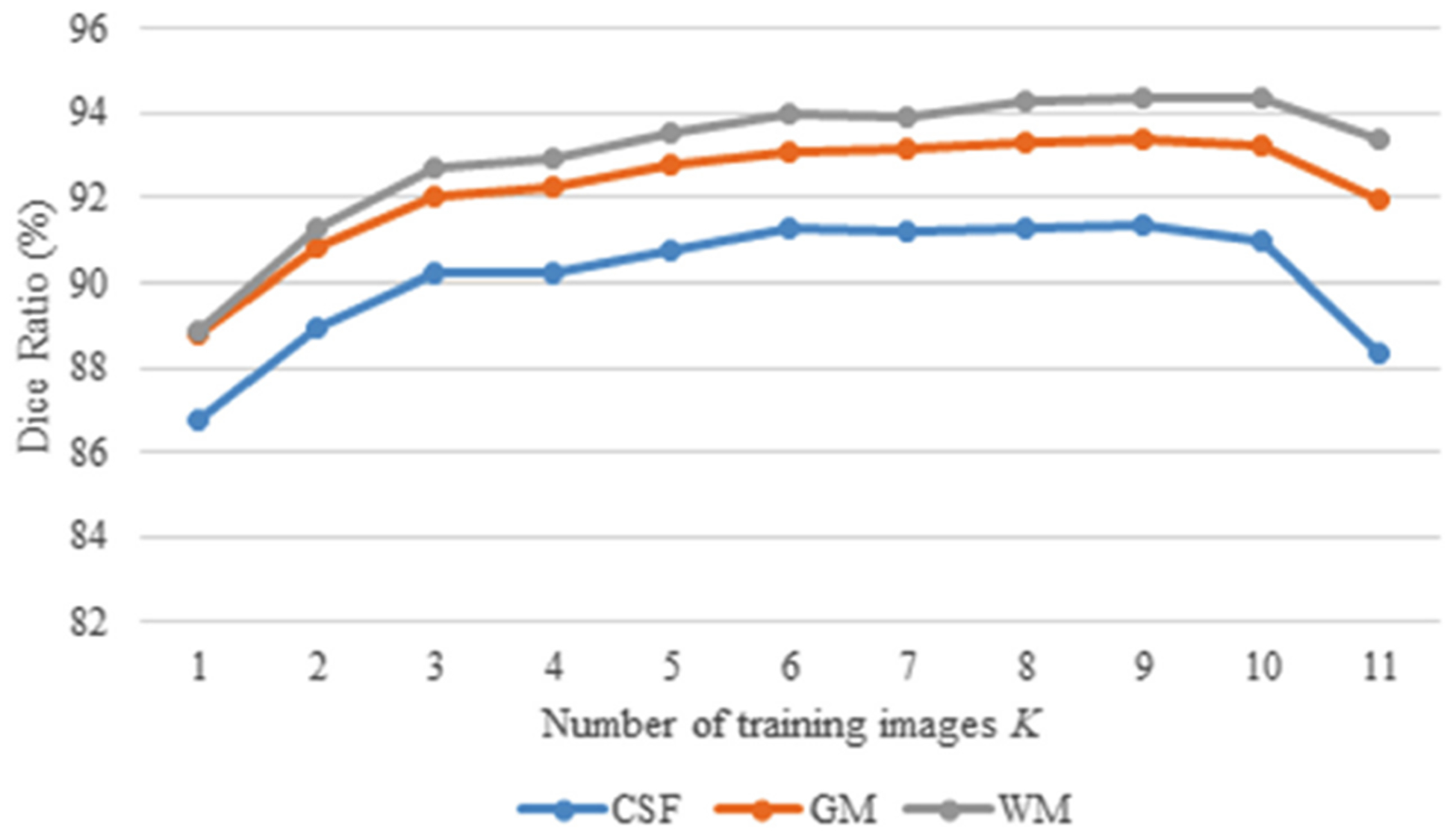
Comparison of different K top-ranked training subjects, in terms of Dice ratio.
Semi-supervised vs. Supervised Learning:
Although the limited number of manual segmentation results for 18-, 12- and 6-month-old subjects are purposely created for validation only, we are wondering whether the performance with the semi-supervised learning is better than the supervised learning or not. To this end, for the supervised learning, we select N subjects with manual labels as the training subjects. Similarly, for the semi-supervised learning, we select the same N training subjects as the supervised learning, plus additional top K-ranked subjects based on the confidence. The same remaining subjects with manual labels are used for testing. The comparison is shown in Fig. 6 in terms of WM segmentation, along with the number of training subjects N. The number of training subjects with manual labels for the supervised learning is N, while N + top K for the semi-supervised learning. As we can see, compared with the supervised learning, the semi-supervised learning (i.e., the proposed method) greatly improves the accuracy of tissue segmentations, especially when the number of training subjects with manual labels is highly limited.
Fig. 6.
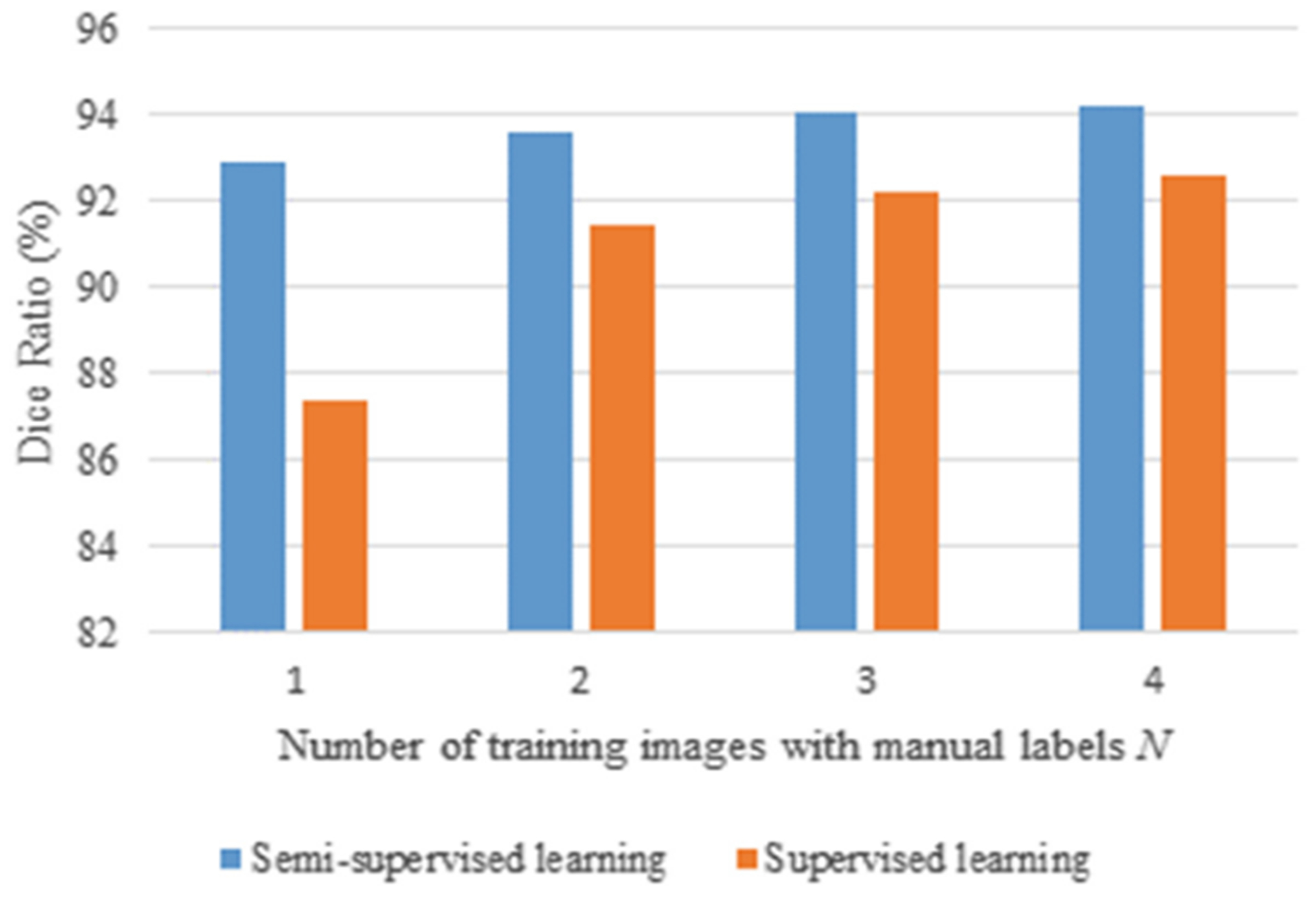
Comparison of the supervised and semi-supervised learning on 6-month-old subjects, in terms of Dice ratio in WM segmentation.
Ablation Study:
To demonstrate the advantage of the proposed spatially-weighted cross-entropy loss, we make a comparison between the results using the cross-entropy loss without/with confidence weights. Figure 7 shows the comparison of WM segmentations, where the first (second) column shows the surface rendering results without (with) confidence weights. We can see that without the guidance of confidence weights, there are many topological and geometric errors as indicated by red arrow in the first column, whereas these errors are alleviated as shown in the second column, which is also more consistent with the corresponding manual labels in the last column.
Fig. 7.
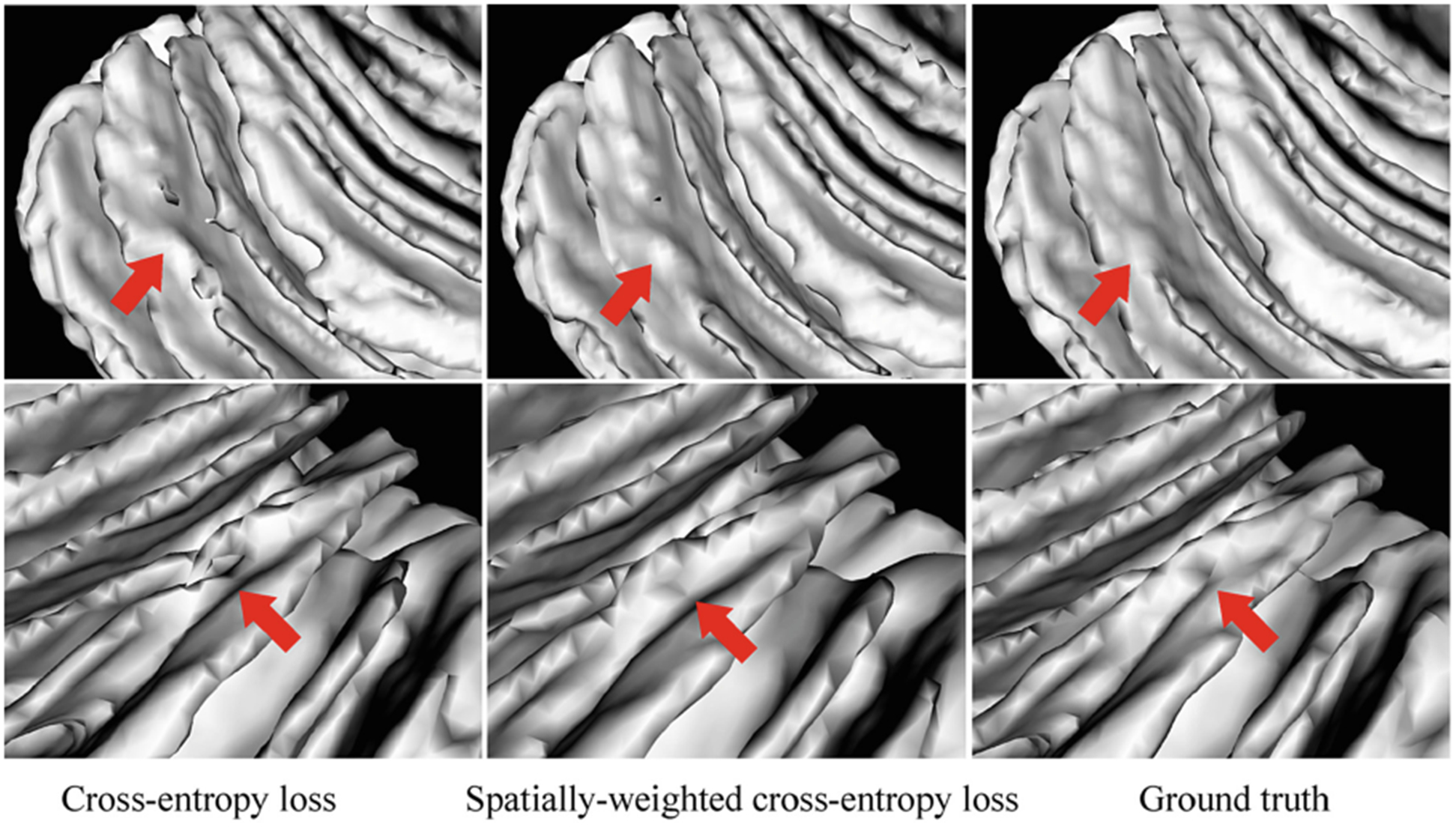
Comparison of the WM segmentations. From left to right: segmentations using cross-entropy loss, the proposed spatially-weighted cross-entropy loss, and the ground truth.
Comparison with the State-of-the-Art Methods:
We make comparisons with volBrain [21] and ADU-Net method [11], where the volBrain is an automated MRI Brain Volumetry System (https://volbrain.upv.es/index.php) and the ADU-Net architecture is the backbone of our segmentation model. In the volBrain system, we choose the CERES pipeline [15] to automatically analyze the cerebellum, which wins a MICCAI cerebellum segmentation challenge. Figure 8 displays the comparison among these methods on 18-, 12-, and 6-month-old testing subjects. Tissue segmentations of the proposed method are much more consistent with the ground truth, which can be observed from both 2D slices and 3D surface rendering results.
Fig. 8.
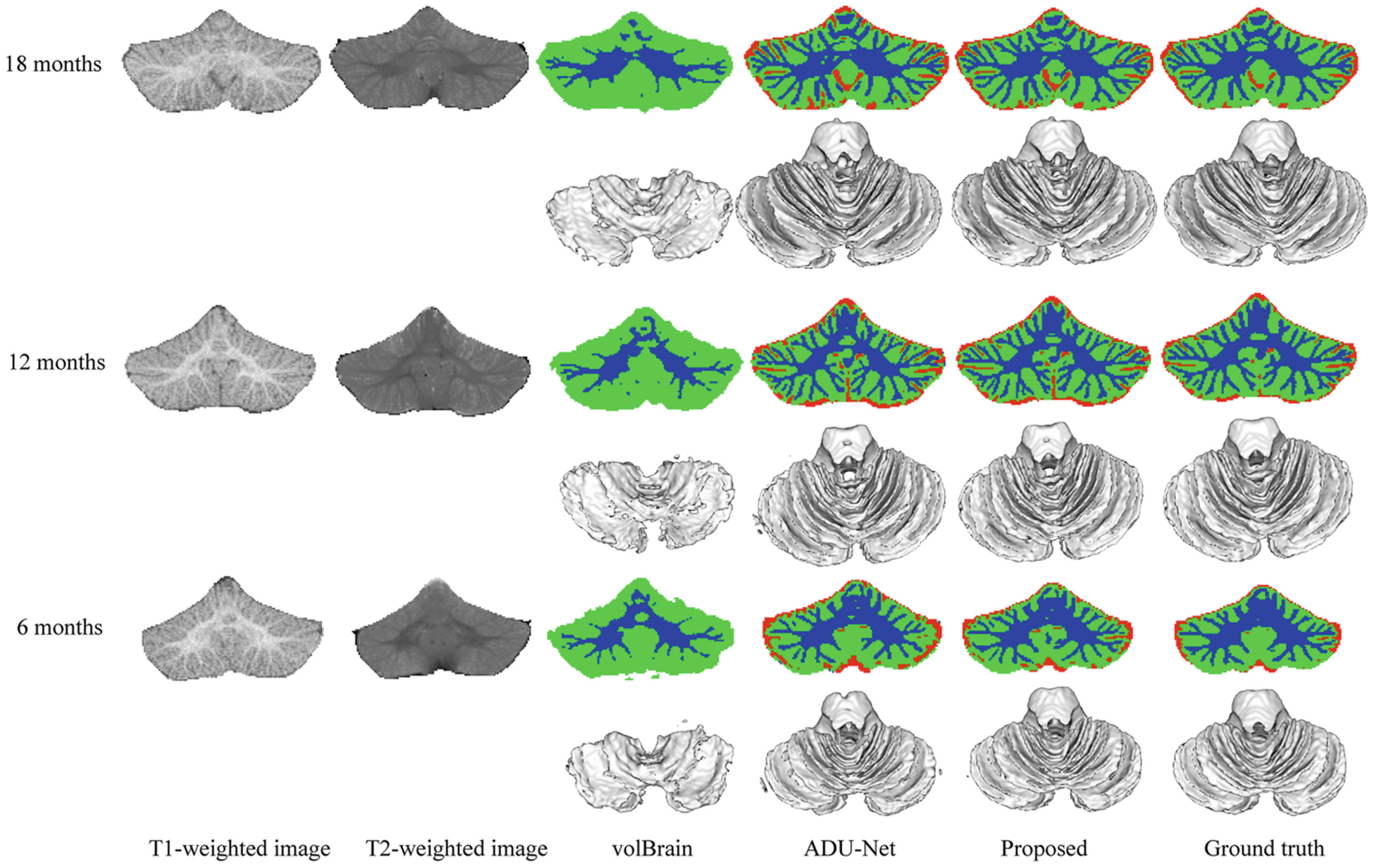
Segmentation results of the volBrain [21], ADU-Net [11] and the proposed method on 18-, 12-, and 6-month-old infant subjects from BCP, with the corresponding 3D WM surface rendering views. The red/green/blue color denotes CSF/GM/WM.
Furthermore, we compute the Dice ratio of tissue segmentation (i.e., CSF, GM, and WM) on 15 infant subjects (i.e., five 18-month-old subjects, five 12-month-old subjects, and five 6-month-old subjects), to evaluate the performance of our method. In order to compare the difference of segmentation results, Wilcoxon signed-rank tests are also calculated for statistical analysis in Table 1. The proposed method achieves a significantly better performance in terms of Dice ratio on CSF and GM for 18-month-old subjects and CSF, GM, and WM for 12- and 6-month-old subjects.
Table 1.
Dice ratio (%) of cerebellum segmentation results on testing subjects at 18 months, 12 months, and 6 months of age.
| Age in month | Method | CSF | GM | WM |
|---|---|---|---|---|
| 18 | volBrain | N/A | 78.23 ± 1.10 | 54.43 ± 3.87 |
| ADU-Net | 87.57 ± 1.70 | 91.57 ± 0.43 | 93.52 ± 0.65 | |
| Proposed | 91.36 ± 1.11+ | 93.40 ± 0.75+ | 94.39 ± 0.69 | |
| 12 | volBrain | N/A | 70.80 ± 13.54 | 49.86 ± 4.70 |
| ADU-Net | 84.06 ± 2.40 | 89.83 ± 0.88 | 91.89 ± 0.68 | |
| Proposed | 90.50 ± 1.25+ | 93.08 ± 0.47+ | 94.14 ± 0.48+ | |
| 6 | volBrain | N/A | 77.76 ± 1.24 | 50.96 ± 3.91 |
| ADU-Net | 76.68 ± 0.76 | 86.30 ± 0.79 | 88.98 ± 1.87 | |
| Proposed | 85.35 ± 0.76+ | 90.23 ± 1.15+ | 92.02 ± 1.92+ |
indicates that our proposed method is significantly better than both volBrain and ADU-Net methods with p-value < 0.05.
5. Conclusion
To deal with the challenging task of infant cerebellum tissue segmentation, we proposed a semi-supervised transfer learning framework guided by the confidence map. We took advantage of 24-month-old subjects with high tissue contrast and effectively transferred the labels from 24-month-old subjects to other younger subjects typically with low tissue contrast. The experiments demonstrate that our proposed method has achieved significant improvement in terms of accuracy. We will further extend our method to newborn subjects and validate on more subjects.
Acknowledgements.
This work was supported in part by National Institutes of Health grants MH109773, MH116225, and MH117943. This work utilizes approaches developed by an NIH grant (1U01MH110274) and the efforts of the UNC/UMN Baby Connectome Project Consortium.
References
- 1.Li G, Wang L, Yap P-T, et al. : Computational neuroanatomy of baby brains: a review. NeuroImage 185, 906–925 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knickmeyer RC, Gouttard S, Kang C, et al. : A structural MRI study of human brain development from birth to 2 years. J. Neurosci 28, 12176–12182 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf U, Rapoport MJ, Schweizer TA: Evaluating the affective component of the cerebellar cognitive affective syndrome. J. Neuropsychiatry Clin. Neurosci 21, 245–253 (2009) [DOI] [PubMed] [Google Scholar]
- 4.Becker EBE, Stoodley CJ: Autism spectrum disorder and the cerebellum. Int. Rev. Neurobiol 113, 1–34 (2013) [DOI] [PubMed] [Google Scholar]
- 5.Wang F, Lian C, Wu Z, et al. : Developmental topography of cortical thickness during infancy. Proc. Nat. Acad. Sci 116, 15855 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan D, Xia S, Rekik I, et al. : Exploring folding patterns of infant cerebral cortex based on multi-view curvature features: methods and applications. NeuroImage 185, 575–592 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Nie J, Wang L, et al. : Measuring the dynamic longitudinal cortex development in infants by reconstruction of temporally consistent cortical surfaces. NeuroImage 90, 266–279 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G, Nie J, Wang L, et al. : Mapping longitudinal hemispheric structural asymmetries of the human cerebral cortex from birth to 2 years of age. Cereb Cortex 24, 1289–1300 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y, Gao K, Wu Z, et al. : Multi-site infant brain segmentation algorithms: The iSeg-2019 Challenge. arXiv preprint arXiv:2007.02096 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Nie D, Li G, et al. : Benchmark on automatic six-month-old infant brain segmentation algorithms: the iSeg-2017 Challenge. IEEE Trans. Med. Imaging 38, 2219–2230 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Li G, Shi F, et al. : Volume-based analysis of 6-month-old infant brain MRI for autism biomarker identification and early diagnosis. Med. Image Comput. Comput. Assist. Interv 11072, 411–419 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cettour-Janet P, et al. : Hierarchical approach for neonate cerebellum segmentation from MRI: an experimental study In: Burgeth B, Kleefeld A, Naegel B, Passat N, Perret B (eds.) ISMM 2019. LNCS, vol. 11564, pp. 483–495. Springer, Cham: (2019). 10.1007/978-3-030-20867-7_37 [DOI] [Google Scholar]
- 13.Bogovic JA, Bazin P-L, Ying SH, Prince JL: Automated segmentation of the cerebellar lobules using boundary specific classification and evolution. Inf. Process Med. Imaging 23, 62–73 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Wang L, Shen D: Cerebellum tissue segmentation with ensemble sparse learning. Proc. Int. Soc. Magn. Reson. Med. Sci. Meet Exhib. Int 25, 0266 (2017) [PMC free article] [PubMed] [Google Scholar]
- 15.Romero JE, Coupé P, Giraud R, et al. : CERES: a new cerebellum lobule segmentation method. NeuroImage 147, 916–924 (2017) [DOI] [PubMed] [Google Scholar]
- 16.Hwang J, Kim J, Han Y, Park H: An automatic cerebellum extraction method in T1-weighted brain MR images using an active contour model with a shape prior. Magn. Reson. Imaging 29, 1014–1022 (2011) [DOI] [PubMed] [Google Scholar]
- 17.Chen J, et al. : Automatic accurate infant cerebellar tissue segmentation with densely connected convolutional network In: Shi Y, Suk H-I, Liu M (eds.) MLMI 2018. LNCS, vol. 11046, pp. 233–240. Springer, Cham: (2018). 10.1007/978-3-030-00919-9_27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie D, Gao Y, Wang L, Shen D: ASDNet: attention based semi-supervised deep networks for medical image segmentation In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 370–378. Springer, Cham: (2018). 10.1007/978-3-030-00937-3_43 [DOI] [Google Scholar]
- 19.Howell BR, Styner MA, Gao W, et al. : The UNC/UMN Baby Connectome Project (BCP): an overview of the study design and protocol development. NeuroImage 185, 891–905 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronneberger O, Fischer P, Brox T: U-Net: convolutional networks for biomedical image segmentation In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham: (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 21.Manjón JV, Coupé P: volBrain: an online MRI brain volumetry system. Front Neuroinform 10, 30 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]


