Culex pipiens mosquitoes are infected with wPip. These endosymbionts induce a conditional sterility called CI resulting from embryonic deaths, which constitutes a cornerstone for Wolbachia antivectorial methods.
KEYWORDS: Culex pipiens, toxin-antitoxin system, Wolbachia, developmental biology, endosymbionts, gene amplification, vectors
ABSTRACT
In arthropods, Wolbachia endosymbionts induce conditional sterility, called cytoplasmic incompatibility (CI), resulting from embryonic lethality. CI penetrance (i.e., embryonic death rate) varies depending on host species and Wolbachia strains involved. All Culex pipiens mosquitoes are infected by the endosymbiotic alphaproteobacteria Wolbachia wPip. CI in Culex, characterized as a binary “compatible/incompatible” phenomenon, revealed an unparalleled diversity of patterns linked to the amplification-diversification of cidA and cidB genes. Here, we accurately studied CI penetrance variations in the light of cid genes divergence by generating a C. pipiens compatibility matrix between 11 lines hosting different phylogenetic wPip groups and exhibiting distinct cid gene repertoires. We showed, as expected, that crosses involving wPip from the same group were mostly compatible. In contrast, only 22% of the crosses involving different wPip groups were compatible, while 54% were fully incompatible. For the remaining 24% of the crosses, “intermediate” compatibilities were reported, and a cytological observation of the first zygotic division confirmed the occurrence of “canonical” CI phenotypes in a fraction of the eggs. Backcross experiments demonstrated that intermediate compatibilities were not linked to host genetic background but to the Wolbachia strains involved. This previously unstudied intermediate penetrance CI was more severe and frequent in crosses involving wPip-IV strains exhibiting cid variants markedly divergent from other wPip groups. Our data demonstrate that CI is not always a binary compatible/incompatible phenomenon in C. pipiens but that intermediate compatibilities putatively resulting from partial mismatch due to Cid proteins divergence exist in this species complex.
INTRODUCTION
In many arthropods, the fertility of two sexual partners undergoes acute reduction due to the presence of the intracellular alphaproteobacteria Wolbachia (1). This conditional sterility, depending on the presence of cytoplasmic factors, is called cytoplasmic incompatibility (CI). CI primarily occurs within crosses between males infected with Wolbachia and uninfected females, thus exhibiting reduced fertility compared to the infected ones. Such “reproductive manipulation” induced by Wolbachia promotes the spread of the infection (2). The loss of fertility for uninfected females, while infected females reproduce well, confers an advantage for Wolbachia transmission, which is the cornerstone of CI evolution. Such loss of fertility does not result from reduced egg production but from a high rate of early embryonic mortality (3, 4). Cytological embryonic observations demonstrated in Culex, Drosophila, and Nasonia that CI induced by Wolbachia is precisely due to defects in paternal chromatin during first zygotic division, suggesting a chromatin modification by some Wolbachia factors (5–9). Such defects during the first embryonic division can be prevented if Wolbachia are present in the eggs. This cytological characterization of the hallmarks of CI has contributed to the formulation of the modification-rescue (mod-resc) model that could putatively be based on toxin-antidote interactions where a toxin (the mod factor) produced by the paternal Wolbachia and introduced in the sperm induces embryonic mortality unless an antidote (the resc factor) is produced by the maternal Wolbachia in the eggs (3).
Recent studies pointed out pairs of adjacent genes called CI factors (cif), within the genomes of CI-inducing Wolbachia, as major molecular actors of CI (10–15). Cif is their general name, while cid or cin are specific names based on their enzymatic domains (deubiquitinase [DUB] for cid and nuclease for cin [16, 17]). The heterologous expression of either cid or cin pairs (each composed of A/B genes) in Drosophila melanogaster males induces early death for a significant number of the embryos when crossed with uninfected females (11, 12, 14). However, the abortive embryo proportion due to CI, also called CI penetrance, varies depending on the cif transgenes. In similar expression conditions, the cid genes induced stronger CI than cin ones (12, 14, 15). Moreover, differences in CI penetrance between the different cid alleles introduced in D. melanogaster have been reported: the cidA/BwPip, which are secreted effectors encoded by the wPip genomes from Culex pipiens (10–12), induced full CI (i.e., null hatching rate [HR], with HR equal to the proportion of hatched eggs) while cidA/BwMel factors (from the wMel genome) only induced a significant decrease in HR (12, 15). Differences in CI penetrance were also reported between wMel and wPip, harboring different cifA and cifB genes, in the natural context of their native hosts. Indeed, in C. pipiens, all wPip strains induced full CI when infected males were crossed with uninfected females (18, 19) while wMel in Drosophila induced a partial HR reduction (12, 20). In C. pipiens, full CI occurs regardless of male age (21, 22), host genetic background (23, 24), or Wolbachia densities (9, 22). The cumulative presence of both functional cid and cin genes (17, 19, 25, 26) and the massive amplification-diversification of cid genes (9, 19, 27) provided putative genomic bases for this full CI induction. Indeed, unlike Wolbachia strains found in other host species where cid genes are monomorphic, each wPip strain encodes a “repertoire” of cid genes, with up to 6 different variants of cidA and cidB genes in a single Wolbachia genome (19, 27).
The strength of wPip-induced CI represents a force that certainly promoted the initial fixation and the maintenance of wPip in the C. pipiens complex (28). All C. pipiens individuals are currently infected with Wolbachia strains belonging to the monophyletic clade of wPip that is diversified into five groups, wPip-I to wPip-V (29). This diversity of wPip strains is responsible for the unparalleled diversity of CI patterns in the C. pipiens complex described as a binary “compatible/incompatible” phenomenon (30, 31). Indeed, hundreds of crosses between C. pipiens lines from different geographical origins all infected with wPip revealed the following two major outcomes based on their HR (21–23, 29, 31–34): (i) compatible crosses, with 80% ≤ mean HR ≤ 100%; in these cases, the number of unhatched eggs is similar to those of intraline crosses; or (ii) fully incompatible crosses, with null HR except for very few eggs (18, 21, 22, 34). In the latter situation, incompatibility can be either unidirectional (one cross direction is incompatible, while the reciprocal cross is compatible) or bidirectional (both cross directions are incompatible) (32, 35, 36). Reconstruction of wPip phylogeny revealed that mosquitoes infected with strains from the same group are more likely to be compatible with each other, while the compatibility between host-harboring wPip strains from different groups is mostly unpredictable (31). Moreover, specific variations in cidB repertoires harbored by males correlated with compatibility/incompatibility variations between C. pipiens lines, suggesting that some specific variants may play a strong role in this “yes-or-no” CI (19, 27). However, few cases were also reported with intermediate HR, i.e., 10% ≤ mean HR ≤ 80%, without knowing if those intermediate HR were linked to the Wolbachia strains involved or other factors such as nuclear incompatibilities (30, 37–43). Indeed, at the time of these intermediate HR observations, no diversity between wPip strains was discovered, and it was not possible to decipher the part of nuclear genetic background versus Wolbachia in the observed intermediate HR.
Our recent reconstruction of wPip phylogenetic groups (29, 31) and discovery of cid genes’ amplification and diversification led us to correlate cid and “yes-or-no” CI diversities in C. pipiens (19, 27). In the present study, we accurately monitored CI penetrance variations in the light of cid genes divergence by generating a C. pipiens compatibility matrix involving 11 lines harboring Wolbachia strains belonging to different wPip groups (wPip-I to wPip-IV) and all harboring different cid repertoires (9, 19). This compatibility matrix is composed of estimated HR obtained from (i) 11 intraline crosses (INTRA), (ii) 12 crosses between lines harboring wPip strains from the same group (INTER-INTRA), and (iii) 83 crosses between lines harboring wPip from different groups (INTER-INTER). We showed, as expected, that all INTRA and INTER-INTRA (except two) crosses were fully compatible. Among the INTER-INTER crosses, 54% were totally incompatible, displaying no hatching, and 22% were considered fully compatible, while 24% of the crosses exhibited mean HRs that can be qualified as intermediate. Backcross experiments demonstrated that such intermediate HRs were not linked to host genetic background but to the Wolbachia strains involved. Moreover, we showed that intermediate HR values were particularly low within crosses involving wPip-IV strains that also present marked phylogenetic difference in their cid repertoires from other wPip groups (19). To visualize the developmental defects responsible for intermediate HR, we monitored the embryonic development and found defects during the first zygotic division and subsequent developmental arrest, which are typical hallmarks of “canonical CI” (9, 14). Altogether, our data demonstrate that CI is not always a “yes-or-no” phenomenon in C. pipiens but that subtle CI variations, referred to as “cryptic CI,” putatively resulting from partial mismatch due to Cif protein divergence, exist in this species complex.
RESULTS
HR in fully compatible crosses.
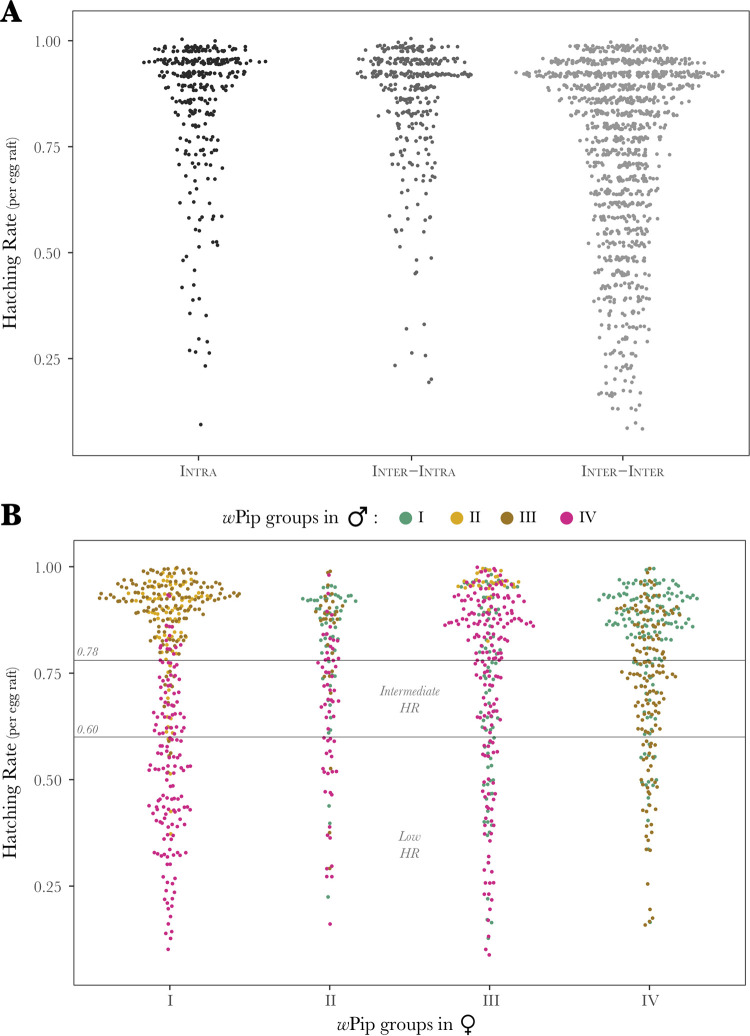
Mean HR of the 11 INTRA crosses were comprised between 0.78 and 0.95, showing that an important part of the eggs (up to 22%) failed to develop even in INTRA crosses. Intermediate HR can thus only refer to crosses with mean HR ≤78% (Fig. 1; Table S1 in the supplemental material; Data Set S1).
FIG 1.
Global hatching rate comparison between INTRA, INTER-INTRA, and INTER-INTER crosses. (A) Distribution of hatching rates (HR) per egg raft in the 11 INTRA crosses (330 egg rafts; 47,504 eggs), the 12 INTER-INTRA crosses (360 egg rafts; 49,461 eggs), and the 38 INTER-INTER crosses in which eggs produced larvae (1,140 egg rafts; 169,215 eggs). (B) Influence of wPip groups present in males and females on hatching rates of INTER-INTER crosses (38 different crosses; 1,140 egg rafts analyzed).
Full matrix of crosses performed in this study. The numbers indicated are mean HR obtained for each cross on 30 eggs rafts with standard deviation. Color scale is an indication of CI penetrance in each cross (green, no CI; red, full CI). Download Table S1, XLSX file, 0.02 MB (16.2KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw data for all INTRA crosses. For each egg raft, the total number of eggs, the total number of hatched larvae, and the calculated HR per egg raft are given. Download Data Set S1, XLSX file, 0.04 MB (42.7KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Depriving lines from Wolbachia did not influence INTRA HR.
To test for the effect of presence/absence of Wolbachia, two C. pipiens lines were tetracycline treated (SlabTC and IstanbulTC). For these “cured lines”, mean HRs were not significantly different from HRs of the corresponding INTRA crosses with infected lines (Wilcoxon W = 356, P = 0.168; and W = 344, P = 0.119 for Slab/SlabTC and Istanbul/IstanbulTC, respectively) (Table S1; Data Set S2).
Raw data for all crosses that involved tetracycline-treated lines. For each egg raft, the total number of eggs, the total number of hatched larvae, and the calculated HR per egg raft are given. Download Data Set S2, XLSX file, 0.02 MB (20.5KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
No influence of host genetic backgrounds on HR.
Crosses involving females harboring the same wPip strain in different genetic backgrounds (i.e. from backcrossed lines [Sl(wPip-I-Tunis) and Sl(wPip-IV-Harash)]) did not differ in their HRs when crossed with males from seven different lines (generalized linear models with mixed effects [GLMM]; χ2 = 2.857, degrees of freedom [df] = 1, P = 0.091). Crosses involving males harboring the same wPip strain in different genetic backgrounds showed similar HRs when crossed with females from five different lines (GLMM; χ2 = 0.414, df = 1, P = 0.520; χ2=0.0137, df= 1, P = 0.907; Table S1). Moreover, reciprocal crosses involving different C. pipiens species (i.e., Culex quinquefasciatus [Slab] versus C. pipiens [Istanbul]) without Wolbachia were not significantly different from corresponding intraspecies crosses (Wilcoxon W = 216, P = 0.764 and W = 185, P = 0.327, respectively; Data Set S3).
Raw data for all crosses that involved backcrossed lines. For each egg raft, the total number of eggs, the total number of hatched larvae, and the calculated HR per egg raft are given. Download Data Set S3, XLSX file, 0.04 MB (43.1KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
INTER-INTER crosses exhibit significantly reduced HR.
The full distribution of HR per egg raft for all the crosses is presented in Fig. 1A. The mean HR (i.e., calculated on 30 egg rafts per cross) of INTRA crosses ranged from 0.78 to 0.95; the mean HR of INTER-INTRA crosses (except for two fully incompatible crosses) ranged from 0.75 to 0.93, while the mean HR of INTER-INTER crosses displayed much more variability, ranging from 0 to 0.96. Fifty-four percent (45/83) of the INTER-INTER crosses were actually fully incompatible, while 46% (38/83) produced numerous larvae (mean HR between 0.48 and 0.96). HR distributions differed significantly among the different cross types (TYPE parameter in the statistical model) that led to larval production, as follows: (i) all the 11 INTRA (330 eggs rafts analyzed for a total of 47,504 eggs), (ii) all the 12 INTER-INTRA (360 egg rafts analyzed for a total of 49,461 eggs), and (iii) 38 out of the 83 INTER-INTER (1,140 egg rafts analyzed for a total of 169,215 eggs [Fig. 1A]). HR from INTER-INTER crosses were significantly lower than others (GLMM; χ2= 8.0371, df = 2, P = 0.018; Fig. 1A). Furthermore, the variance in HR per egg raft was significantly higher in INTER-INTER crosses (Levene’s test, P < 0.001), while it did not differ between INTRA and INTER-INTRA (Levene’s test, P = 0.65; Fig. 1A).
The INTER-INTER crosses category shows a higher occurrence of intermediate HR.
Among the 38 INTER-INTER crosses in which eggs hatched (Data Set S4), 20 crosses displayed a mean HR below 78%, referred to as intermediate HR, while only 1 cross out of 12 in the INTER-INTRA showed such intermediate values. INTER-INTER crosses showed significantly more intermediate HR crosses than other types (chi-square test; χ2 = 7.346, df = 1, P = 0.006; Table S1).
Raw data for all INTER-INTRA (between different lines that harbor wPip strains for the same wPip group) and INTER-INTER crosses (between lines that harbor wPip strains form different wPip groups). For each egg raft, the total number of eggs, the total number of hatched larvae and the calculated HR per egg raft are given. Download Data Set S4, XLSX file, 0.1 MB (98.3KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The lowest HRs were observed in INTER-INTER crosses involving wPip-IV strains.
For the 38 INTER-INTER crosses which were not fully incompatible, global models did not reveal any significant effect of the wPip group hosted by either female or male lines (GLMM; χ2 = 0.268, df = 3, P = 0.966; χ2 = 2.742, df = 3, P = 0.433, respectively) but pointed out a significant interaction effect between the wPip groups involved in the crosses (generalized linear models [GLM]; χ2 = 113.764, df = 13, P < 0.001; for detailed statistics, see Text S1). Careful inspection of the HR matrix revealed that 8 INTER-INTER crosses out of 38 showed a mean HR below 60%, here called low HR (Fig. 1B; Table S1). All these eight INTER-INTER crosses with low HR involved wPip-IV strains (see HR per egg raft full distribution in Fig. 1B; pink dots show HR obtained in crosses involving males infected with wPip-IV strains). INTER-INTER crosses with backcrossed line Sl(wPip-IV-Harash) did not differ from crosses involving Harash lines (GLM; χ2 = 0.0137, df = 1, P = 0.907), demonstrating that it was the wPip-IV strain harbored in the cytoplasm and not the host genetic background that explained such a low HR.
Explanations on GLM and GLMM models performed in this study. Download Text S1, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Intermediate HR results from cryptic but canonical CI.
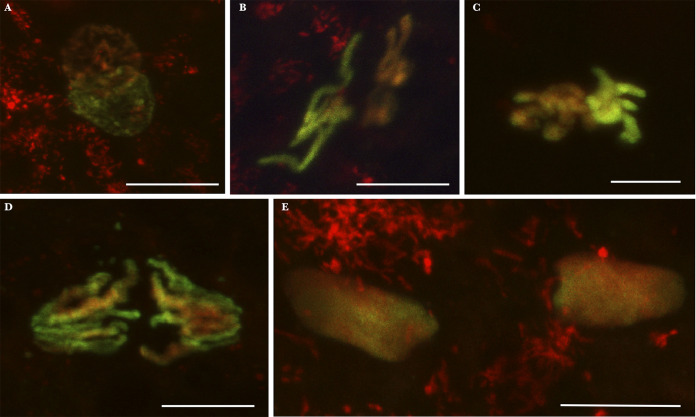
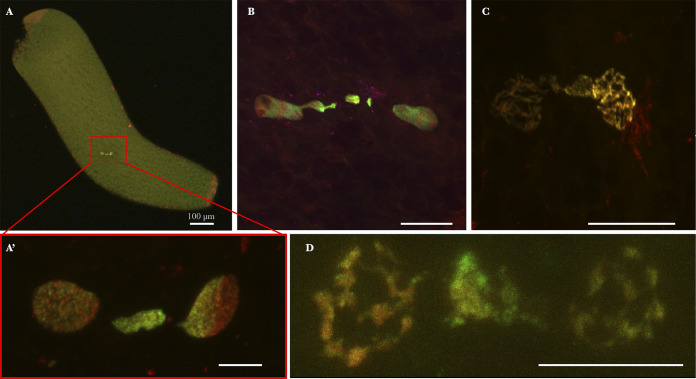
As low HRs (mean HR under 0.6) were only observed in INTER-INTER crosses involving wPip-IV strains, we (i) studied the first zygotic division resulting from these crosses, and (ii) in an attempt to quantify putative CI defects, compared them with INTER-INTRA and INTRA crosses at 5 h (Table 1 and Table S2). To verify whether intermediate HRs were due to previously described canonical CI cellular mechanisms (5–9), we visualized the first zygotic division with paternal and maternal chromatin labeled in green/yellow and red, respectively. In INTER-INTER crosses with intermediate HR, an important proportion of eggs normally hatched. Such normal embryogenesis, as documented in Fig. 2, is similar to what was observed for all INTRA embryos previously documented (9). After fertilization, maternal and paternal pronuclei migrated toward each other and apposed (documented embryos with confocal microscopy images, n = 3; Fig. 2A). Then, paternal and maternal chromatins condensed and entered into the first zygotic division (n = 1; Fig. 2B). During the first mitotic division, paternal and maternal chromosomes aligned in separate regions at the metaphase plate (n = 2, Fig. 2C). Both sets of chromosomes segregated equally during anaphase (n = 2; Fig. 2D) to produce two diploid nuclei (n = 1; Fig. 2E). Although our observations of first zygotic division events are not quantitative due to the technical challenge to monitor the different steps of this fast process, observations of embryos’ early development in INTER-INTER crosses with intermediate HR enabled us to document the presence of first zygotic division defects (n = 4; Fig. 3) that were previously observed in fully incompatible INTER-INTER crosses and absent in INTRA ones (9). As it was the green-labeled chromatin that exhibited such defects, it can be concluded that paternal chromatin is affected (Fig. 3A, A′, B, C, and D).
TABLE 1.
Proportion of embryos that did not reach normal blastoderm stage 5 h postoviposition in one INTRA, one INTER-INTRA, and one INTER-INTER cross
| Cross (male × female) | Cross type | No. of blastoderm-stage embryos | No. of embryos with abnormal development | No. of embryos with no sign of development | Total no. of embryos | % of embryos that did not reach blastoderm stage (5 h postoviposition) |
|---|---|---|---|---|---|---|
| Tunis × Tunis | INTRA | 94 | 0 | 5 | 99 | 5 |
| Ichkeul-13 × Harash | INTER-INTRA | 46 | 0 | 1 | 47 | 2 |
| Ichkeul-13 × SI(wPip-I-Tunis) | INTER-INTER | 36 | 5 | 4 | 45 | 20 |
| Ichkeul 13 × Tunis | INTER-INTER | 105 | 20 | 20 | 145 | 28 |
FIG 2.
Culex pipiens embryos from INTER-INTER crosses exhibiting normal first division. Paternal chromatin appears in green/yellow (acetylated histone H4 labeling is dominant), and maternal chromatin appears in red (propidium iodide labeling is dominant). These embryos have been collected and fixed 30 min to 1 h postoviposition. (A) Apposition of maternal and paternal pronuclei; (B) chromatin under condensation; (C) condensed chromatin; (D) first mitotic division anaphase; (E) two nuclei following the first division. Scale bar represents 10 μm.
FIG 3.
Culex pipiens embryos from INTER-INTER crosses exhibiting CI in first division. Paternal chromatin appears in green/yellow (acetylated histone H4 labeling is dominant), and maternal chromatin appears in red (propidium iodide labeling is dominant). These embryos have been collected and fixed in the first hour postoviposition. (A) Global view of a C. pipiens embryo undergoing a first mitotic division. (A′) Magnification of panel A showing paternal chromatin failed to segregate properly and form a chromatin spot between segregated nuclei. (B, C, and D) Other kinds of failed first divisions observed. Confocal stacks were obtained on embryos from several INTER-INTER crosses. Scale bar represents 10 μm.
Crosses performed to study cytological embryogenesis in intermediate situations. Bold letters represent crosses that were documented for developmental defects during the first zygotic division at 30 min to 1 h postoviposition; asterisk shows crosses that were compared for developmental defects at 5 h postoviposition. Download Table S2, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
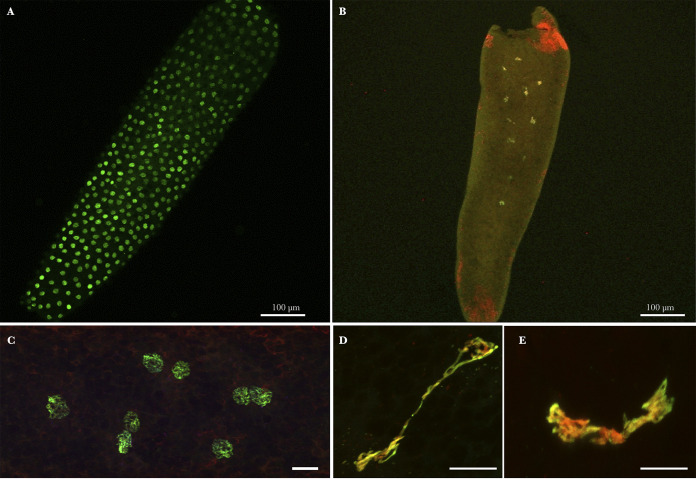
For only three crosses involving wPip-I and wPip-IV strains (one INTRA between Tunis wPip-I infected individuals; one INTER-INTRA between Ichkeul-13 wPip-IV males and Harash wPip-IV strain females; one INTER-INTER between Harash wPip-IV males and Tunis wPip-I females), we were able to produce enough observable embryos to assess the proportion of embryos with abnormal development 5 h postoviposition as presented in Table 1. At this time, embryos should have reached the syncytial blastoderm stage (∼3,200 “normal” nuclei; Fig. 4A and C), while embryos considered “abnormal” only presented few nuclei (less than 50; Fig. 4B). Moreover, atypical mitotic features were observed in these abnormal embryos (Fig. 4D and E). The proportion of abnormal embryos was less than 6% in INTRA and INTER-INTRA crosses while reaching at least 20% in the INTER-INTER cross with intermediate HR (Table 1; chi-square test; χ2 = 29.998, df = 3, P < 0.001).
FIG 4.
Culex pipiens embryos 5 h postoviposition in INTER-INTER crosses. Green/yellow (acetylated histone H4 labeling) and red (propidium iodide labeling). (A) Global view of a normal C. pipiens embryo having reached the expected syncytial stage. (B) Global view of an abnormal C. pipiens embryo exhibiting only few (less than 15) nuclei 5 h postoviposition. (C) Normal nuclei in a syncytial embryo. (D and E) Atypical mitotic features observed in abnormal embryos. Confocal stacks were obtained on embryos from several INTER-INTER crosses. Red dots (especially visible at the embryo’s poles in panel B) are propidium iodide-labeled Wolbachia in the embryo’s cytoplasm. Scale bar represents 10 μm.
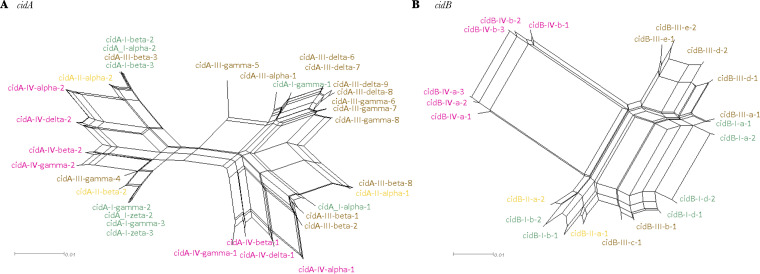
cid variants from wPip-IV repertoires are divergent from those of other wPip groups.
The phylogenetic cidA and cidB networks constructed with wPip strains repertoires showed that wPip strains from the wPip-IV group exhibited markedly divergent cidA and cidB variants. For both cidA and cidB variants, wPip-IV variants clustered remotely from other groups’ variants (Fig. 5; Tables S3 and S4). Two well-separated clusters of wPip-IV cidA variants appeared on the network, while all cidB variants clustered altogether (Fig. 5). For other wPip groups, no clear wPip-group-based clustering was observed (Fig. 5).
FIG 5.
Phylogenetic networks of the cidA and cidB genes. Networks obtained with 34 cidA (A) and 21 cidB (B) nucleotide variants present in the repertoires of the 11 strains from the four phylogenetic wPip groups studied here. The networks were obtained using the neighbor-net method. Each edge (or set of parallel edges) corresponds to a split in the data set and has a length equal to the weight of the split. Incompatible splits produced by recombination are represented by boxes in the network. wPip-I cid variants are in green, wPip-II cid variants are in yellow, wPip-III cid variants in brown, and wPip-IV cid variants are in pink.
cidA and cidB repertoires of the wPip strains studied here. Each wPip strain harbors a specific combination of different cidA and cidB variants in its genome. Each combination is called a repertoire. Download Table S3, DOCX file, 0.02 MB (21.6KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers for all cidA and cidB wPip variants sequenced to date. All these variants were included in phylogenetic networks analyses. Download Table S4, DOCX file, 0.02 MB (17.6KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In arthropods in which CI is mainly studied between infected males and uninfected females, including major insect models such as Drosophila and Nasonia, CI penetrance was proved to depend on Wolbachia strains, their densities, host genetic background, age of the males, and environmental factors such as temperature (44–59). On the contrary, in Culex pipiens s.l. mosquitoes, these factors did not affect CI penetrance (19, 22–24, 33): full CI (hatching rate [HR], 0) is reported between infected males and uninfected females (cured of Wolbachia with antibiotics) whatever their geographical origin, age, or genetic background (9, 18, 23, 33). However, hundreds of crosses between lines infected with Wolbachia revealed unparalleled variations in CI patterns in C. pipiens. Two main opposite outcomes were observed: either the crosses were compatible (mean HR ≥ 80%) or incompatible, producing almost no larvae (mean HR < 0.01%) (18, 21–23, 31, 32, 34, 40). Early in the study of CI in Culex, backcross experiments demonstrated that the host genome does not influence the outcome of a given cross (9, 24, 32, 60). In the present study, we conducted backcross experiments for two of our lines and also performed crosses between cured individuals from different C. pipiens species, which again confirmed that host genetic background does not impact compatibility.
Most crosses and backcrosses showed that CI in C. pipiens is a binary compatible/incompatible phenotype under the sole control of Wolbachia. However, in the numerous articles that presented results of interline C. pipiens crosses from different parts of the world, rare cases of intermediate HR were reported (30, 37–43). At the time of these publications, all the wPip were considered clonal due to monomorphic genetic markers available (34, 61). Intermediate HRs were thus attributed to putative undiscovered Wolbachia variability (including different wPip sublines in the same laboratory line) and most probably to putative host “restorer” nuclear factors counteracting Wolbachia CI induction (40, 41, 62). In the present paper, we investigated these intermediate HR situations in light of our present knowledge of wPip genomes (19, 27, 29, 31). To that extent, we studied 106 crosses between 11 C. pipiens isofemale lines infected with different wPip strains from different groups (I to IV), each exhibiting different cidA-cidB repertoires (9, 19) (Table S3 in the supplemental material). Different types of crosses were performed, including (i) INTRA crosses between mosquitoes from the same line, (ii) INTER-INTRA crosses between mosquitoes infected with different strains from the same wPip group, and (iii) INTER-INTER crosses between mosquitoes infected with different wPip groups.
For the 11 INTRA crosses performed in this study, mean HRs were all comprised between 78% and 95%, showing that a significant proportion of eggs never hatched even in fully compatible crosses. Previous cytological observations of C. pipiens early development in seven INTRA crosses with or without Wolbachia (i.e., after antibiotics treatment) did not detect any CI typical defects (9). Here, we reported no difference in HR in the same lines with or without Wolbachia, confirming that CI induced by Wolbachia is not responsible for the 5% to 22% of the eggs that did not reach the larval stage. Abortive eggs in INTRA crosses certainly resulted from imperfect fertilization and/or intrinsic mortality during development from eggs to larvae (9, 18, 40). The 12 INTER-INTRA crosses, involving lines from different locations but harboring the same wPip group, exhibited HRs similar to INTRA crosses, except for two cases of unidirectional incompatibility, again demonstrating that the wPip group is a major predictor of compatibility between C. pipiens lines (Fig. 1A; Table S1) (31).
Heterogeneity in compatibility clearly increased in INTER-INTER crosses (Table S1). Among the 83 performed here, we found that 54% of them were fully incompatible, while the other 46% (38/83) were fertile and exhibited HR comprised between 48% and 96%. Global HR statistical analyses, including all fertile crosses (11 INTRA, 12 INTER-INTRA, and 38 INTER-INTER crosses) showed that HR was significantly lower in INTER-INTER crosses and that variance in HRs among egg rafts was significantly higher in INTER-INTER than INTER-INTRA and INTRA crosses (Fig. 1A). Moreover, we found that 53% of the fertile INTER-INTER crosses actually exhibited HRs that were low enough to be characterized as intermediate. We also found that the interaction between the wPip groups infecting the male and female lines significantly influenced HR. Careful inspection of the HR matrix revealed that the crosses with a low HR below 60% (8 crosses out of the 20 with intermediate HR) were only observed in INTER-INTER crosses involving wPip-IV strains (Fig. 1B and Table S1). Atyame et al. (31) had already shown that wPip-IV group-infected C. pipiens lines exhibited markedly different crossing types from lines infected with other wPip groups. Network phylogenetic analyses of all the 34 cidA and 21 cidB different variants characterized in the wPip strains studied here revealed that cid-IV variants (especially cidB) were divergent, gathering in specific clusters, while other wPip groups are mixed altogether. This suggests that Cid proteins that are considered major effectors of CI (15, 17) are divergent in wPip-IV strains compared to other wPip groups (Fig. 5).
To investigate whether intermediate HR resulted from canonical CI, i.e., paternal chromatin defects during first zygotic division, we monitored the first stages of embryonic development in embryos from INTER-INTER crosses. In these crosses, even with low HR, many embryos exhibited normal development into larvae (Fig. 2). However, in a few embryos, we were able to document imperfect paternal chromatin segregation during the first zygotic division (Fig. 3). These embryonic defects, which were never observed in INTRA crosses (9), were similar to those reported in fully incompatible crosses (9). Such defects in the first zygotic division likely produced aneuploid nuclei which might disrupt further development or even arrest the embryogenesis. The proportion of embryos that did not reach blastoderm stage 5 h postoviposition, but presented instead few nuclei only, can be considered a quantitative proxy for the occurrence of CI defects during the first division. We observed a larger amount of abnormal developmental stages, 5 h postoviposition, in INTER-INTER crosses than the INTRA and INTER-INTRA crosses (Table 1). Abnormal embryos, which represented 20% of the embryos in the INTER-INTER crosses studied and 5% in the INTRA one (Table 1), displayed very few (or no) nuclei (Fig. 4B). These observations suggest that embryonic defects during the first division are responsible for the intermediate HR observed in the analyzed INTER-INTER cross (Table 1). The intermediate HR observed in INTER-INTER crosses could be attributed to cryptic CI (in that it has a weak penetrance) but canonical CI (in that it translates into the same cytological defects).
In the light of the toxin-antidote model of CI, penetrance would depend on the interaction between CidA, CidB, and their specific substrates, eventually leading to paternal chromatin defects or its rescue (15, 16, 63). In C. pipiens, as all wPip genomes encode a repertoire of several polymorphic variants of CidA and CidB (19, 27), full compatibility could result from multiple interactions between different CidA and CidB variants even in INTRA or INTER-INTRA crosses. In every C. pipiens male, several CidB proteins differing in their amino acid sequences might be introduced in the sperm and then in the egg during fertilization where several CidA proteins might also be present. Full compatibilities reported here in some INTER-INTER crosses involving different wPip groups with totally different CidA/CidB repertoires (Fig. 1) suggest (i) that strict specific interactions between cognate variants are not required for full compatibility, and (ii) a potential redundancy in the interaction between CidA/CidB variants. The intermediate HR resulting from cryptic CI in a given INTER-INTER cross can hypothetically result from partial rescue due to imperfect interactions between the CidA and the CidB from the two wPip strains repertoires. Since most of the embryos from intermediate HR crosses developed into living larvae, it certainly means that, in those individuals, CidB toxicity has been efficiently counteracted. On the contrary, in embryos exhibiting CI, CidB toxicity would not have been counteracted properly. This heterogeneity could be explained if embryo rescue depends on one or a few matching CidA variants which might be required in a larger quantity for the rescue to occur. However, it is possible that in certain eggs, the expression of the(se) CidA variant(s) would be too low to counteract the CidB toxicity. This would be especially true for neutralizing CidB proteins encoded by wPip-IV strains that show striking differences in their sequences from other wPip groups (Fig. 5). Less efficacy in the interactions between CidB-IV proteins and CidA from other groups could explain their higher probability to be involved in both (i) full incompatibility as reported in reference 31, and (ii) cryptic CI as reported here.
The interactions between the CidA and CidB repertoires encoded by wPip strains determine the developmental fate of each embryo of a given cross, normal development versus CI. CI penetrance (i.e., the proportion of embryos undergoing CI) in a given cross could then be determined by the diversity of cidA/cidB genes of the different wPip genomes hosted by the different C. pipiens lines, their expression levels, and the affinity between the resulting proteins.
MATERIALS AND METHODS
Culex pipiens lines.
Eleven isofemale lines were used (Table S5 in the supplemental material). They differed in (i) their geographical origins, (ii) the species they belong to, (iii) the wPip group (I, II, III, or IV), and (iv) their cid repertoires. The Wolbachia group was checked by performing a pk1 PCR-restriction fragment length polymorphism (RFLP) test (64) on DNA extracted using cetyltrimethylammonium bromide (CTAB) protocol (65). Tetracycline-treated Wolbachia-free lines (TC lines), named SlabTC and IstanbulTC, were obtained from Slab- and Istanbul-infected lines as described in reference 33. The absence of Wolbachia was checked by PCR on a fragment of the wsp gene using the primers designed in reference 66. TC-treated lines were raised at least four generations without tetracycline before experiments. The wPip-I strain from the Tunis line and wPip-IV strain from the Harash line were independently introgressed into the Slab line nuclear genetic background through 8 backcrosses as described in reference 9.
Information on the Culex lines and the Wolbachia strains studied. Download Table S5, DOCX file, 0.02 MB (26.1KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hatching rate estimations.
To test for a putative effect of Wolbachia on basic intraline HR, we performed two intraline crosses between males and females from cured lines (SlabTC and IstanbulTC) and compared them with infected intraline crosses (Slab and Istanbul, respectively). To test for a putative impact of the different wPip groups on HR, we carried out 106 different crosses between males and females, including (i) from the same line (11 intraline crosses, called INTRA crosses), (ii) from two distinct mosquito lines infected with wPip strains from the same group (12 interline-intragroup crosses, called INTER-INTRA crosses), and (iii) from two distinct mosquito lines infected with wPip strains from different groups (83 interline-intergroup crosses, called INTER-INTER crosses).
To test for a potential impact of host genetic background on HR, we performed 23 extra crosses involving the two backcrossed lines [Sl(wPip-I-Tunis) or Sl(wPip-IV-Harash)]. Moreover, to study the putative effect of interspecies crosses (i.e., C. pipiens versus C. quinquefasciatus) on HR, we performed the reciprocal crosses between SlabTC (C. quinquefasciatus) and IstanbulTC (C. pipiens) lines. To perform each of these 135 different crosses, 2-day-old males (n = 50) and females (n = 100) were put together in cages. After 6 days, females were blood fed with turkey blood using a Hemotek feeding system (Discovery Workshops). After 5 days, egg rafts were collected. After the death of all the larvae (i.e., about 5 days after hatching), pictures of both eggs and larvae for 30 egg rafts per cross were taken. Eggs and larvae were counted manually on ImageJ (67). HR was calculated per egg raft as the ratio between the total number of larvae and the total number of eggs.
Cellular study of embryogenesis.
To search for putative embryonic defects that might confirm the involvement of canonical CI in INTER-INTER crosses resulting in intermediate HR, several crosses involving males from different lines infected with wPip-IV strains were performed (Table S2). To that extent, cages containing 50 males and 100 females were put into a closet where the day-night cycle was inverted to allow collection of eggs during the day. After 6 days in these cages, females were fed with turkey blood, and waterpots were placed into the cages for 30 min to 1 h to allow females to lay egg rafts. For C. pipiens embryos, at 25°C, the meiosis is approximately completed 30 min postoviposition and the first mitotic nucleus division 15 min after, while 5 h after oviposition, the embryos reach the blastoderm stage (9). Freshly collected eggs (30 min to 5 h) were fixed, dechorionated, and observed as previously described in reference 9.
Statistical analysis.
We used generalized linear models (GLM) or generalized linear models with mixed effects (GLMM) with a logit link function (see Text S1). To test for potential impact of Wolbachia presence/absence and host species, Wilcoxon tests were performed (68). To compare the proportion of (i) intermediate HR between different types (INTRA, INTER-INTRA, and INTER-INTER), and (ii) abnormal embryos between INTRA, INTER-INTRA, and INTER-INTER crosses, χ2 tests were performed. The differences in variance among INTRA, INTER-INTRA, and INTER-INTER crosses were analyzed using Levene’s test (69). All computations were performed using R version 3.4.4 (70).
Phylogenetic networks of the cidA and cidB genes.
All the cidA and cidB repertoires of the Wolbachia strains hosted by the 11 crossed lines were already published except for Brazil that has been obtained by PCR cloning followed by Sanger sequencing as previously described in references 9 and 19. Sequenced variants (accession numbers given in Table S4) were aligned using the Muscle algorithm implemented in SeaView 6.4.1 software (71) and then analyzed within a phylogenetic network framework from uncorrected P distances by the neighbor-net method implemented in SplitsTree4 (72) to account for potentially conflicting signals due to recombination.
ACKNOWLEDGMENTS
This work was funded by the French ANR (project CIAWOL, ANR-16-CE02-0006-01). Sequencing data were generated on the GenSeq platform. Confocal microscopy was performed in the MRI-CRBM platform. Embryos optical observations were performed at the CytoEvol facilities.
Footnotes
Citation Sicard M, Namias A, Perriat-Sanguinet M, Carron E, Unal S, Altinli M, Landmann F, Weill M. 2021. Cytoplasmic incompatibility variations in relation with Wolbachia cid genes divergence in Culex pipiens. mBio 12:e02797-20. https://doi.org/10.1128/mBio.02797-20.
REFERENCES
- 1.Yen JH, Barr AR. 1971. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 232:657–658. doi: 10.1038/232657a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann AA, Turelli M, Harshman LG. 1990. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics 126:933–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst GDD, Frost CL. 2015. Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harb Perspect Biol 7:a017699. doi: 10.1101/cshperspect.a017699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landmann F 2019. The Wolbachia endosymbionts. Microbiol Spectr 7. doi: 10.1128/microbiolspec.BAI-0018-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaini G, Riparbelli MG, Giordano R, Dallai R. 1996. Mitotic defects associated with cytoplasmic incompatibility in Drosophila simulans. J Invertebr Pathol 67:55–64. doi: 10.1006/jipa.1996.0009. [DOI] [Google Scholar]
- 6.Lassy CW, Karr TL. 1996. Cytological analysis of fertilization and early embryonic development in incompatible crosses of Drosophila simulans. Mech Dev 57:47–58. doi: 10.1016/0925-4773(96)00527-8. [DOI] [PubMed] [Google Scholar]
- 7.Tram U, Sullivan W. 2002. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296:1124–1126. doi: 10.1126/science.1070536. [DOI] [PubMed] [Google Scholar]
- 8.Landmann F, Orsi GA, Loppin B, Sullivan W. 2009. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog 5:e1000343. doi: 10.1371/journal.ppat.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonneau M, Landmann F, Labbé P, Justy F, Weill M, Sicard M. 2018. The cellular phenotype of cytoplasmic incompatibility in Culex pipiens in the light of cidB diversity. PLoS Pathog 14:e1007364. doi: 10.1371/journal.ppat.1007364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckmann JF, Fallon AM. 2013. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem Mol Biol 43:867–878. doi: 10.1016/j.ibmb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann JF, Ronau JA, Hochstrasser M. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol 2:1–7. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LePage DP, Metcalf JA, Bordenstein SR, On J, Perlmutter JI, Shropshire JD, Layton EM, Funkhouser-Jones LJ, Beckmann JF, Bordenstein SR. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shropshire JD, On J, Layton EM, Zhou H, Bordenstein SR. 2018. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc Natl Acad Sci U S A 115:4987–4991. doi: 10.1073/pnas.1800650115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Ronau JA, Beckmann JF, Hochstrasser M. 2019. A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc Natl Acad Sci U S A 116:22314–22321. doi: 10.1073/pnas.1914571116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckmann JF, Sharma GD, Mendez L, Chen H, Hochstrasser M. 2019. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. Elife 8:e50026. doi: 10.7554/eLife.50026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beckmann JF, Bonneau M, Chen H, Hochstrasser M, Poinsot D, Merçot H, Weill M, Sicard M, Charlat S. 2019. The toxin–antidote model of cytoplasmic incompatibility: genetics and evolutionary implications. Trends Genet 35:175–185. doi: 10.1016/j.tig.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Zhang M, Hochstrasser M. 2020. The biochemistry of cytoplasmic incompatibility caused by endosymbiotic bacteria. Genes (Basel) 11:852. doi: 10.3390/genes11080852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duron O, Weill M. 2006. Wolbachia infection influences the development of Culex pipiens embryo in incompatible crosses. Heredity (Edinb) 96:493–500. doi: 10.1038/sj.hdy.6800831. [DOI] [PubMed] [Google Scholar]
- 19.Bonneau M, Atyame CM, Beji M, Justy F, Cohen-Gonsaud M, Sicard M, Weill M. 2018. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat Commun 9:319. doi: 10.1038/s41467-017-02749-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shropshire JD, Bordenstein SR. 2019. Two-by-one model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet 15:e1008221. doi: 10.1371/journal.pgen.1008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasgon JL, Scott TW. 2003. Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics 165:2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duron O, Fort P, Weill M. 2007. Influence of aging on cytoplasmic incompatibility, sperm modification and Wolbachia density in Culex pipiens mosquitoes. Heredity (Edinb) 98:368–374. doi: 10.1038/sj.hdy.6800948. [DOI] [PubMed] [Google Scholar]
- 23.Atyame CM, Duron O, Tortosa P, Pasteur N, Fort P, Weill M. 2011. Multiple Wolbachia determinants control the evolution of cytoplasmic incompatibilities in Culex pipiens mosquito populations. Mol Ecol 20:286–298. doi: 10.1111/j.1365-294X.2010.04937.x. [DOI] [PubMed] [Google Scholar]
- 24.Duron O, Bernard J, Atyame CM, Dumas E, Weill M. 2012. Rapid evolution of Wolbachia incompatibility types. Proc Biol Sci 279:4473–4480. doi: 10.1098/rspb.2012.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsey ARI, Rice DW, Bordenstein SR, Brooks AW, Bordenstein SR, Newton ILG. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol 10:434–451. doi: 10.1093/gbe/evy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez J, Klasson L, Welch JJ, Jiggins FM. 2020. Life and death of selfish genes: comparative genomics reveals the dynamic evolution of cytoplasmic incompatibility. Mol Biol Evol doi: 10.1093/molbev/msaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonneau M, Caputo B, Ligier A, Caparros R, Unal S, Perriat-Sanguinet M, Arnoldi D, Sicard M, Weill M. 2019. Variation in Wolbachia cidB gene, but not cidA, is associated with cytoplasmic incompatibility mod phenotype diversity in Culex pipiens. Mol Ecol 28:4725–4736. doi: 10.1111/mec.15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumas E, Atyame CM, Milesi P, Fonseca DM, Shaikevich EV, Unal S, Makoundou P, Weill M, Duron O. 2013. Population structure of Wolbachia and cytoplasmic introgression in a complex of mosquito species. BMC Evol Biol 13:181. doi: 10.1186/1471-2148-13-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atyame CM, Delsuc F, Pasteur N, Weill M, Duron O. 2011. Diversification of Wolbachia endosymbiont in the Culex pipiens mosquito. Mol Biol Evol 28:2761–2772. doi: 10.1093/molbev/msr083. [DOI] [PubMed] [Google Scholar]
- 30.Laven H 1951. Crossing experiments with Culex strains. Evolution (N Y) 5:370–375. doi: 10.2307/2405682. [DOI] [Google Scholar]
- 31.Atyame CM, Labbé P, Dumas E, Milesi P, Charlat S, Fort P, Weill M. 2014. Wolbachia divergence and the evolution of cytoplasmic incompatibility in Culex pipiens. PLoS One 9:e87336. doi: 10.1371/journal.pone.0087336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laven H 1967. Speciation and evolution in Culex pipiens, p 251–275. In Wright J, Pal R (ed), Genetics of insect vectors of disease. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 33.Duron O, Bernard C, Unal S, Berthomieu A, Berticat C, Weill M. 2006. Tracking factors modulating cytoplasmic incompatibilities in the mosquito Culex pipiens. Mol Ecol 15:3061–3071. doi: 10.1111/j.1365-294X.2006.02996.x. [DOI] [PubMed] [Google Scholar]
- 34.Guillemaud T, Pasteur N, Rousset F. 1997. Contrasting levels of variability between cytoplasmic genomes and incompatibility types in the mosquito Culex pipiens. Proc Biol Sci 264:245–251. doi: 10.1098/rspb.1997.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neill SL, Karr TL. 1990. Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature 348:178–180. doi: 10.1038/348178a0. [DOI] [PubMed] [Google Scholar]
- 36.Breeuwer JAJ, Werren JH. 1990. Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature 346:558–560. doi: 10.1038/346558a0. [DOI] [PubMed] [Google Scholar]
- 37.Sasa M, Shirasaka A, Kurihara T. 1966. Crossing experiments between fatigans, pallens and molestus colonies of the mosquito Culex pipiens s. l. from Japan and Southern Asia, with special reference to hatchability of hybrid eggs. Appl Exp Med 36:187–210. [PubMed] [Google Scholar]
- 38.French WL 1978. Genetic and phenogenetic studies on the dynamic nature of the cytoplasmic inheritance system in Culex pipiens. Genetics 88:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irving-Bell RJ 1983. Cytoplasmic incompatibility within and between Culex molestus and Cx. quinquefasciatus (Diptera: Culicidae). J Med Entomol 20:44–48. doi: 10.1093/jmedent/20.1.44. [DOI] [Google Scholar]
- 40.Magnin M, Pasteur N, Raymond M. 1987. Multiple incompatibilities within populations of Culex pipiens L. in southern France. Genetica 74:125–130. doi: 10.1007/BF00055223. [DOI] [PubMed] [Google Scholar]
- 41.O'Neill SL, Paterson HE. 1992. Crossing type variability associated with cytoplasmic incompatibility in Australian populations of the mosquito Culex quinquefasciatus Say. Med Vet Entomol 6:209–216. doi: 10.1111/j.1365-2915.1992.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 42.Barr AR 1966. Cytoplasmic incompatibility as a means of eradication of Culex pipiens L. Proc Calif Mosq Cont Assoc 34:32–35. [PubMed] [Google Scholar]
- 43.Magnin M, Pasteur N. 1987. Phénomènes d’incompatibilité cytoplasmique chez le moustique Culex pipiens L. (Diptera: Culicidae) du sud de la France. Effet de la tétracycline. Cah ORSTOM 25:21–25. [Google Scholar]
- 44.Merçot H, Charlat S. 2004. Wolbachia infections in Drosophila melanogaster and D. simulans: polymorphism and levels of cytoplasmic incompatibility. Genetica 120:51–59. doi: 10.1023/B:GENE.0000017629.31383.8f. [DOI] [PubMed] [Google Scholar]
- 45.Bordenstein SR, Werren JH. 2007. Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia. Heredity (Edinb) 99:278–287. doi: 10.1038/sj.hdy.6800994. [DOI] [PubMed] [Google Scholar]
- 46.Kittayapong P, Mongkalangoon P, Baimai V, O'Neill SL. 2002. Host age effect and expression of cytoplasmic incompatibility in field populations of Wolbachia-superinfected Aedes albopictus. Heredity (Edinb) 88:270–274. doi: 10.1038/sj.hdy.6800039. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds KT, Hoffmann AA. 2002. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res 80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- 48.McGraw EA, Merritt DJ, Droller JN, O'Neill SL. 2001. Wolbachia-mediated sperm modification is dependent on the host genotype in Drosophila. Proc Biol Sci 268:2565–2570. doi: 10.1098/rspb.2001.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann AA, Turelli M, Simmons GM. 1986. Unidirectional incompatibility between populations of Drosophila simulans. Evolution (N Y) 40:692–701. doi: 10.2307/2408456. [DOI] [PubMed] [Google Scholar]
- 50.Sicard M, Bouchon D, Ceyrac L, Raimond R, Thierry M, Le Clec'h W, Marcadé I, Caubet Y, Grève P. 2014. Bidirectional cytoplasmic incompatibility caused by Wolbachia in the terrestrial isopod Porcellio dilatatus. J Invertebr Pathol 121:28–36. doi: 10.1016/j.jip.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 51.Sinkins SP, Braig HR, O'Neill SL. 1995. Wolbachia pipientis: bacterial density and unidirectional cytoplasmic incompatibility between infected populations of Aedes albopictus. Exp Parasitol 81:284–291. doi: 10.1006/expr.1995.1119. [DOI] [PubMed] [Google Scholar]
- 52.Bordenstein SR, Werren JH. 1998. Effects of A and B Wolbachia and host genotype on interspecies cytoplasmic incompatibility in Nasonia. Genetics 148:1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rousset F, Solignac M. 1995. Evolution of single and double Wolbachia symbioses during speciation in the Drosophila simulans complex. Proc Natl Acad Sci U S A 92:6389–6393. doi: 10.1073/pnas.92.14.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vavre F, Dedeine F, Quillon M, Fouillet P, Fleury F, Boulétreau M. 2001. Within-species diversity of Wolbachia-induced cytoplasmic incompatibility in haplodiploid insects. Evolution (N Y) 55:1710–1714. doi: 10.1554/0014-3820(2001)055[1710:WSDOWI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 55.Boyle L, O'Neill SL, Robertson HM, Karr TL. 1993. Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science 260:1796–1799. doi: 10.1126/science.8511587. [DOI] [PubMed] [Google Scholar]
- 56.Clancy DJ, Hoffmann AA. 1998. Environmental effects on cytoplasmic incompatibility and bacterial load in Wolbachia-infected Drosophila simulans. Entomol Exp Appl 86:13–24. doi: 10.1046/j.1570-7458.1998.00261.x. [DOI] [Google Scholar]
- 57.Noda H, Koizumi Y, Zhang Q, Deng K. 2001. Infection density of Wolbachia and incompatibility lever in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem Mol Biol 31:727–737. doi: 10.1016/S0965-1748(00)00180-6. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda T, Ishikawa H, Sasaki T. 2003. Infection density of Wolbachia and level of cytoplasmic incompatibility in the Mediterranean flour moth, Ephestia kuehniella. J Invertebr Pathol 84:1–5. doi: 10.1016/s0022-2011(03)00106-x. [DOI] [PubMed] [Google Scholar]
- 59.Jamnongluk W, Kittayapong P, Baisley KJ, O'Neill SL. 2000. Wolbachia infection and expression of cytoplasmic incompatibility in Armigeres subalbatus (Diptera: Culicidae). J Med Entomol 37:53–57. doi: 10.1603/0022-2585-37.1.53. [DOI] [PubMed] [Google Scholar]
- 60.Ghelelovitch S 1952. Sur le déterminisme génétique de la stérilité dans les croisements entre différentes souches de Culex autogenicus Roubaud. C R Hebd Seances Acad Sci 234:2386–2388. [PubMed] [Google Scholar]
- 61.Ruang-Areerate T, Kittayapong P, Baimai V, O'Neill SL. 2003. Molecular phylogeny of Wolbachia endosymbionts in Southeast Asian mosquitoes (Diptera: Culicidae) based on wsp gene sequences. J Med Entomol 40:1–5. doi: 10.1603/0022-2585-40.1.1. [DOI] [PubMed] [Google Scholar]
- 62.Rousset F, Raymond M, Kjellberg F. 1991. Cytoplasmic incompatibilities in the mosquito Culex pipiens: how to explain a cytotype polymorphism? J Evolution Biol 4:69–81. doi: 10.1046/j.1420-9101.1991.4010069.x. [DOI] [Google Scholar]
- 63.Beckmann JF, Bonneau M, Chen H, Hochstrasser M, Poinsot D, Merçot H, Weill M, Sicard M, Charlat S. 2019. Caution does not preclude predictive and testable models of cytoplasmic incompatibility: a reply to Shropshire et al. Trends Genet 35:399–400. doi: 10.1016/j.tig.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altinli M, Gunay F, Alten B, Weill M, Sicard M. 2018. Wolbachia diversity and cytoplasmic incompatibility patterns in Culex pipiens populations in Turkey. Parasit Vectors 11:198. doi: 10.1186/s13071-018-2777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers SO, Bendich AJ. 1994. Extraction of total cellular DNA from plants, algae and fungi, p 183–190. In Gelvin SB, Schilperoort RA (ed), Plant molecular biology manual. Springer, Dordrecht, Netherlands. [Google Scholar]
- 66.Berticat C, Rousset F, Raymond M, Berthomieu A, Weill M. 2002. High Wolbachia density in insecticide-resistant mosquitoes. Proc Biol Sci 269:1413–1416. doi: 10.1098/rspb.2002.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauer DF 1972. Constructing confidence sets using rank statistics. J Am Stat Assoc 67:687–690. doi: 10.1080/01621459.1972.10481279. [DOI] [Google Scholar]
- 69.Levene H 1960. Robust tests for equality of variances, p 278–292. In Olkin I (ed), Contributions to probability and statistics essays in honor of Harold Hotelling. Stanford University Press, Palo Alto, CA. [Google Scholar]
- 70.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 71.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 72.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full matrix of crosses performed in this study. The numbers indicated are mean HR obtained for each cross on 30 eggs rafts with standard deviation. Color scale is an indication of CI penetrance in each cross (green, no CI; red, full CI). Download Table S1, XLSX file, 0.02 MB (16.2KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw data for all INTRA crosses. For each egg raft, the total number of eggs, the total number of hatched larvae, and the calculated HR per egg raft are given. Download Data Set S1, XLSX file, 0.04 MB (42.7KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw data for all crosses that involved tetracycline-treated lines. For each egg raft, the total number of eggs, the total number of hatched larvae, and the calculated HR per egg raft are given. Download Data Set S2, XLSX file, 0.02 MB (20.5KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw data for all crosses that involved backcrossed lines. For each egg raft, the total number of eggs, the total number of hatched larvae, and the calculated HR per egg raft are given. Download Data Set S3, XLSX file, 0.04 MB (43.1KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw data for all INTER-INTRA (between different lines that harbor wPip strains for the same wPip group) and INTER-INTER crosses (between lines that harbor wPip strains form different wPip groups). For each egg raft, the total number of eggs, the total number of hatched larvae and the calculated HR per egg raft are given. Download Data Set S4, XLSX file, 0.1 MB (98.3KB, xlsx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Explanations on GLM and GLMM models performed in this study. Download Text S1, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Crosses performed to study cytological embryogenesis in intermediate situations. Bold letters represent crosses that were documented for developmental defects during the first zygotic division at 30 min to 1 h postoviposition; asterisk shows crosses that were compared for developmental defects at 5 h postoviposition. Download Table S2, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
cidA and cidB repertoires of the wPip strains studied here. Each wPip strain harbors a specific combination of different cidA and cidB variants in its genome. Each combination is called a repertoire. Download Table S3, DOCX file, 0.02 MB (21.6KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers for all cidA and cidB wPip variants sequenced to date. All these variants were included in phylogenetic networks analyses. Download Table S4, DOCX file, 0.02 MB (17.6KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Information on the Culex lines and the Wolbachia strains studied. Download Table S5, DOCX file, 0.02 MB (26.1KB, docx) .
Copyright © 2021 Sicard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.