Abstract
Chlorpyrifos (CPF) is an organophosphate insecticide commonly used to treat fruit and vegetable crops. CPF can cause severe adverse effects on body organs including the liver and central nervous system. This study investigated the CPF-induced inflammation in mice and explored the role of intestinal flora changes in liver inflammation. Adult C57BL/6 male mice were exposed to a CPF of 0.01-, 0.1-, 1- and 10-mg/kg bodyweight for 12 weeks. The mice in experimental group given CPF solution dissolved in corn oil vehicle by gavage, was administered by intraoral gavage for 5 days per week for 12 weeks. Histopathological examination and inflammatory factor detection were performed on mice liver tissue. Faeces were used for 16S ribosomal RNA high-throughput sequencing to explore the impact of CPF on intestinal flora structure and diversity. The results showed that 1- and 10-mg/kg CPF caused different degrees of liver focal inflammation. The structure of intestinal flora changed significantly in mice including the decreased beneficial bacteria (Akkermansia, Prevotella and Butyricimonas) and increased pathogenic bacteria (Helicobacter and Desulfovibrio). Meanwhile, the results of Q-RT-PCR showed that there was more total bacterial DNA in the liver tissue of the mice treated with 10-mg/kg groups. In conclusion, the imbalance of intestinal flora, the decreased abundance of beneficial bacteria and the increased abundance of pathogenic bacteria, as well as the increase of total bacterial DNA in the liver tissues, maybe associated with the liver focal inflammation induced by CPF.
Keywords: chlorpyrifos, liver inflammation, intestinal flora, 16S rRNA
Introduction
The widespread use of pesticides and improper release not only increases the yield of crops but also brings a series of environmental health problems. Organophosphorus pesticides (OPs) are the most widely used insecticides in agricultural activities because of their low environmental persistence and high efficiency [1]. Since most OPs are highly lipophilic, they can easily enter through the animal’s skin and accumulate in their body and milk [2]. In addition, OPs residues were found in fruits, vegetables, grains and even groundwater [3]. OPs poisoning is a global health problem resulting in ~100 000 deaths every year in Asian countries alone [4]. Chlorpyrifos (CPF) is a broad-spectrum organophosphorus insecticide widely used for pest control [5, 6]. The main ways of human exposure to CPF (from the soil, air, water and food) are ingestion, inhalation and dermal contact [7]. China’s “National food safety standard-Maximum residue limits for pesticides in food” set the standard for the maximum residues limits of CPF, which is 0.1 mg/kg in cabbage and soybean, 0.5 mg/kg in rice and wheat and 1 mg/kg in beans and cauliflower. The acceptable daily intake (ADI) for CPF was set at 0.01 mg/kg [8]. Excessive use and residues of CPF are associated with high morbidity and mortality of related diseases caused by acute and chronic exposure in humans [9]. CPF elicits several toxic effects including reproductive toxicity [10], neurotoxicity [11], cardiotoxicity [12] and hepatic dysfunction [13]. CPF is metabolized by cytochrome P450 in hepatocytes and converted to CPF-oxon, which is the main toxic metabolite of CPF [14]. Liver is the organ where the activation and detoxification of CPF takes place [15].
In a normal physiological state, the human intestinal microbiota consists of ~100 trillion microorganisms living in the gastrointestinal tract [16]. Intestinal flora and human body have a symbiotic relationship and maintain a dynamic balance in a certain proportion. Once this balance is broken, the occurrence of bacterial imbalance may lead to pathological changes [17] and the occurrence of many diseases such as inflammatory bowel disease [18], Parkinson’s disease [19] and non-alcoholic fatty liver [20]. Studies have shown that overgrowth of bacterial in the small intestine is closely related to the severity of alcoholic cirrhosis [21]. The liver and intestine originate from the same germ layer and have many important anatomical and functional connections [22]. When the intestine barrier is compromised, intestinal microorganisms can translocate to the liver through the portal system, causing inflammation and hepatic injury [23]. Some translocated intestinal products might also directly interact with host factors and contribute to exacerbation of liver disease [24]. The crosstalk between the gut and liver is increasingly recognized. CPF was reported to cause hepatic lipid metabolism disorders that are associated with gut oxidative stress and microbiota dysbiosis in adult zebrafish [25]. In another study, mice exposed to CPF (1 mg/kg) for 30 day can perturb the gut microbiota composition and urine metabolites [26]. In addition, exposure to CPF during gestation had the potential to affect maturation in the pups’ intestinal development, intestinal microbial dysbiosis and influence the development of immune system [27]. Therefore, we hypothesized that CPF could induce the alteration of intestinal flora which in turn contribute on CPF-induced liver inflammation.
In this study, we exposed C57BL/6 male mice to CPF and investigated structure of intestinal flora the inflammatory changes in liver and analyzed the association between structures of intestinal flora the inflammatory changes in liver of mice exposed to CPF. This study explored the role of intestinal flora in CPF-induced liver focal inflammation from a new perspective, which has important significance for reducing the health risks of human exposure to pesticide.
Materials and Methods
Test materials and animals
Chlorpyrifos (97.4% pure) obtained from ShengBang Green Chemical, Shandong, and corn oil is commercially available, was used for experimentation.
Wild-type male mice (C57BL/6, 8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). During the whole experiment, mice were housed in an experimental environment with a temperature of 22 ± 2°C, relative humidity of 50 ± 5% and artificial lighting in a 12-h/12-h light/dark cycle, and the water (Reverse Osmosis pure water) and basic diet (60Co irradiation sterilization feed, Keao Xieli Feed Co., Ltd., Beijing, China) were always available. After 7 days of acclimation, mice were divided randomly into five groups: four exposure groups and a control group. According to ADI of CPF and dose conversion between animals and human [28], the mice were exposed to CPF dissolved in corn oil by oral gavage at dosages of 0.01 mg/kg (CPF1), 0.1 mg/kg (CPF2), 1 mg/kg (CPF3) and 10 mg/kg (CPF4) 5 days per week for 12 weeks (n = 10 for each group). Throughout the treatment period, each animal was observed at least once daily for clinical signs of toxicity related to CPF exposure. Freshly voided fecal pellets were collected after 12 weeks of exposure, by placing each mice in an autoclaved plastic container. The fresh fecal pellets were placed into sterilized microcentrifuge tubes with forceps and frozen at −80°C. The liver was rapidly excised and washed in ice-cold phosphate-buffered saline. The organ coefficient was calculated as organ weight/body weight × 100%. Then, a part of the liver tissue was immediately stored at −80°C, and the remaining liver tissue was fixed in 4% paraformaldehyde. The protocol was approved by the Committee on the Ethics of Animal Experiments of Shandong Academy of Occupational Health and Occupational Medicine. All surgery was performed under sodium pentobarbital euthanasia, and all efforts were made to minimize suffering.
Histological analysis
The liver collected from CPF1, CPF2, CPF3, CPF4 and control groups were fixed in 4% paraformaldehyde, embedded in paraffin wax, sectioned at 4-μm thickness and stained with hematoxylin and eosin (H&E) for microscopic observation. The scoring criteria by Ishak [29] were used to evaluate the degree of liver inflammation in mice (Table 1).
Table 1.
score criteria for liver inflammation
| Numerical grade or stage | Score |
|---|---|
| Periportal or periseptal interface hepatitis (piecemeal necrosis) | |
| Absent | 0 |
| Mild (focal, few portal areas) | 1 |
| Mild/moderate (focal, most portal areas) | 2 |
| Moderate (continuous ~60% of tracts or septa) | 3 |
| Severe (continuous around >50% of tracts or septa) | 4 |
| Confluent necrosis | 0 |
| Absent | 0 |
| Focal confluent necrosis | 1 |
| Zone 3 necrosis in some areas | 2 |
| Zone 3 necrosis in most areas | 3 |
| Zone 3 necrosis+occasional portal-central (P-C) bridging | 4 |
| Zone 3 necrosis+multiple P-C bridging | 5 |
| Focal confluent necrosis | 6 |
| Focal (spotty) lytic necrosis, apoptosis and focal inflammation | |
| Absent | 0 |
| One focus or less per 10 × objective | 1 |
| Two to four foci per 10 × objective | 2 |
| Five to 10 foci per 10 × objective | 3 |
| More than 10 foci per 10 × objective | 4 |
| Portal inflammation | |
| None | 0 |
| Mild, some or all portal areas | 1 |
| Moderate, some or all portal areas | 2 |
| Moderate/marked, all portal areas | 3 |
| Marked, all portal areas | 4 |
Measurement of levels of inflammatory factors in liver
We selected the liver tissue of the same part, and the tissue samples were homogenized according to the concentration of 10%. The expression levels of TNF-α, IL-6 and IL-1β in liver were measured by enzyme-linked immunosorbent assay (ELISA). All of these liver indexes were measured by using commercial reagent kits from Biokits Technologies Institute (Beijing, China) according to the manufacturer’s instructions.
DNA extraction, PCR amplification and 16S rRNA gene sequencing
Microbial DNA was extracted from fecal samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to manufacturer’s protocols. The final DNA concentration and purification were determined by NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′- ACTCCTACGGGAGGCAGCAG-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system (GeneAmp 9700, ABI, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database using confidence threshold of 70%. Alpha diversity was calculated to estimate the microbial communities’ diversity, including the index of ace, chao, shannon, simpon and observed species (Sobs).
Gene expression analysis
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The reverse transcription reaction was carried out with reverse transcription enzyme (Toyobo, Shanghai, China). Real-time PCR was carried out on an ABI7300 real-time PCR system and the specific primers are shown in Table 2.
Table 2.
the primer sequences for Q-RT-PCR
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| 16S | Eub338 | ACTCCTACGGGAGGCAGCAG |
| Eub806 | GGACTACHVGGGTWTCTAAT |
Statistical analysis
The data were analyzed by SPSS 22.0 software and analyzed using one-way analysis of variance followed by Dunnett’s post hoc test with a confidence interval of 95% (IBM Corp, Chicago, USA). Differential abundances of genera were determined by non-parametric tests including Kruskal–Wallis H test. Values are expressed as the mean ± SD, P < 0.05, was accepted as significance.
Results
Body weight and organ coefficient are affected by CPF treatment
During the exposure period, CPF did not cause the abnormal mortality of mice. Compared with the control group, except for the significant decrease of body weight in the CPF4-treated group (P < 0.05), there was no significant change in all the other exposure groups (P > 0.05) (Fig. 1A). In addition, there was a trend toward increased liver coefficients with increased CPF dosage (Fig. 1B).
Figure 1.
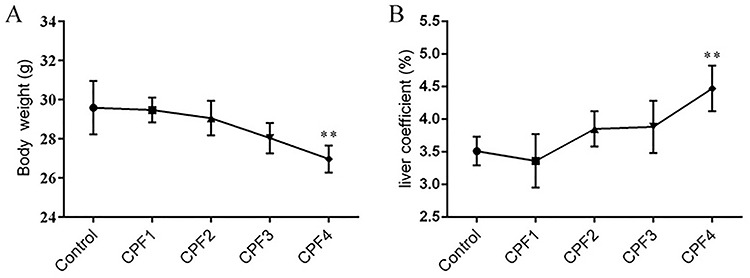
body weight (A) and the liver coefficient (B) of mice; each value is expressed as a mean ± SD, n = 10. *P < 0.05, **P < 0.01 vs. control (0 mg/kg).
Histopathological changes of livers in CPF-exposed mice
Histological changes were examined in the livers of the mice. In the control, CPF1- and CPF2-treated groups, the livers of mice exhibited normal hepatocyte and central vein architecture, whereas CPF3-treated group mice showed a small amount of inflammatory cell infiltration (Fig. 2D), and CPF4-treated group mice showed focal inflammation, hepatocytes with different degrees of necrosis (Fig. 2E). Histopathological change of mice from the control and CPF1-treated group was 0 by the hepatic numerical scores (Table 3). Compared with the control, there was no significant change in hepatic numerical scores in CPF2-treated group (P > 0.05) and significantly increased in CPF3- and CPF4-treated groups (P < 0.05).
Figure 2.
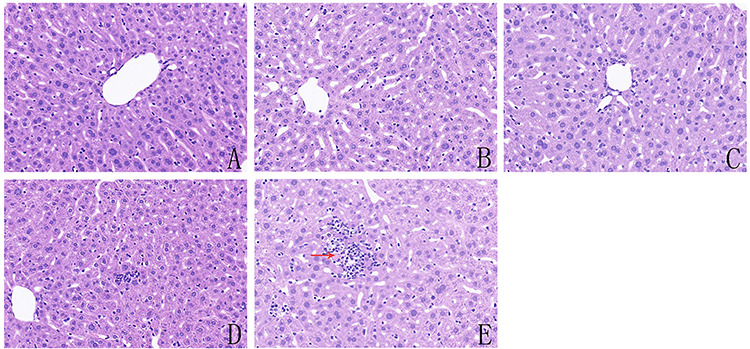
histological evaluation of the livers of mice from control and CPF-exposed groups: (A) mice from control group; (B) mice treated with 0.01 mg/kg CPF; (C) mice treated with 0.1 mg/kg CPF; (D) mice treated with 1 mg/kg CPF and (E) mice treated with 10 mg/kg CPF; the red arrows indicate necrotic hepatocytes, and the photomicrographs were taken at ×200 magnification after H&E staining, n = 4.
Table 3.
the comparison of liver inflammation score between control and CPF-exposed mice
| Group | Control | CPF1 | CPF2 | CPF3 | CPF4 |
|---|---|---|---|---|---|
| Score | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.50 ± 0.58 | 2.25 ± 0.50** | 3.75 ± 0.96** |
* P < 0.05, **P < 0.01 vs. control (0 mg/kg), n = 4.
Effect of CPF on the levels of TNF-α, IL-1β and IL-6 in the livers of mice
To further confirm inflammatory changes in the livers of mice, we detected the level of three representative pro-inflammatory cytokines TNFα, IL-1β and IL-6 in livers by ELISA assay kits. As shown in Fig. 3, the levels of TNFα, IL-1β and IL-6 in livers of mice from CPF3- and CPF4-treated groups were significantly increased (P < 0. 05) compared with that in mice from control group, but low level of CPF (CPF1 and 2) did not cause significant changes of these cytokines (P > 0.05). Significant difference in these cytokines between mice from CPF4- and CPF3-treated group has been found (P < 0.05).
Figure 3.
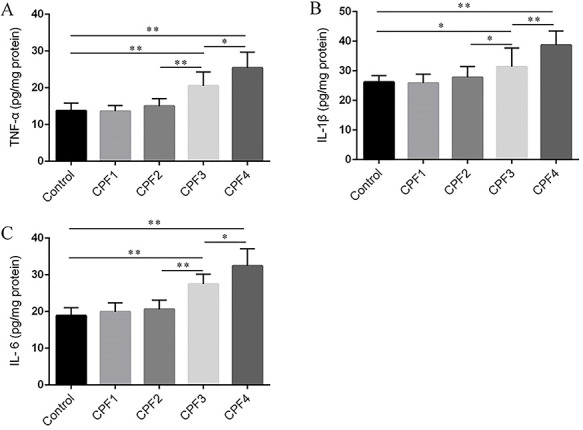
the levels of TNF-a (A), IL-1β (B) and IL-6 (C) in the livers of mice were measured by ELISA; each value is expressed as a mean ± SD, n = 6. *P < 0.05, **P < 0.01.
Overall structure and community diversity changes of fecal microbiota
In order to compare the similarity and repeatability of intestinal flora composition between the control group and CPF-treated (CPF3, CPF4) group more intuitively, we first analyze the composition of fecal microbiota at the OTU classification level by using Venn diagram (Fig. 4A). Circles of different colors represent different groups, and numbers represent the number of shared or unique species between groups. The Venn diagram shows that there are 582 OTUs, ~62% of which are shared by all groups. Compared with the control group, there were 190 unshared OTUs in CPF3- and CPF4-treated groups. Next, we introduce the method of multivariate statistics to analyze the relative abundance information of OTUs of three groups by PCoA (principal co-ordinates analysis). As shown in Fig. 4B, CPF-treated mice are clearly separated from controls. The above results show that CPF can significantly affect the composition of intestinal flora. In order to reflect the diversity and richness of intestinal microbial community, we calculated the sobs index, shannon index, simpson index, ace index and chao index of each group (Table 4). Shannon index demonstrated species richness and evenness, which increased significantly in CPF3- and CPF4-treated groups (P < 0.05). Meanwhile, the simpson index of CPF4-treated group decreased significantly, indicating higher micorbiota diversity of mice from CPF4-treated group.
Figure 4.
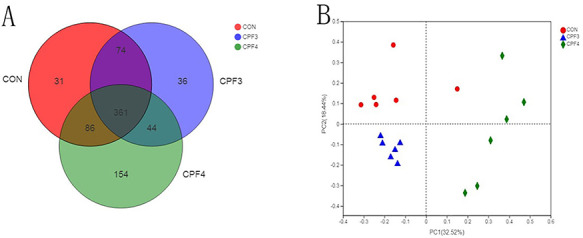
effects of CPF on the gut microbiota composition of the fecal, n = 6; (A) Venn diagram of OTUs among each group, and each symbol represents one sample; (B) analysis of β diversity of intestinal flora in mice, and PCoA was performed to calculate the distances between fecal samples from the mice of control and CPF groups, and each point represents a sample. A clear separation is observed between the samples of control and CPF3 and CPF4 groups.
Table 4.
the comparison of α-diversity indexes between control and CPF-exposed mice
| Group | Sobs | Shannon | Simpson | Ace | Chao |
|---|---|---|---|---|---|
| Control | 289.00 ± 43.59 | 3.15 ± 0.24 | 0.12 ± 0.04 | 352.22 ± 52.19 | 357.02 ± 57.50 |
| CPF3 | 320.50 ± 54.05 | 3.62 ± 0.14** | 0.06 ± 0.01 | 399.26 ± 61.89 | 399.91 ± 73.87 |
| CPF4 | 355.50 ± 76.34 | 3.81 ± 0.35** | 0.06 ± 0.02* | 453.47 ± 99.00 | 435.86 ± 100.77 |
* P < 0.05, **P < 0.01 vs. control (0 mg/kg), n = 6.
Intestinal flora changes of CPF-treated mice
Next, we compared the relative abundances of the dominant taxa present in the guts of the three groups. At the phylum level, the Bacteroidetes, Firmicutes and Actinobacteria accounted for more than 90% of the total sequence, and they were the dominant groups (Fig. 5). As shown in Fig. 5, with the increase of CPF concentration, the relative abundance of Verrucomicrobia decreased significantly. To compare the gut microbiota composition of the three groups, we conducted a statistical analysis of gut microbiota at the genus level using the Kruskal–Wallis H test (Fig. 6A). CPF significantly changed the relative abundance of Prevotellaceae_UCG-001, Faecalibaculum, Akkermansia, Desulfovibrio and Helicobacter. Importantly, the relative abundance of Akkermansia was negatively correlated with CPF concentration. The relative abundance of Akkermansia in mice from CPF4-treated group was about 1/10 of that from control. Helicobacter was not found in control group mice, but it was found in mice from CPF3- and CPF4-treated groups (Fig. 6B); especially, the relative abundance of Helicobacter was positively correlated with CPF concentration.
Figure 5.
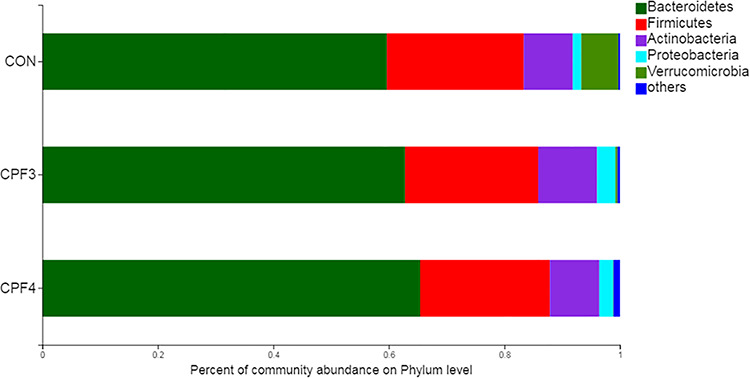
the composition of gut microbiota at the phylum level after exposure to CPF, n = 6.
Figure 6.
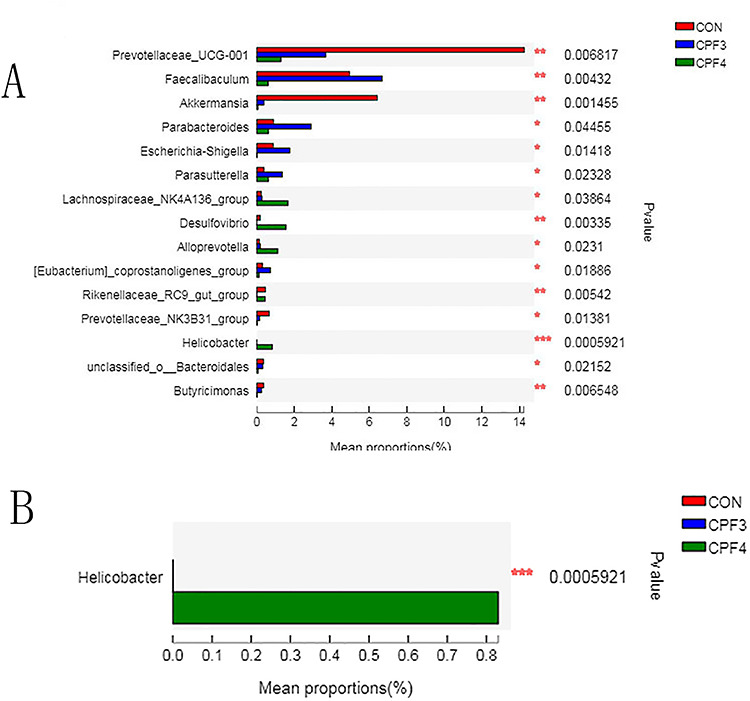
(A) significantly differences of the gut microbiota induced by CPF between CPF3, CPF4 and control groups at genus level; (B) significantly differences of Helicobacter induced by CPF between CPF3, CPF4 and control groups. Data were showed as relative abundance (%) of genus in each group, and statistical analysis was performed by the Kruskal–Wallis H test, n = 6, *P < 0.05, **P < 0.01, ***P < 0.001.
Effect of CPF on the total bacterial DNA in the liver
We detected the bacterial 16S rRNA gene in livers of mice by Q-RT-PCR to evaluate whether gut bacteria had translocated to the liver. The result showed that the levels of total bacterial DNA significantly increased in the livers of mice from CPF4-treated group only compared with control (P < 0.05) (Fig. 7). There was no significant difference of the total bacterial DNA between the mice from CPF3- and CPF4-treated groups (P = 0.062).
Figure 7.
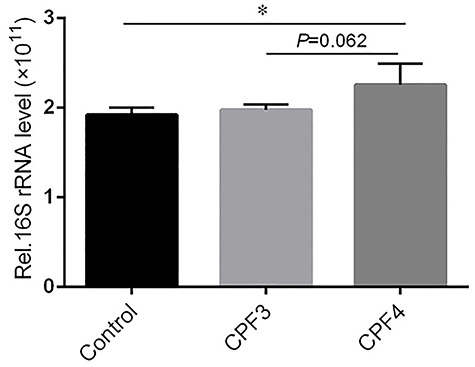
the total bacterial load in the livers of mice from CPF3, CPF4 and control groups, and each value is expressed as a mean ± SD, n = 3, *P < 0.05, **P < 0.01.
Discussion
Human exposure to CPF is mainly through the intake of its residues from food, especially in fruits and vegetables. The liver is the detoxification organ of the body. When the detoxification ability of the liver is not enough to solve excessive internal or external poisons, it will cause poisoning or pathological changes [30]. It has been demonstrated that CPF (5.4 mg/kg) could cause hepatotoxicity via changing the profile of liver marker enzymes such as ALP, AST, LDH and organizational structure [31, 32]. In this study, we found that CPF at 1 and 10 mg/kg could cause obvious dose-dependent effects on the livers, namely, CPF increased the levels of TNFα, IL-1β and IL-6 and inflammation with the increase of dosage.
In recent years, more and more attention has been paid to the influence of structural changes of intestinal flora on human diseases. Studies found that CPF can alter significantly the composition of intestinal flora, including the abundance of bacteria associated with diabetes and obesity phenotypes, leading to an obesity phenotype in rats with normal diet [33]. In another study, mice exposed to 1-mg/kg CPF for 30 days showed imbalance in intestinal flora and changes in urinal metabolites, such as amino acids, short-chain fatty acids and bile acids, and led to intestinal inflammation and abnormal intestinal permeability [26]. Studies have shown that the gut microbiomes of patients with advanced liver disease and cirrhosis are characterized by an increase in potentially pathogenic bacteria, along with reduced numbers of bacteria with beneficial properties [34]. Alcoholic liver disease patients also displayed reduced fungal diversity and Candida overgrowth, presenting the evidence of the role of the gut microbiomes in pathogenesis of liver disease [35].
In this study, Illumina MiSeq sequencing was used to study the intestinal flora diversity of mice exposed to CPF and control group mice. Through Alpha diversity analysis, we found that the intestinal flora diversity of CPF-exposed mice was higher than that of the control, which may be related to the over reproduction of intestinal bacteria caused by intestinal flora imbalance. In addition, the beta diversity showed that the CPF3- and CPF4-treated group was significantly separated from the control. This indicated that CPF can cause intestinal flora imbalance in mice. It has been reported that mice exposed to CPF (1 mg/kg) for 30 day could perturb the gut microbiota composition [26], which is consistent with our study. Intestinal flora could regulate modulates plasma levels of lipopolysaccharide (LPS). The disturbed intestinal flora may induce chronic low-grade inflammation through LPS [36]. Therefore, we speculated that CPF-induced intestinal flora imbalance may contribute on observed the inflammation in the liver of the mice exposed to CPF. After analyzing the sequencing data, we found that the fecal intestinal flora of the three groups of samples belonged to five phylum, including Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Verrucomicrobia. Akkermansia was a gram-negative anaerobic bacteria, which was inversely related to many health problems. Individuals with fewer Akkermansia were more likely to have obesity, inflammation and type 2 diabetes [37, 38]. One study has indicated that Akkermansia was absolutely dominant in Verrucomicrobia, accounting for ~83% [39]. But our results showed that the relative abundance of Akkermansia decreased significantly in CPF-treated mice. Interestingly, the relative abundance of Akkermansia in mice from the CPF4-treated group was about 1/10 of that from the control. It has been reported that decreased Akkermansia abundance was associated with thinning of the mucus layer, destruction of intestinal barrier integrity and increased inflammation, which could promote alcoholic liver disease and non-alcoholic liver injury [40, 41]. Therefore, we speculated that the significant reduction of Akkermansia in CPF4-treated group may be associated with CPF-induced damage of liver.
Symbiotic bacteria in the gut have a dual function, with some having anti-inflammatory properties and others inducing inflammation under certain circumstances [42]. Studies have shown that Helicobacter is closely related to non-alcoholic fatty liver disease, which can stimulate inflammatory and immune response, excessive release inflammatory factors such as IL-6, IL-8, TNF-α and IL-β, thus aggravating inflammatory response and insulin resistance [43]. Helicobacter is a kind of gram-negative bacilli, which can be colonized not only in gastric mucosa but also in liver and other extra-gastrointestinal organs and tissues [44]. Helicobacter DNA has been found in the liver of patients with a number of chronic liver diseases, including hepatitis C virus-related chronic hepatitis and cirrhosis [45]. Interestingly, in our result, Helicobacter was not detected in the intestinal flora of mice from the control, but Helicobacter appeared in CPF3- and CPF4-treated groups and showed increasing significantly, which has the same trend as the liver inflammatory including histopathological changes and the levels of TNFα, IL-1β and IL-6. Therefore, we speculated that CPF exposure altered the intestinal microecology of mice, and created conditions for Helicobacter colonization in the gastrointestinal tract. Helicobacter may play an important role in the CPF-induced damage of liver. Hence, the regulator mechanism of Helicobacter in the CPF-induced liver inflammatory may be worth further study.
Moreover, we found that the abundance of Prevotellacea_UCG-001, Prevotellacea _NK3B31_group and Butyricimonas was significantly decreased in CPF3- and CPF4-treated groups. It has been reported that Prevotella and Butyricimonas were significantly decreased in patients with nonalcoholic steatohepatitis and chronic hepatitis B, respectively [46, 47]. Furthermore, the abundance of Desulfovibrio significantly increased in CPF3- and CPF4-treated groups. Desulfovibrio was a Gram-negative endotoxin-producing bacterium that was known to lead to an increase in intestinal permeability and circulating gut-derived antigens, primarily LPS [48]. Meanwhile, Desulfovibrio has been shown to increase significantly in relative abundance in non-alcoholic fatty liver disease mice [49]. In conclusion, the genera of Akkermansia, Helicobacter, Prevotella, Butyricimonas and Desulfovibrio were significantly changed by CPF exposure. Meanwhile, there was obviously dose effect relationship between intestinal flora disorder and liver inflammation degree. These results suggested that liver inflammation caused by CPF in mice may be closely associated with intestinal flora disorder characterized by the decrease of beneficial bacteria and the increase of pathogenic bacteria.
It has been confirmed that CPF could lead to the damage of intestinal barrier and the increase of intestinal permeability [50]. The increase of intestinal permeability can make pro-inflammatory factors that enter the intestinal mucosa continuously, activate the inflammatory cascade reaction and even cause the translocation of intestinal bacteria or metabolites to the parenteral tissue, thus promoting the inflammatory process [51, 52]. The liver is located at the intersection between the host and the gut commensal microbiota, and the bacterial products translocated from the gut lumen were easily exposed in the liver when intestinal epithelial barrier functions were disrupted [53, 54]. In this study, we examined the bacterial 16S rRNA gene in livers of mice by Q-RT-PCR to evaluate whether increased total bacterial DNA in the liver. The result showed that the levels of total bacterial DNA increased significantly in the livers of mice from CPF4-treated group compared with control. And the levels of total bacterial DNA in CPF4-treated group showed an increasing trend compared with CPF3-treated, which has the same trend with the liver inflammation. It has been reported that bacterial translocation is an important event in the pathological process from stable liver cirrhosis toward acute-on-chronic liver failure in chronic liver disease [55]. Basing on the CPF-induced damage to intestinal barrier and increase of intestinal permeability [50], we speculated that CPF may cause the translocation of gut bacteria translocation to liver leading to observed inflammatory. This study investigated the role of intestinal flora in CPF-induced liver inflammation from a new perspective. However, the detailed mechanism of intestinal flora regulating liver inflammation still need to be further studied.
Conclusion
CPF could induce significant inflammatory response in mice liver tissues. Meanwhile, CPF also induced significant perturbations on the abundance of intestinal flora resulting in the decrease of beneficial bacteria and the increase of pathogenic bacteria, especially arisen Helicobacter. In addition, there was a significant increase of total bacterial DNA in the liver tissue of the mice from CPF4-treated group. The imbalance of intestinal flora and the increase of total bacterial DNA in the liver suggested a potential mechanism of CPF-induced liver inflammation, namely CPF-induced gut bacteria translocation leaded to liver inflammation. However, the detailed mechanism underlying how the changed intestinal bacteria affect the liver needs to be further studied. Additionally, whether the increase of total bacterial DNA in the liver tissues of the CPF4-treated group was related to intestinal bacterial translocation needs to be further verified. Our research provides new insights of hepatotoxicity induced by CPF.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81573198), the Department of Science and Technology of Shandong Province (2017GSF18142), the Shandong First Medical University and Shandong Academy of Medical Sciences (2019QL001) and the Innovation Project of Shandong Academy of Medical Sciences.
References
- 1. Das S, Adhya TK. Degradation of chlorpyrifos in tropical rice soils. J Environ Manage 2015;152:36–42. [DOI] [PubMed] [Google Scholar]
- 2. Dar MA, Kaushik G, Villarreal-Chiu JF. Pollution status and bioremediation of chlorpyrifos in environmental matrices by the application of bacterial communities: a review. J Environ Manage 2019;239:124–36. [DOI] [PubMed] [Google Scholar]
- 3. Singh BK, Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 2006;30:428–71. [DOI] [PubMed] [Google Scholar]
- 4. Rambabu L, Megson IL, Eddleston M. Does oxidative stress contribute to toxicity in acute organophosphorus poisoning?- a systematic review of the evidence. Clin Toxicol (Phila) 2020;58:437–52. [DOI] [PubMed] [Google Scholar]
- 5. Das S, Hageman KJ, Taylor M et al. Fate of the organophosphate insecticide, chlorpyrifos, in leaves, soil, and air following application. Chemosphere 2020;243:125194. [DOI] [PubMed] [Google Scholar]
- 6. Banaee M, Akhlaghi M, Soltanian S et al. Combined effects of exposure to sub-lethal concentration of the insecticide chlorpyrifos and the herbicide glyphosate on the biochemical changes in the freshwater crayfish Pontastacus leptodactylus. Ecotoxicology 2020;29:1500–15. [DOI] [PubMed] [Google Scholar]
- 7. Eaton DL, Daroff RB, Autrup H et al. Review of the toxicology of chlorpyrifos with an emphasis on human exposure and neurodevelopment. Crit Rev Toxicol 2008;38 Suppl 2:1–125. [DOI] [PubMed] [Google Scholar]
- 8. GB National food safety standard-maximum residue limits for pesticides in food: GB 2763-2019. Beijing, China (in Chinese) 2019. [Google Scholar]
- 9. Abhilash PC, Singh N. Pesticide use and application: an Indian scenario. J Hazard Mater 2009;165:1–12. [DOI] [PubMed] [Google Scholar]
- 10. Pallotta MM, Barbato V, Pinton A et al. In vitro exposure to CPF affects bovine sperm epigenetic gene methylation pattern and the ability of sperm to support fertilization and embryo development. Environ Mol Mutagen 2019;60:85–95. [DOI] [PubMed] [Google Scholar]
- 11. Mie A, Ruden C, Grandjean P. Safety of safety evaluation of pesticides: developmental neurotoxicity of chlorpyrifos and chlorpyrifos-methyl. Environ Health 2018;17:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zafiropoulos A, Tsarouhas K, Tsitsimpikou C et al. Cardiotoxicity in rabbits after a low-level exposure to diazinon, propoxur, and chlorpyrifos. Hum Exp Toxicol 2014;33:1241–52. [DOI] [PubMed] [Google Scholar]
- 13. Goel A, Dani V, Dhawan DK. Chlorpyrifos-induced alterations in the activities of carbohydrate metabolizing enzymes in rat liver: the role of zinc. Toxicol Lett 2006;163:235–41. [DOI] [PubMed] [Google Scholar]
- 14. Tanvir EM, Afroz R, Chowdhury M et al. A model of chlorpyrifos distribution and its biochemical effects on the liver and kidneys of rats. Hum Exp Toxicol 2016;35:991–1004. [DOI] [PubMed] [Google Scholar]
- 15. Deng Y, Zhang Y, Lu Y et al. Hepatotoxicity and nephrotoxicity induced by the chlorpyrifos and chlorpyrifos-methyl metabolite, 3,5,6-trichloro-2-pyridinol, in orally exposed mice. Sci Total Environ 2016;544:507–14. [DOI] [PubMed] [Google Scholar]
- 16. Lozupone CA, Stombaugh JI, Gordon JI et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Biedermann L, Rogler G. The intestinal microbiota: its role in health and disease. Eur J Pediatr 2015;174:151–67. [DOI] [PubMed] [Google Scholar]
- 18. Franzosa EA, Sirota-Madi A, Avila-Pacheco J et al. Author correction: gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:898. [DOI] [PubMed] [Google Scholar]
- 19. Cirstea MS, Yu AC, Golz E et al. Microbiota composition and metabolism are associated with gut function in Parkinson's disease. Mov Disord 2020;35:1208–17. [DOI] [PubMed] [Google Scholar]
- 20. Chen J, Vitetta L. Gut microbiota metabolites in NAFLD pathogenesis and therapeutic implications. Int J Mol Sci 2020;21:5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartmann P, Seebauer CT, Schnabl B. Alcoholic liver disease: the gut microbiome and liver cross talk. Alcohol Clin Exp Res 2015;39:763–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Angelo JR, Guerrero-Zayas MI, Tremblay KD. A fate map of the murine pancreas buds reveals a multipotent ventral foregut organ progenitor. PLoS One 2012;7:e40707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elamin EE, Masclee AA, Dekker J et al. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev 2013;71:483–99. [DOI] [PubMed] [Google Scholar]
- 24. Tripathi A, Debelius J, Brenner DA et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 2018;15:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X, Shen M, Zhou J et al. Chlorpyrifos disturbs hepatic metabolism associated with oxidative stress and gut microbiota dysbiosis in adult zebrafish. Comp Biochem Physiol C Toxicol Pharmacol 2019;216:19–28. [DOI] [PubMed] [Google Scholar]
- 26. Zhao Y, Zhang Y, Wang G et al. Effects of chlorpyrifos on the gut microbiome and urine metabolome in mouse (Mus musculus). Chemosphere 2016;153:287–93. [DOI] [PubMed] [Google Scholar]
- 27. Joly Condette C, Bach V, Mayeur C et al. Chlorpyrifos exposure during perinatal period affects intestinal microbiota associated with delay of maturation of digestive tract in rats. J Pediatr Gastroenterol Nutr 2015;61:30–40. [DOI] [PubMed] [Google Scholar]
- 28. Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm 2016;7:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishak K, Baptista A, Bianchi L et al. Histological grading and staging of chronic hepatitis. J Hepatol 1995;22:696–9. [DOI] [PubMed] [Google Scholar]
- 30. Almazroo OA, Miah MK, Venkataramanan R. Drug metabolism in the liver. Clin Liver Dis 2017;21:1–20. [DOI] [PubMed] [Google Scholar]
- 31. Uzun FG, Kalender Y. Chlorpyrifos induced hepatotoxic and hematologic changes in rats: the role of quercetin and catechin. Food Chem Toxicol 2013;55:549–56. [DOI] [PubMed] [Google Scholar]
- 32. Mansour SA, Mossa AH. Adverse effects of lactational exposure to chlorpyrifos in suckling rats. Hum Exp Toxicol 2010;29:77–92. [DOI] [PubMed] [Google Scholar]
- 33. Fang B, Li JW, Zhang M et al. Chronic chlorpyrifos exposure elicits diet-specific effects on metabolism and the gut microbiome in rats. Food Chem Toxicol 2018;111:144–52. [DOI] [PubMed] [Google Scholar]
- 34. Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol 2017;14:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang AM, Inamine T, Hochrath K et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest 2017;127:2829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blaut M Gut microbiota and energy balance: role in obesity. Proc Nutr Soc. 2015;74:227–34. [DOI] [PubMed] [Google Scholar]
- 37. Plovier H, Everard A, Druart C et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017;23:107–13. [DOI] [PubMed] [Google Scholar]
- 38. Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog 2017;106:171–81. [DOI] [PubMed] [Google Scholar]
- 39. Naito Y, Uchiyama K, Takagi T. a next-generation beneficial microbe: Akkermansia muciniphila. J Clin Biochem Nutr 2018;63:33–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grander C, Adolph TE, Wieser V et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 2018;67:891–901. [DOI] [PubMed] [Google Scholar]
- 41. Everard A, Belzer C, Geurts L et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004;4:478–85. [DOI] [PubMed] [Google Scholar]
- 43. Okushin K, Tsutsumi T, Ikeuchi K et al. Helicobacter pylori infection and liver diseases: epidemiology and insights into pathogenesis. World J Gastroenterol 2018;24:3617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smyk DS, Koutsoumpas AL, Mytilinaiou MG et al. Helicobacter pylori and autoimmune disease: cause or bystander. World J Gastroenterol 2014;20:613–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rocha M, Avenaud P, Ménard A et al. Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut 2005;54:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boursier J, Mueller O, Barret M et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J, Wang Y, Zhang X et al. Gut microbial Dysbiosis is associated with altered hepatic functions and serum metabolites in chronic hepatitis B patients. Front Microbiol 2017;8:2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moreno-Indias I, Torres M, Sanchez-Alcoholado L et al. Normoxic recovery mimicking treatment of sleep apnea does not reverse intermittent hypoxia-induced bacterial dysbiosis and low-grade endotoxemia in mice. Sleep 2016;39:1891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao W, Xiao M, Yang J et al. The combination of Ilexhainanoside D and ilexsaponin A1 reduces liver inflammation and improves intestinal barrier function in mice with high-fat diet-induced non-alcoholic fatty liver disease. Phytomedicine 2019;63:153039. [DOI] [PubMed] [Google Scholar]
- 50. Liang Y, Zhan J, Liu D et al. Organophosphorus pesticide chlorpyrifos intake promotes obesity and insulin resistance through impacting gut and gut microbiota. Microbiome 2019;7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manfredo Vieira S, Hiltensperger M et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018;359:1156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Soderholm JD, Peterson KH, Olaison G et al. Epithelial permeability to proteins in the noninflamed ileum of Crohn's disease? Gastroenterology 1999;117:65–72. [DOI] [PubMed] [Google Scholar]
- 53. Balmer ML, Slack E, de Gottardi A et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med 2014;6:237ra66. [DOI] [PubMed] [Google Scholar]
- 54. Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol 2012;590:447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology 2005;41:422–33. [DOI] [PubMed] [Google Scholar]


